Introduction
Liver cirrhosis is a global health challenge,
responsible for ~1.47 million deaths (2.4% of all deaths) in 2019,
with the greatest burden occurring in Southeast Asia and the
Western Pacific regions (1).
Chronic hepatitis B virus (HBV) infection, affecting an estimated
296 million people worldwide, remains the leading cause of
cirrhosis and hepatocellular carcinoma, accounting for ~331,000
cirrhosis-related deaths in 2019 and contributing to more than half
of hepatocellular carcinoma cases in endemic areas (2). The most recent WHO estimates further
indicate that HBV caused ~1.1 million deaths in 2022, mainly
attributable to cirrhosis and hepatocellular carcinoma (3). Pathologically, HBV-associated
cirrhosis is characterized by progressive fibrosis, distortion of
hepatic architecture and sustained infiltration of diverse immune
cell populations, including neutrophils, hepatic macrophages, T and
B lymphocytes and natural killer cells, accompanied by a
pro-inflammatory cytokine milieu rich in IL-1, IL-6, IL-17, IL-22,
IL-35, TGF-β and TNF-α (4,5). Persistent hepatic inflammation
promotes stellate cell activation and extracellular matrix
deposition, perpetuating fibrogenesis and facilitating the
transition to decompensated disease (6).
Gut-derived microbiota and their metabolites,
together with nutrients and other signals, reach the liver via the
portal vein, a process referred to as the gut-liver axis (7,8).
Inflammatory cytokines and immune responses mediated by
infiltrating cells are key contributors to the initiation and
progression of liver fibrosis (9).
In recent years, growing attention (10-12)
has been directed toward the gut microbiota as a key regulator of
liver physiology, particularly via the gut-liver axis. These
complex microbial communities regulate both physiological and
immunological functions of the host, influencing not only local
intestinal processes but also systemic responses. Numerous
microbiome-wide association studies have demonstrated a link
between gut microbial dysbiosis and chronic diseases, including
metabolic syndrome, inflammatory bowel disease and liver
pathologies (13,14). Microbiota-derived metabolites and
pathogen-associated molecular patterns directly influence hepatic
immune responses and metabolic processes. In liver diseases such as
non-alcoholic fatty liver disease (NAFLD), alcoholic liver disease
and cirrhosis, consistent shifts in microbial composition have been
documented (15,16). Notably, in cirrhosis, enrichment of
taxa such as Fusobacterium, Veillonella and members
of the Enterobacteriaceae family is associated with
heightened systemic inflammation and greater disease severity, with
several studies also indicating that these changes are more
pronounced in decompensated patients (17,18).
Mechanistic studies have begun to clarify how
microbial alterations influence liver health (7,19). The
gut microbiota produces a broad repertoire of bioactive
metabolites, including short-chain fatty acids (SCFAs),
lipopolysaccharides (LPS), bile acids, choline derivatives, indole
compounds, vitamins, lipids and niacin, which that regulate hepatic
immune signaling and metabolic balance (19,20).
Among these, SCFAs such as butyrate, acetate and propionate are
particularly notable for their ability to strengthen epithelial
tight junctions, enhance mucosal barrier integrity, suppress
intestinal inflammation and limit overgrowth of pathogenic
bacteria, in part via activation of free FA receptors 2 and
3(20). In experimental NAFLD
models, enrichment of SCFA-producing taxa is associated with
reduced hepatic lipid accumulation and attenuation of inflammatory
responses (21,22). Other microbial products such as bile
acids, tryptophan metabolites and LPS further modulate macrophage
polarization, antimicrobial peptide production and fibrotic
remodeling through receptors such as farnesoid X Receptor),
TGR5(Takeda G-protein coupled receptor 5) and AhR(Aryl Hydrocarbon
receptor), thereby linking gut dysbiosis to the pathophysiology of
both cirrhosis and hepatocellular carcinoma (8,23,24).
In parallel with these metabolic and
immunomodulatory functions, the gut microbiota is key for
preserving intestinal homeostasis. It achieves this by maintaining
the integrity of the mucus layer, sustaining the expression and
spatial organization of tight junction proteins and coordinating
mucosal immune responses. In cirrhosis, this equilibrium is
disrupted, characterized by diminished tight junction protein
expression, increased intestinal permeability and translocation of
bacterial products, which collectively exacerbate hepatic
inflammation, accelerate fibrogenesis and foster conditions that
facilitate pathogenic biofilm formation, aberrant immune activation
and chronic inflammation (25-27).
Histopathological analyses of cirrhotic livers typically
demonstrate dense infiltration by neutrophils, macrophages, T and B
lymphocytes and dendritic cells, indicative of a sustained
pro-inflammatory microenvironment (28,29).
Within this immune landscape, CD4+ T helper (Th) cell
subsets, including Th1, Th2 and Th17, and regulatory T cells serve
a central role in orchestrating immune responses: They regulate
CD8+ T cell expansion, promote B cell activation and
modulate multiple immune effectors throughout disease progression
(30,31). Although evidence links gut microbial
dysbiosis to immune activation in cirrhosis, most studies are
observational, and the precise mechanistic interplay between
microbiota alterations, systemic inflammation and barrier
dysfunction, particularly in HBV-associated cirrhosis, remains to
be elucidated (32-34).
The present study aimed to systematically
characterize the composition, diversity and predicted functions of
the gut microbiota in patients with HBV-associated liver cirrhosis.
By integrating microbial profiling with histological assessment of
immune infiltration and intestinal barrier integrity, the present
study aimed to elucidate the mechanistic underpinnings of the
gut-liver axis in cirrhosis progression and provide a basis for
microbiome-based diagnostic and therapeutic strategies.
Materials and methods
Study population and sample
collection
A total of 31 individuals with HBV-associated liver
cirrhosis (21 male, 10 female; median age, 48 years; age range,
28-65 years) were enrolled between April 2023 and April 2024,
before receiving any therapeutic intervention. Inclusion criteria
were as follows: age between 18 and 70 years; a confirmed diagnosis
of liver cirrhosis based on histology or imaging as described
above; chronic HBV infection; no prior receipt of antiviral therapy
for HBV. Exclusion criteria included: hepatocellular carcinoma;
human immunodeficiency virus infection; pregnancy or severe
cardiac; pulmonary; renal diseases. Cirrhosis was diagnosed by
liver biopsy or by concordant findings from at least two imaging
modalities (ultrasound, computed tomography, or magnetic resonance
imaging). Disease severity was assessed using the Child-Pugh
classification (35), which
evaluates serum total bilirubin and albumin, prothrombin time,
ascites and hepatic encephalopathy, categorizing patients as class
A (5-6 points), B (7-9 points) or C (≥10 points). The control group
included 15 healthy volunteers (8 male, 7 female; median age, 46
years; age range, 30-58 years) with no history of physical or
psychological disorder, confirmed through annual comprehensive
health evaluations including blood, urine and fecal analyses, liver
function and biochemical tests, hepatitis virus markers, chest
radiography and abdominal ultrasound. All participants were
recruited from the Third Affiliated Hospital of Sun Yat-Sen
University (Guangzhou, China).
Fecal sample collection was performed using a
TinyGene fecal collection box according to the manufacturer's
instructions. Boxes were placed on ice and stored at -80˚C before
further analysis.
A total of six normal liver tissue sections were
from para-hemangioma sites of patients with hepatic hemangioma
without hepatitis (three male, three female; median age, 45 years;
age range, 32-60 years). Paired samples of liver cirrhosis were
obtained from six separate patients with HBV-infected liver
cirrhosis during operations (four male, 2 female; median age, 48
years; age range, 30-65 years) before any therapeutic intervention.
Colonic mucosal specimens of 10 patients with untreated liver
cirrhosis (6 male, 4 female; median age, 47 years; age range, 27-63
years) and 10 healthy volunteers (5 male, 5 female; median age, 45
years; age range, 27-57 years) were obtained from the Endoscopic
Center of the Third Affiliated Hospital of Sun Yat-Sen University.
All tissues were verified by histopathology. The acquisition of
these tissues was approved by the Clinical Research Ethics
Committee of the Third Affiliated Hospital of Sun Yat-Sen
University (approval no. RG2023-033-01).
16S rRNA sequencing
The 16S rRNA gene was amplified using primers
as follows: Forward, 5'-GTGCCAGCMGCCGCGGTAA-3' and reverse,
5'-CCGTCAATTCMTTTGAGTTT-3' targeting the V5-V6 hyper-variable
regions (M were degenerate bases, representing A or C) and
sequenced using the Illumina, Inc. platform following the
manufacturer's protocols. The library was constructed by using the
two-step PCR amplification method with the following thermocycling
conditions: Initial preamplification stage at 94˚C for 2 min and
then 22 cycles at 94˚C for 30 sec, 55˚C for 30 sec, and 72˚C for 30
sec. The second step preamplification stage at 94˚C for 2 min and
then 8 cycles at 94˚C for 30 sec, 55˚C for 30 sec, and 72˚C for 30
sec. QIAamp DNA Stool Mini Kit (51604, QIAGEN) and
Phusion™ High-Fidelity DNA Polymerase (M0530L, New
England Biolabs were used. The quality and integrity of the
extracted DNA were verified using agarose gel electrophoresis and a
Bioanalyzer 2100 (Agilent Technologies). Sequencing libraries were
quantified using a Qubit fluorometer (Thermo Fisher Scientific) and
the final loading concentration was adjusted to 4 nM. Paired-end
sequencing (2x300 bp) was performed on an Illumina MiSeq platform
using the MiSeq Reagent Kit v3 (600-cycl, MS-102-300, Illumin). Raw
reads were processed using Trimmomatic v0.39 (usadellab.org/cms/?page=trimmomatic),
FLASH v1.2.11 (ccb.jhu.edu/software/FLASH/), and Mothur v1.39.5
(mothur.org/). After demultiplexing, dereplication, and
filtering, operational taxonomic units (OTUs) were clustered at 97%
similarity using UCLUST v1.2.22q (drive5.com/usearch/) against the SILVA v128
(arb-silva.de/) and Greengenes v13_8 databases
(greengenes.secondgenome.com).
α-diversity (Chao1, ACE, Shannon, Simpson) was calculated using
Mothur v1.39.5 and QIIME v1.9.1 (qiime.org/),
while β-diversity (Jaccard, unweighted UniFrac, weighted UniFrac)
was assessed and visualized by PCoA with FastTree v2.1.3
(microbesonline.org/fasttree/).
Differentially abundant taxa were identified using MetaStats
(metastats.cbcb.umd.edu/), and LEfSe
(huttenhower.sph.harvard.edu/lefse/) was used to detect
taxa differing significantly between groups. β-diversity clustering
was evaluated by analysis of similarity (ANOSIM) and correlations
between distance matrices were examined with the Mantel test.
Functional metagenome prediction
The USEARCH v9.2.64 global alignment command
(drive5.com/usearch/) was used to capture
OTU representative sequences from the Greengenes database v13_8
(greengenes.secondgenome.com).
Functional metagenome reconstruction was then performed using
PICRUSt v1.1.4 (picrust.github.io/picrust/). The predicted metagenome
functions were annotated against the Kyoto Encyclopedia of Genes
and Genomes (KEGG) orthology (kegg.jp/).
Histological staining
Hematoxylin-eosin (H&E), Sirius red,
immunohistochemical and immunofluorescence staining were performed
as previously described (36-38).
The cell index was determined by dividing the number of
histological staining signal-positive cells by the total number of
cells in ≥20 randomly selected fields of view (magnification,
x200). The antibodies were as follows: Anti-myeloperoxidase (MPO;
cat. no. A1374, Abclonal), -CD4 (cat. no. sc-19641), -CD19 (cat.
no. sc-19650; both Santa Cruz Biotechnology, Inc.), -CD68 (cat. no.
ab31630), -α-smooth muscle actin (cat. no. ab5694; both Abcam),
-claudin-1 (cat. no. 13255S), -zonula occludens-1 (ZO-1; cat. no.
8193T) and -E-cadherin (cat. no. 14472S; all Cell Signaling
Technology, Inc.).
ELISA
DAO and endotoxin levels from peripheral superficial
vein blood were examined with ELISA kits (cat. nos. JL-T0829 and
JL52002D, respectively; both J&L Biological) to estimate
intestinal permeability and bacterial translocation according to
the manufacturer's instructions.
Statistical analysis
Data were analyzed using R software (version 3.4.1)
and GraphPad Prism 6 (Dotmatics) software, including richness
estimators, the Ace, Chao and Shannon index, a diversity estimator
and rank dissimilarity and abundance distribution. Microbiome
compositional dissimilarity was represented by PCoA plots, and
quantified by unweighted or weighted UniFrac distance values. The
significant separation of the microbiome composition was determined
by ANOSIM and the significance differences in α and β diversity and
taxonomy between groups were analyzed using Wilcox test. Continuous
data are expressed as the mean ± SEM of ≥3 independent experimental
repeats and analyzed by Student's two-tailed paired t-test or one-
or two-way ANOVA followed by Bonferroni correction as described
previously (39). P<0.05 was
considered to indicate a statistically significant difference.
Results
Gut microbiota profile differs between
healthy volunteers and patients with liver cirrhosis
The present study compared the gut microbiota of
healthy volunteers with that of patients with liver cirrhosis using
Illumina MiSeq high-throughput sequencing of the 16S rRNA
gene. In total, ~1,466,285 sequence reads of 16S rRNA genes,
with an average length of ~410 bp, and 432 OTUs were obtained after
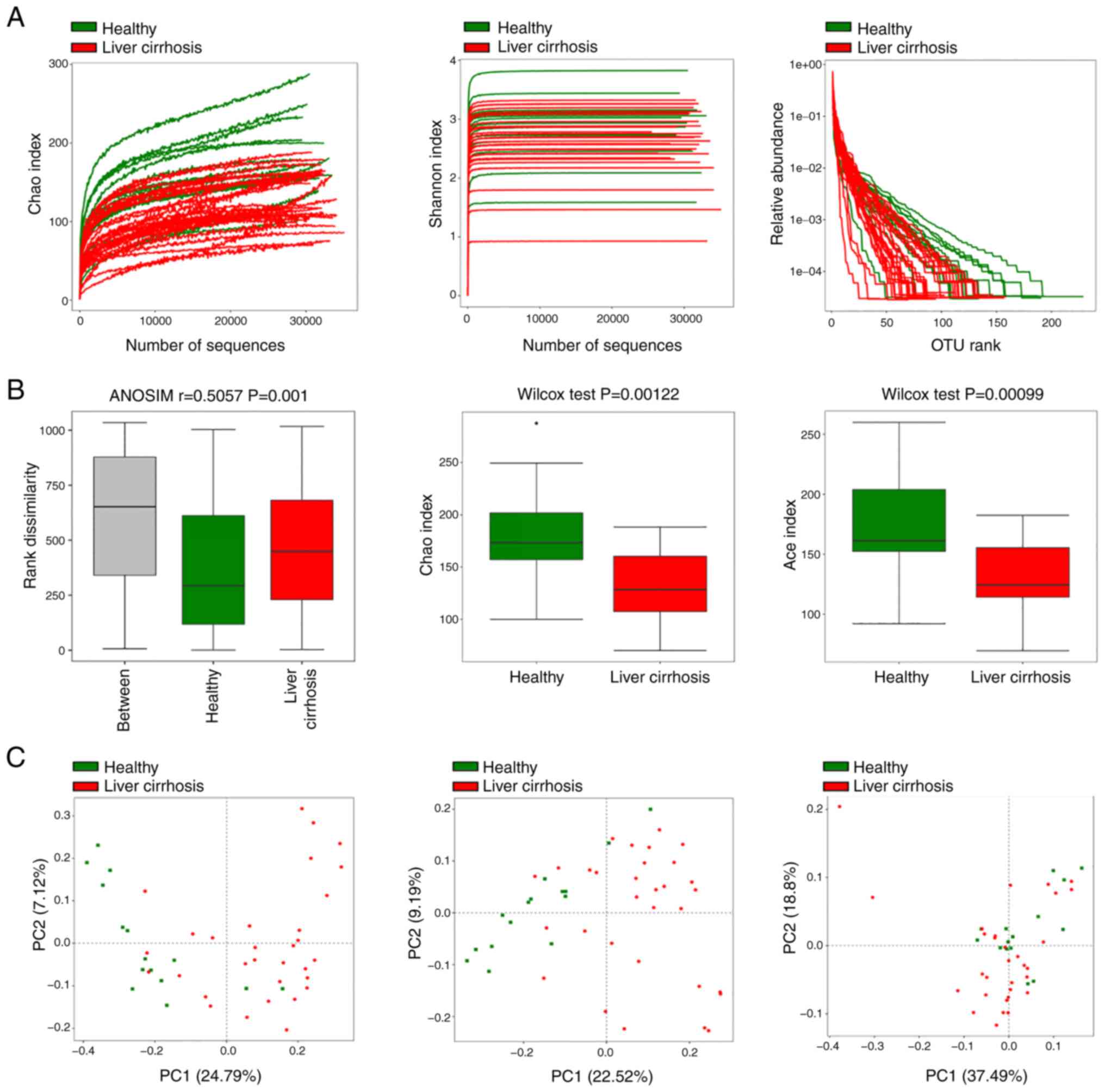
sequencing and quality filtering (Fig.
1A). On average, healthy volunteers had more OTUs than patients
with liver cirrhosis (371 vs. 362 OTUs, respectively; 301 core OTUs
were shared). The average number of reads was not significantly
different between healthy and cirrhosis group (31,643 vs. 31,988
reads, respectively).
To evaluate the alterations in the microbiota
structure between healthy volunteers and patients with liver
cirrhosis, α diversity was measured. Patients with liver cirrhosis
had notably decreased microbial species richness and evenness in
comparison with volunteers, based on the Chao and Shannon index and
OTU rank abundance analysis (Fig.
1A). Furthermore, rank dissimilarity revealed that the
between-group difference was greater than the within-group
difference (Fig. 1B), and the Chao
and Shannon indices analyzed using the Wilcoxon test showed that
patients with liver cirrhosis had significantly lower gut microbial
diversity than healthy volunteers (Fig.
1B). β diversity based on OTUs or phylogenetic analysis was
calculated using Jaccard and unweighted and weighted UniFrac
phylogenetic distance matrices; microbial composition of patients
with liver cirrhosis was different from that of healthy volunteers
(Fig. 1C). Core bacterial genus
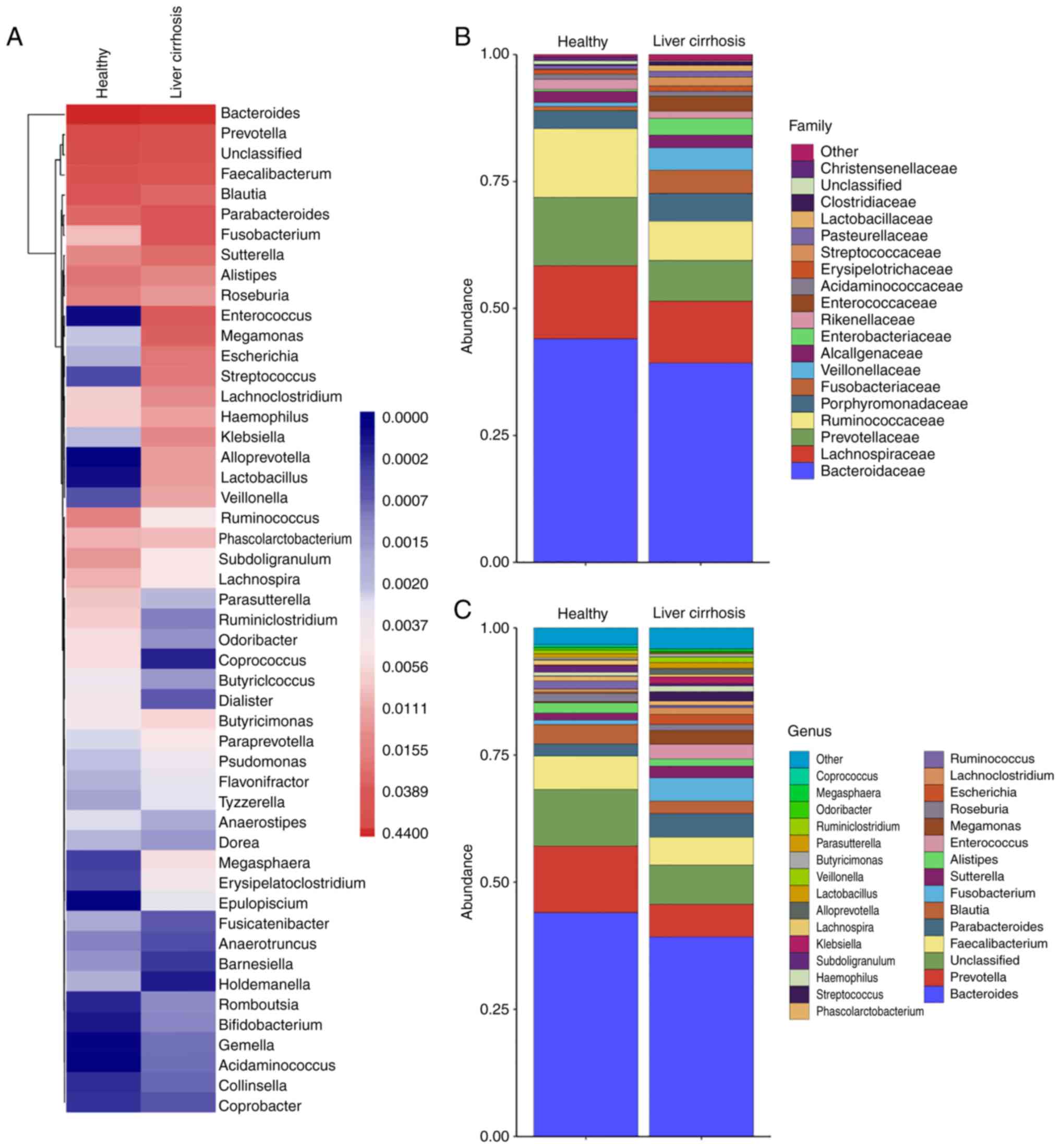
analysis using a heatmap demonstrated that the two groups had
similar microbiota structures or communities, but different
richness and abundance (Fig. 2A).
The relative bacterial community richness and abundance at the
family and genus levels were similar between groups (Fig. 2B and C).
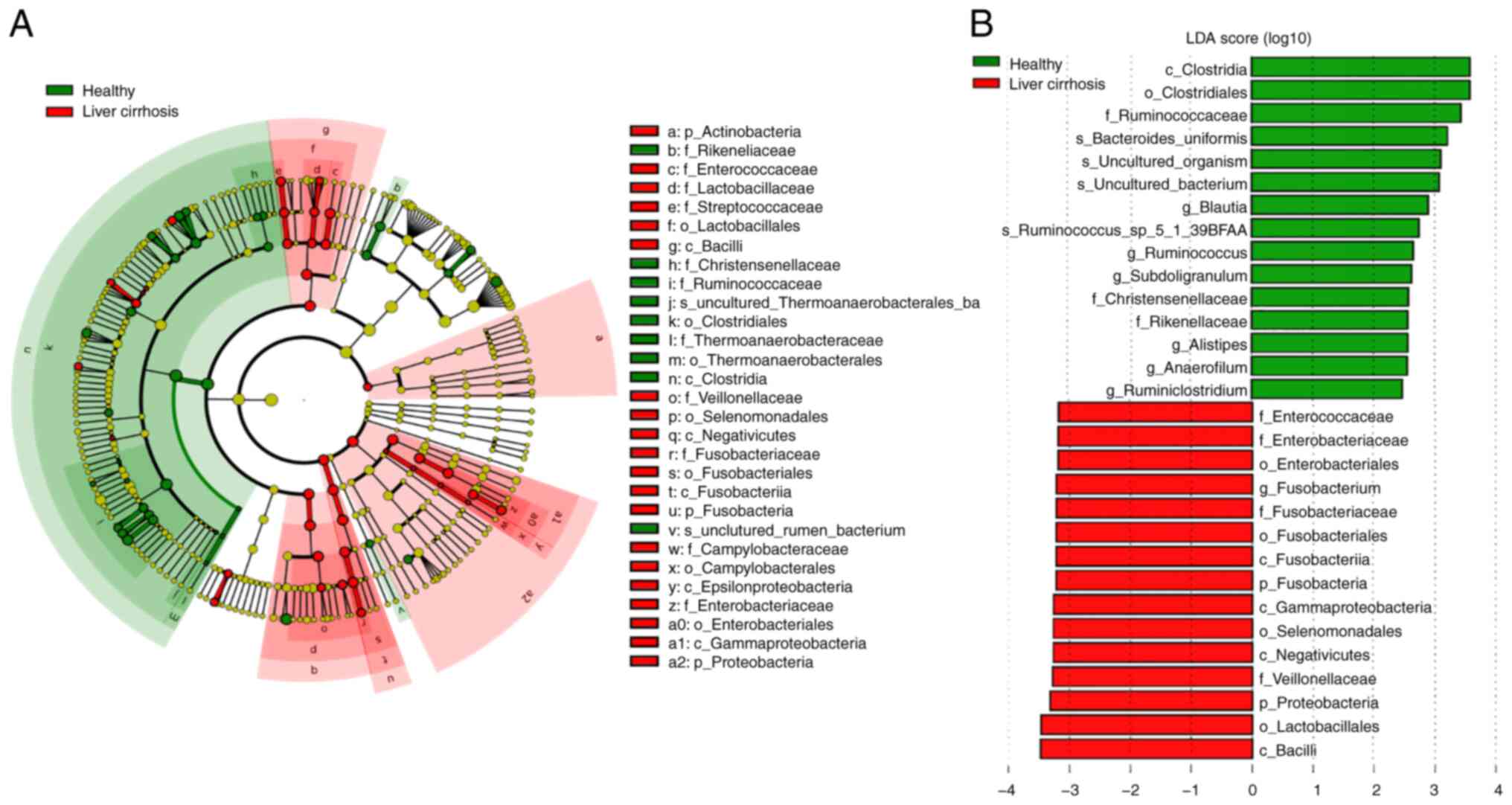
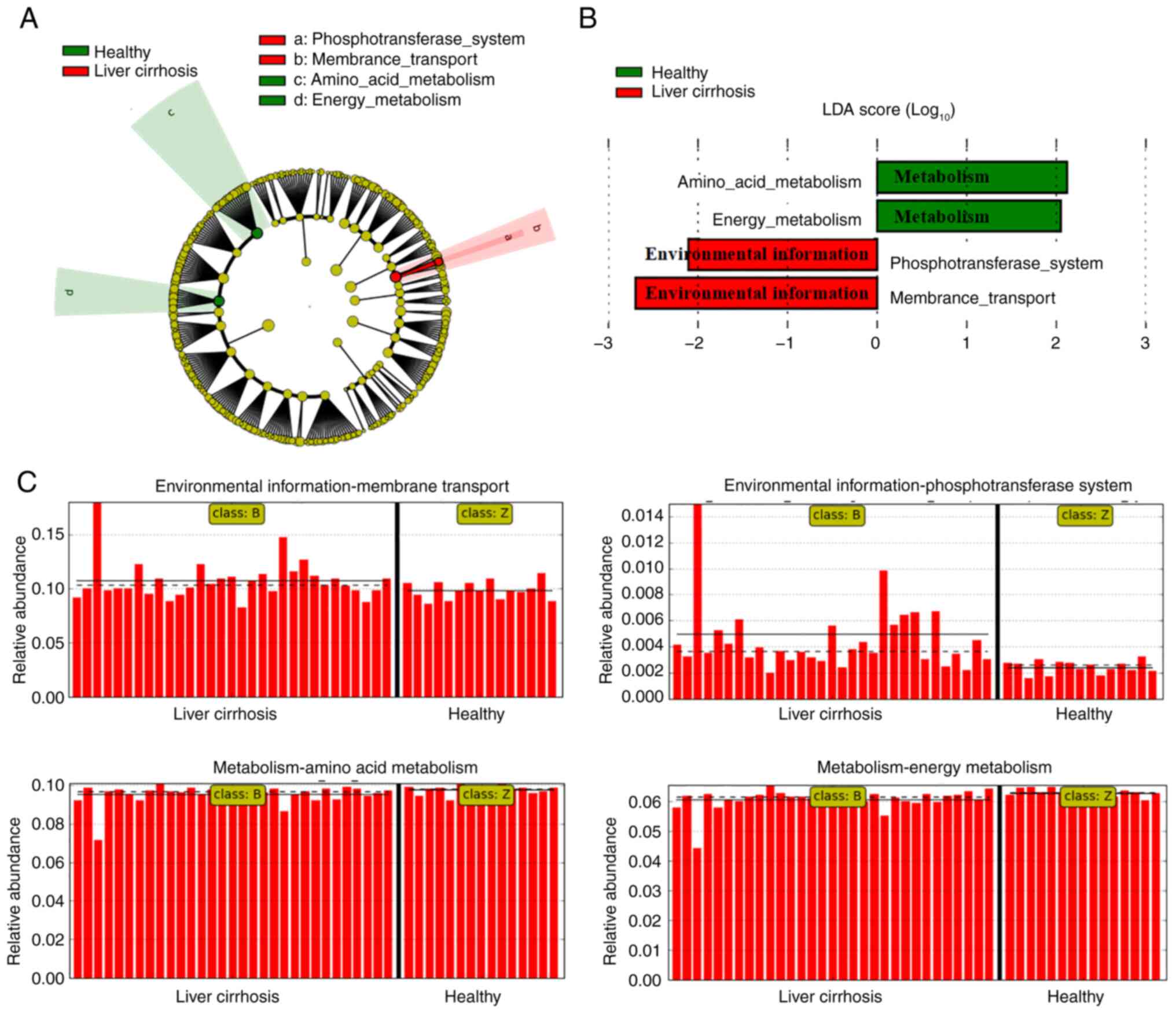
LEfSe was conducted to determine the most relevant
microbial taxa responsible for the differences between the groups
(Fig. 3A). This analysis identified
29 taxa that were differentially abundant between the two groups.
The microbiota of patients with liver cirrhosis was enriched in
Fusobacterium (genus), Veillonellaceae (family),
Lactobacillales (family), Negativicutes (class),
Gammaproteobacteria (class), and Enterobacteriaceae
(family), whereas healthy the microbiota of healthy volunteers was
enriched in Clostridiales (order), Ruminococcaceae
(family), Bacteroides (species), Subdoligranulum
(genus) and Subdoligranulum (genus; Fig. 3A and B). Moreover, the differentially abundant
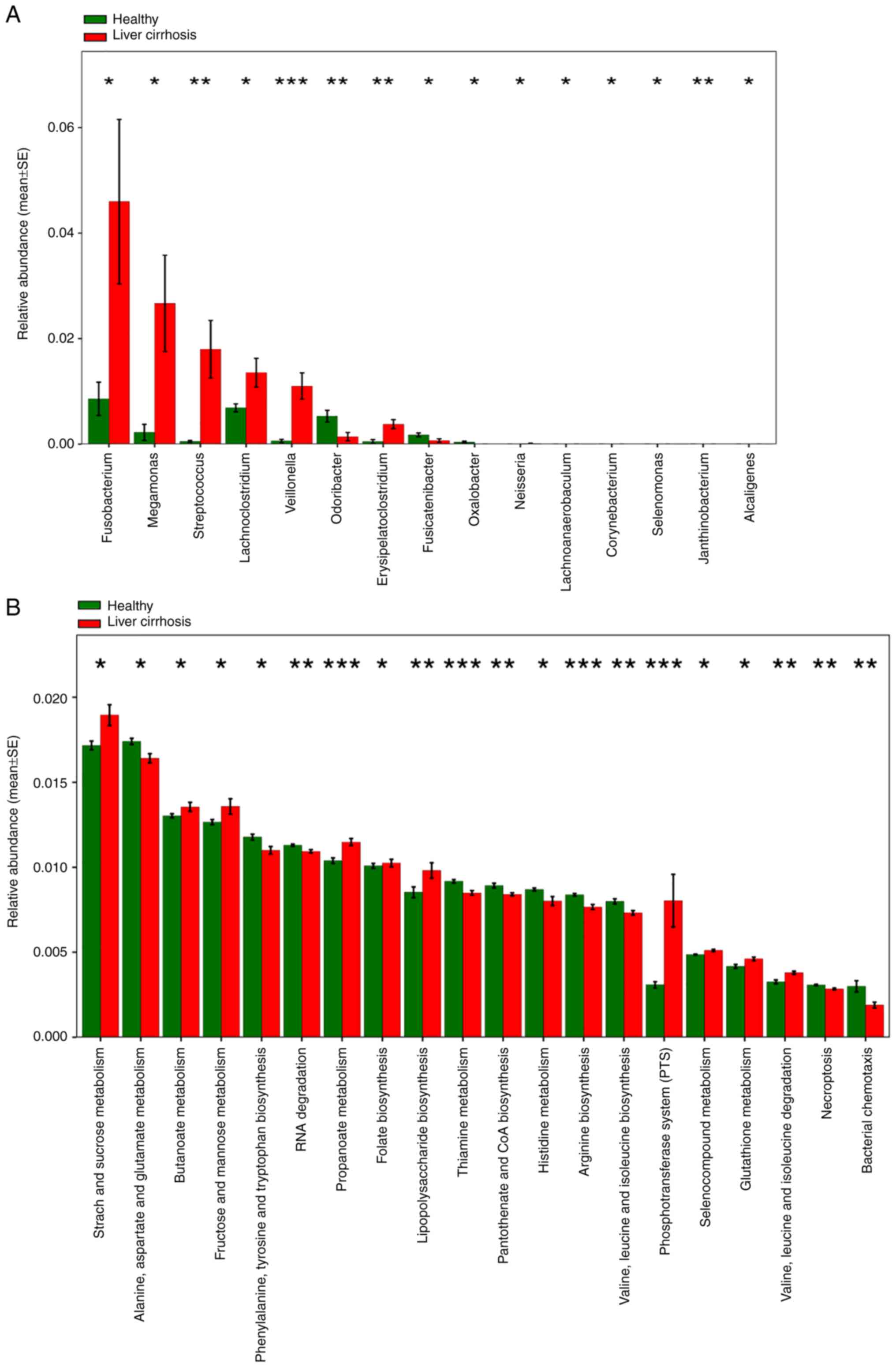
features of the microbial taxa at the genus level verified that the
abundance of Fusobacterium, Megamonas,
Streptococcus, Lachnoclostridium, Veillonella
and other bacteria was increased in the fecal samples of patients
with liver cirrhosis (Fig. 4A).
Based on the differences in community richness and
abundance, metagenome functional content based on the 16S
rRNA gene sequences was predicted using PICRUSt. Predicted KEGG
orthology pathways significantly enriched in liver cirrhosis
included ‘starch and sucrose metabolism’, ‘butanoate metabolism’,
‘fructose and mannose metabolism’, ‘propanoate metabolism’ and
‘phosphotransferase system (PTS)’, indicating that these metabolic
pathways participated in the progression of liver cirrhosis
(Fig. 4B). Moreover, to verify the
most relevant functional pathways based on the microbial
differences between healthy volunteers and patients with liver
cirrhosis, LEfSe analysis was performed. Functional composition of
the total gut microbiota in patients with liver cirrhosis was
primarily associated with ‘phosphotransferase system (PTS)’ and
‘membrane transport’ involved in environmental information
processing, in contrast to ‘amino acid metabolism’ and ‘energy
metabolism’ functions enriched in healthy volunteers (Fig. 5A-C). Collectively, these data
demonstrated the gut microbiota profile differs between healthy
volunteers and patients with liver cirrhosis, and that a specific
microbial community, diversity, and associated metagenome function
are present in those with liver cirrhosis.
Gut microbiota of cirrhotic patients
influences the host immune response and the gut barrier
The bacterial species with increased abundance in
liver cirrhosis contribute to environmental information and
metabolic pathway components, which may instigate inflammation of
the host (17). Moreover, the gut
microbiota participates in the preservation of tolerance and
immunity of mucosal surfaces by regulating organic inflammatory
responses (40). Based on these
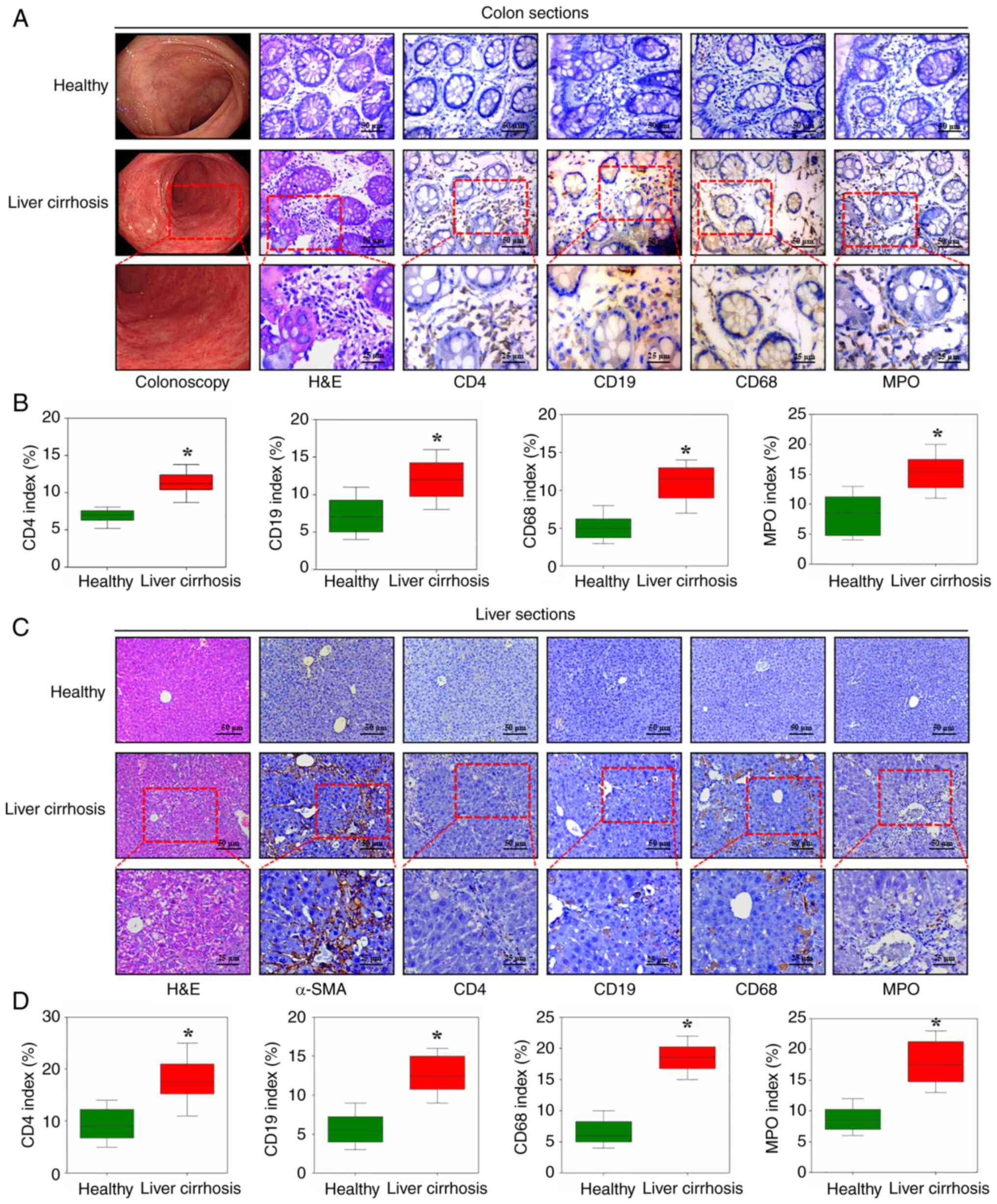
findings, the colonic mucosal specimens from patients with liver
cirrhosis and healthy volunteers were analyzed, which demonstrated
increased infiltration of inflammatory cells, including T
lymphocytes (CD4), B lymphocytes (CD19), macrophages (CD68) and
neutrophils (MPO), in the colonic tissues of patients with liver
cirrhosis. However, there was no notable histological architectural
alterations between the two groups based on H&E staining
(Fig. 6A and B). As expected, upregulated T and B
lymphocyte, macrophage and neutrophil infiltration, as well as
excess collagen deposition, was found in the liver tissues of
cirrhotic patients compared with those of healthy volunteers
(Fig. 6C and D). On this basis, the present study
investigated how gut microbial changes relate to barrier
dysfunction and inflammatory responses in cirrhosis.
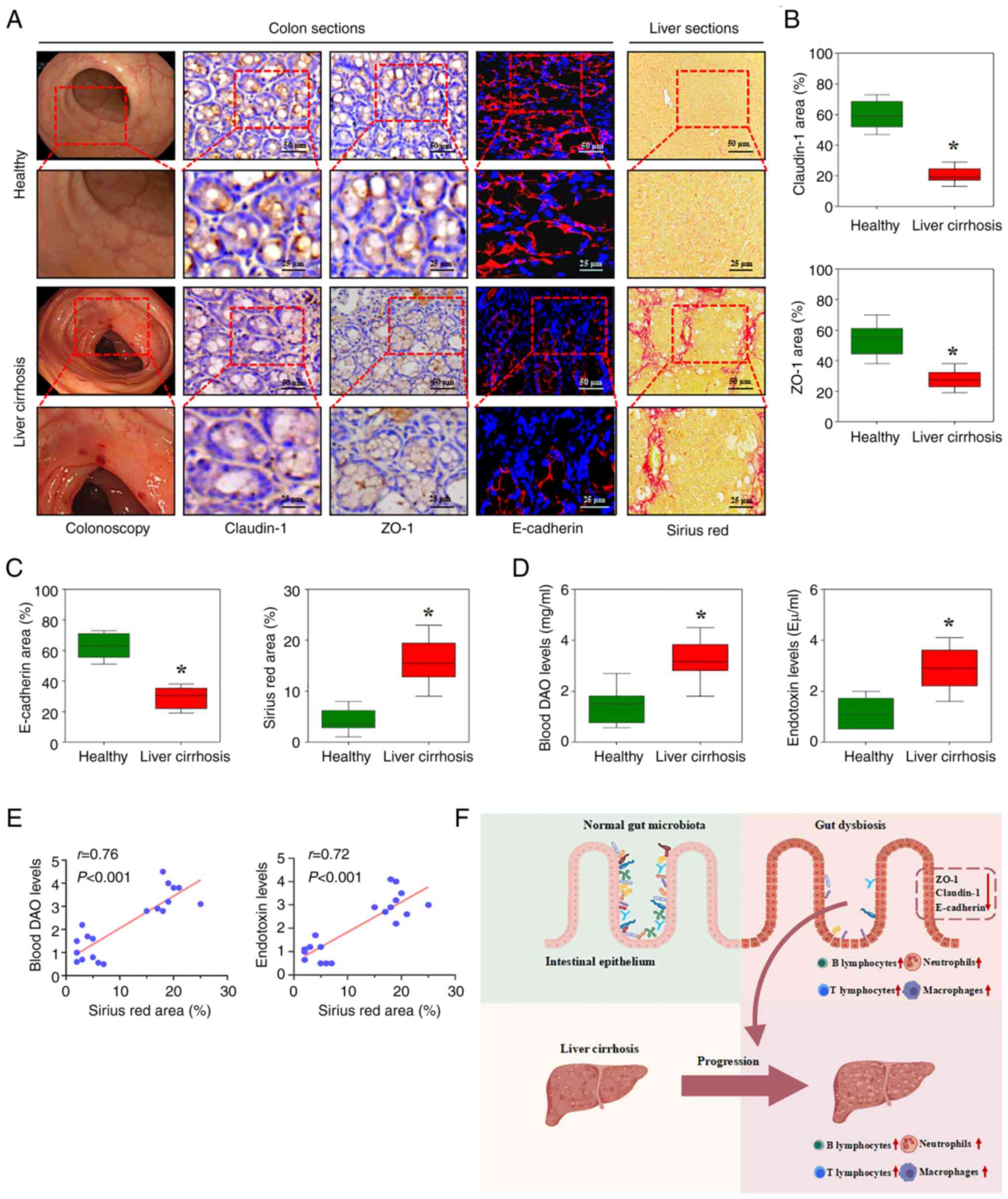
The functional composition of the total gut
microbiota in patients with liver cirrhosis primarily involved
‘phosphotransferase system (PTS)’ and ‘membrane transport’. Changes
in the gut microbiota modulate systemic microbe-derived metabolite
levels by altering intestinal permeability and the gut barrier,
especially tight junctions between epithelial cells (16). Claudin-1, ZO-1 and E-cadherin were
downregulated in colonic samples from patients with cirrhosis whose
liver tissues showed excess collagen deposition by Sirius red
staining (Fig. 7A-C). Intestinal
permeability, based on blood DAO determination, was significantly
increased, and blood endotoxin detection showed enhanced bacterial
translocation analysis in cirrhotic liver sections (Fig. 7D). In addition, the blood DAO and
endotoxin levels showed a positive correlation with collagen
deposition, as indicated by Sirius red staining of the liver tissue
(Fig. 7E). The results demonstrated
that the gut microbiota influences colonic and hepatic immune
responses, intestinal permeability and the gut barrier, which would
contribute to the progression of liver cirrhosis (Fig. 7F).
Discussion
The gut microbiome serves a key role in immune
development and function in the host, and in determining the host
metabolic state. An increasing number of studies have suggested
that the gut-derived microbiota and their components, metabolites,
nutrients and other signals can be delivered to the liver via the
portal vein circulation to serve as a bioreactor for autonomous
metabolic and immunological regulation and regulate responses
within the host environment (8,23,24).
Here, patients with HBV-associated cirrhosis exhibit distinct gut
microbial profiles compared with healthy individuals, characterized
by decreased diversity, enrichment of pro-inflammatory taxa,
functional shifts toward environmental information processing
pathways, compromised intestinal barrier function and marked
hepatic immune cell infiltration.
By comparing the gut microbiota between healthy
volunteers and patients with liver cirrhosis using high-throughput
sequencing of the 16S rRNA gene, the present study
demonstrated that patients with liver cirrhosis had decreased
microbial species richness, evenness, and diversity, although they
had similar microbial structures and communities according to core
bacterial genus analysis. This is supported by several studies
(41,42) indicating a disordered gut microbial
community and decreased diversity and species richness associated
with cirrhosis. Decreased microbial diversity is now recognized as
a feature of disease states, including inflammatory disorder,
colorectal cancer, and gastric carcinoma (43-46).
Furthermore, LEfSe analysis showed that the gut microbiota of
patients with liver cirrhosis was enriched in Fusobacterium,
Veillonellaceae, Lactobacillales,
Negativicutes, Gammaproteobacteria,
Enterobacteriaceae, whereas the proportion of phylum
Bacteroides, Clostridiales, Ruminococcaceae,
and Subdoligranulum was decreased compared with that of the
healthy volunteers. These results align with a previous study that
observed a marked loss of Bacteroides and significant
increases in Veillonella, Enterobacteriaceae and
Fusobacterium abundance in cirrhosis (47). This suggest that dysbiosis, or an
unfavorable change in the composition of the microbiome in liver
cirrhosis, is central to the pathophysiology of the onset,
progression and development of complications of liver cirrhosis.
Nonetheless, inconsistencies remain; for example, Chen et al
(47) reported decreased
Bacteroidetes and increased Proteobacteria and
Fusobacteria, whereas Sarangi et al (48) observed no significant differences in
microbial abundance. The differences in findings emphasize the need
for more research to clarify the specific roles of microbial taxa
in cirrhosis progression and treatment.
To explore functional consequences, KEGG orthology
analysis was performed on metagenomic sequencing data and found
that the total microbiota of patients with liver cirrhosis was
predominantly enriched in ‘phosphotransferase system (PTS)’ and
‘membrane transport’, pathways involved in environmental
information processing, whereas the microbiota of healthy controls
showed enrichment in ‘amino acid metabolism’ and ‘energy
metabolism’. Consistent with the present data, a previous study
also reported enhanced transport- and metabolism-associated
pathways, along with a marked loss of cell cycle-associated gene
functions, in cirrhotic patients (7,47).
Notably, the depletion of SCFA-producing taxa, such as
Faecalibacterium, Eubacterium hallii,
Ruminococcus and Agathobacter, has also been observed
in cirrhotic patients (49). These
butyrate-producing bacteria are key suppliers of energy for
colonocytes and contribute to maintaining mucosal integrity. Their
loss may compromise epithelial energy metabolism, attenuate
butyrate-mediated anti-inflammatory signaling and disrupt the
regulation of immune homeostasis. This depletion may aggravate the
decrease in tight junction proteins and the increased intestinal
permeability, thereby weakening barrier function and facilitating
microbial translocation to the liver (22,40).
These findings highlight the association between the gut microbiota
and host metabolism in liver cirrhosis, with the microbial balance
shifted toward a dysbiosis during the process of liver
cirrhosis.
The activation of inflammatory cells to produce
inflammatory cytokines and components is a key contributor to the
initiation and development of liver cirrhosis (28,29).
Increased T and B lymphocyte, macrophage and neutrophil
infiltration was detected in patients with liver cirrhosis compared
with healthy volunteers, and this was accompanied by excess
collagen deposition in liver tissue. These data indicated that
various inflammatory cells and responses are affected by
gut-derived microbiota, and their components, metabolites or
signals participate in the initiation and development of liver
fibrosis.
The dysbiotic microbial community associated with
liver cirrhosis is essential for the development and regulation of
the immune and metabolism systems of the host (7,19,20).
Changes in the gut microbiota modulate systemic microbe-derived
metabolite levels or signals by altering intestinal permeability
and the gut barrier, thus contributing to disease development
(23).
A critical mechanistic link between the dysbiotic
microbial community and the immune and metabolic systems of the
host is the structural compromise of the intestinal epithelial
barrier, which relies on tight junction proteins such as claudin-1
and ZO-1 and the adherens junction protein E-cadherin. To identify
potential associations between tight junctions in colonic
epithelial cells and microbial dysbiosis, the present study
analyzed colonic sections and found that claudin-1, ZO-1 and
E-cadherin levels were suppressed in colonic samples from cirrhotic
patients; this was associated with enhanced intestinal permeability
and bacterial translocation, suggesting that the tight junctions
between colonic epithelial cells were damaged during liver
fibrogenesis. In radiation-induced enteritis, gut dysbiosis
disrupts the localization of claudin-1, occludin and ZO-1,
weakening epithelial cohesion (50). Therapeutic modulation of the
microbiota can mitigate such injury; for example, Yu-Ping-Feng-San
(a traditional Chinese medicine decoction) treatment restores ZO-1,
occludin and claudin-1 expression in LPS-induced barrier damage
while decreasing inflammatory responses (51). At the molecular level, claudin-1
expression is regulated via the Piezo1/ROCK1/2 pathway, whereby
Piezo1 activation by mechanical or inflammatory cues decreases
claudin-1 protein expression and thereby impairs junctional
integrity (52). Similarly, loss of
SLC26A3 results in downregulation of ZO-1, occludin and E-cadherin
alongside microbial imbalance, underscoring the reciprocal
association between junctional protein expression and microbiota
composition (53). In metabolic
liver diseases such as NAFLD and NASH, breakdown of tight junctions
allows endotoxin influx to the liver, promoting inflammatory and
fibrotic changes (54). These data
highlight that gut barrier impairment, mediated by altered
junctional proteins in the context of dysbiosis, is a key driver of
microbial translocation and hepatic injury in cirrhosis.
While the present findings provide insights into the
gut microbiota-immune-barrier axis in HBV-associated cirrhosis,
several limitations should be noted. First, the cross-sectional
design precludes causal inference between microbiota dysregulation
and inflammation or barrier dysfunction, and the contribution of
HBV infection cannot be excluded; interventional approaches such as
probiotics, fecal microbiota transplantation or other
microbiota-targeted strategies are needed to test whether restoring
microbial balance can ameliorate these changes. Second, the sample
size may have limited the detection of subtle associations, and
data on diet, medication and antiviral therapy, important
microbiome modulators, were incomplete; these should be collected
in future longitudinal and interventional studies. Finally,
functional predictions based on 16S rRNA sequencing lack the
resolution of shotgun metagenomics, metatranscriptomics or targeted
metabolomics, which should be integrated to define microbiota-host
interactions in HBV-related cirrhosis. To address these issues,
future studies should adopt longitudinal designs with larger and
well-characterized cohorts, integrate clinical and dietary
information with multi-omics approaches, and evaluate
interventional strategies such as probiotic or prebiotic
supplementation, dietary modulation and fecal microbiota
transplantation to determine whether restoring microbial balance
can mitigate inflammation, reinforce barrier function and slow
disease progression.
In conclusion, the gut microbiota profile in
patients with HBV-associated liver cirrhosis differs markedly from
that of healthy individuals, showing enrichment of taxa such as
Fusobacterium, Veillonella and members of the
Enterobacteriaceae family, alongside depletion of beneficial
butyrate-producing genera including Faecalibacterium,
Ruminococcus and Agathobacter. These compositional
shifts, together with decreased overall diversity and altered
functional potential, particularly enrichment in
‘phosphotransferase system (PTS)’ and ‘membrane transport’ and
depletion of ‘amino acid metabolism’ and ‘energy metabolism’, are
closely associated with impaired colonic and hepatic immune
responses, increased intestinal permeability and compromised gut
barrier integrity. Gut microbiota dysbiosis contributes to the
pathophysiology of HBV-associated cirrhosis. Understanding these
microbial alterations may provide a basis for microbiota-targeted
strategies to mitigate hepatic inflammation and preserve barrier
function in affected patients. Future longitudinal and
interventional studies are warranted to establish causality and
evaluate the therapeutic potential of microbiota-targeted
interventions in HBV-associated cirrhosis.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Natural Science
Foundation of Guangdong Province (grant no. 2023A1515011204), the
National Natural Science Foundation of China (grant no. 82170569)
and the Science and Technology Planning Projects of Guangzhou City
(grant nos. 2024A04J6565 and 2025A03J3193).
Availability of data and materials
The data generated in the present study may be found
in the ScienceDB under accession number 31253.11.sciencedb.28999 or
at the following URL: doi.org/10.57760/sciencedb.28999.
Authors' contributions
KX and YZ designed the methodology and analyzed
data. KX and YZ confirm the authenticity of all the raw data. ShT,
XO and JL designed the methodology. SiT conceived the study and
wrote and edited the manuscript. All authors have read and approved
the final manuscript.
Ethics approval and consent to
participate
Informed consent was obtained from all participants
before the initiation of the study. The present study protocol was
approved by the Institute Research Ethics Committee of the Third
Affiliated Hospitals of Sun Yat-sen University (Guangzhou, China;
approval no. RG2023-033-01).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Huang DQ, Terrault NA, Tacke F, Gluud LL,
Arrese M, Bugianesi E and Loomba R: Global epidemiology of
cirrhosis-aetiology, trends and predictions. Nat Rev Gastroenterol
Hepatol. 20:388–398. 2023.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Hsu YC, Huang DQ and Nguyen MH: Global
burden of hepatitis B virus: Current status, missed opportunities
and a call for action. Nat Rev Gastroenterol Hepatol. 20:524–537.
2023.PubMed/NCBI View Article : Google Scholar
|
|
3
|
World Health Organization. Hepatitis B.
Fact sheet. Geneva, 2025. Available from: https://www.who.int/news-room/fact-sheets/detail/hepatitis-b.
|
|
4
|
Suh YG, Kim JK, Byun JS, Yi HS, Lee YS,
Eun HS, Kim SY, Han KH, Lee KS, Duester G, et al: CD11b(+) Gr1(+)
bone marrow cells ameliorate liver fibrosis by producing
interleukin-10 in mice. Hepatology. 56:1902–1912. 2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kornek M, Popov Y, Libermann TA, Afdhal NH
and Schuppan D: Human T cell microparticles circulate in blood of
hepatitis patients and induce fibrolytic activation of hepatic
stellate cells. Hepatology. 53:230–242. 2011.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Hammerich L and Tacke F: Hepatic
inflammatory responses in liver fibrosis. Nat Rev Gastroenterol
Hepatol. 20:633–646. 2023.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Usami M, Miyoshi M and Yamashita H: Gut
microbiota and host metabolism in liver cirrhosis. World J
Gastroenterol. 21:11597–11608. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Levy M, Blacher E and Elinav E:
Microbiome, metabolites and host immunity. Curr Opin Microbiol.
35:8–15. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Luedde T and Schwabe RF: NF-kappaB in the
liver-linking injury, fibrosis and hepatocellular carcinoma. Nat
Rev Gastroenterol Hepatol. 8:108–118. 2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Pabst O, Hornef MW, Schaap FG, Cerovic V,
Clavel T and Bruns T: Gut-liver axis: Barriers and functional
circuits. Nat Rev Gastroenterol Hepatol. 20:447–461.
2023.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Hov JR and Karlsen TH: The microbiota and
the gut-liver axis in primary sclerosing cholangitis. Nat Rev
Gastroenterol Hepatol. 20:135–154. 2023.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Anand S and Mande SS: Host-microbiome
interactions: Gut-Liver axis and its connection with other organs.
NPJ Biofilms Microbiomes. 8(89)2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Lynch SV and Pedersen O: The human
intestinal microbiome in health and disease. N Engl J Med.
375:2369–2379. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Lavelle A, Lennon G, Winter DC and
O'Connell PR: Colonic biogeography in health and ulcerative
colitis. Gut Microbes. 7:435–442. 2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Maslennikov R, Poluektova E, Zolnikova O,
Sedova A, Kurbatova A, Shulpekova Y, Dzhakhaya N, Kardasheva S,
Nadinskaia M, Bueverova E, et al: Gut microbiota and bacterial
translocation in the pathogenesis of liver fibrosis. Int J Mol Sci.
24(16502)2023.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Liu Y, Chen Z, Li C, Sun T, Luo X, Jiang
B, Liu M, Wang Q, Li T, Cao J, et al: Associations between changes
in the gut microbiota and liver cirrhosis: A systematic review and
meta-analysis. BMC Gastroenterol. 25(16)2025.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kang EJ, Cha MG, Kwon GH, Han SH, Yoon SJ,
Lee SK, Ahn ME, Won SM, Ahn EH and Suk KT: Akkermansia muciniphila
improve cognitive dysfunction by regulating BDNF and serotonin
pathway in gut-liver-brain axis. Microbiome. 12(181)2024.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Sharma SP, Gupta H, Kwon GH, Lee SY, Song
SH, Kim JS, Park JH, Kim MJ, Yang DH, Park H, et al: Gut microbiome
and metabolome signatures in liver cirrhosis-related complications.
Clin Mol Hepatol. 30:845–862. 2024.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Hackstein CP, Assmus LM, Welz M, Klein S,
Schwandt T, Schultze J, Förster I, Gondorf F, Beyer M, Kroy D, et
al: Gut microbial translocation corrupts myeloid cell function to
control bacterial infection during liver cirrhosis. Gut.
66:507–518. 2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Qin N, Yang F, Li A, Prifti E, Chen Y,
Shao L, Guo J, Le Chatelier E, Yao J, Wu L, et al: Alterations of
the human gut microbiome in liver cirrhosis. Nature. 513:59–64.
2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Shashni B, Tajika Y, Ikeda Y, Nishikawa Y
and Nagasaki Y: Self-assembling polymer-based short chain fatty
acid prodrugs ameliorate non-alcoholic steatohepatitis and liver
fibrosis. Biomaterials. 295(122047)2023.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Mao T, Zhang C, Yang S, Bi Y, Li M and Yu
J: Semaglutide alters gut microbiota and improves NAFLD in db/db
mice. Biochem Biophys Res Commun. 710(149882)2024.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Spiljar M, Merkler D and Trajkovski M: The
immune system bridges the gut microbiota with systemic energy
homeostasis: Focus on TLRs, mucosal barrier, and SCFAs. Front
Immunol. 8(1353)2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Acharya C and Bajaj JS: Gut microbiota and
complications of liver disease. Gastroenterol Clin North Am.
46:155–169. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Jandl B, Dighe S, Baumgartner M,
Makristathis A, Gasche C and Muttenthaler M: Gastrointestinal
biofilms: Endoscopic detection, disease relevance, and therapeutic
strategies. Gastroenterology. 167:1098–1112.e5. 2024.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wang J, He M, Yang M and An X: Gut
microbiota as a key regulator of intestinal mucosal immunity. Life
Sci. 345(122612)2024.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Kuo CH, Wu LL, Chen HP, Yu J and Wu CY:
Direct effects of alcohol on gut-epithelial barrier: Unraveling the
disruption of physical and chemical barrier of the gut-epithelial
barrier that compromises the host-microbiota interface upon alcohol
exposure. J Gastroenterol Hepatol. 39:1247–1255. 2024.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Jung Y, Witek RP, Syn WK, Choi SS,
Omenetti A, Premont R, Guy CD and Diehl AM: Signals from dying
hepatocytes trigger growth of liver progenitors. Gut. 59:655–665.
2010.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Seki E and Schwabe RF: Hepatic
inflammation and fibrosis: Functional links and key pathways.
Hepatology. 61:1066–1079. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Du YN, Tang XF, Xu L, Chen WD, Gao PJ and
Han WQ: SGK1-FoxO1 signaling pathway mediates Th17/Treg imbalance
and target organ inflammation in angiotensin II-induced
hypertension. Front Physiol. 9(1581)2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Muranski P and Restifo NP: Essentials of
Th17 cell commitment and plasticity. Blood. 121:2402–2414.
2013.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Liu R, Kang JD, Sartor RB, Sikaroodi M,
Fagan A, Gavis EA, Zhou H, Hylemon PB, Herzog JW, Li X, et al:
Neuroinflammation in murine cirrhosis is dependent on the gut
microbiome and is attenuated by fecal transplant. Hepatology.
71:611–626. 2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhao B, Jin Y, Shi M, Yu L, Li G, Cai W,
Lu Z and Wei C: Gut microbial dysbiosis is associated with
metabolism and immune factors in liver fibrosis mice. Int J Biol
Macromol. 258(129052)2024.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Santiago A, Sanchez E, Clark A, Pozuelo M,
Calvo M, Yañez F, Sarrabayrouse G, Perea L, Vidal S, Gallardo A, et
al: Sequential changes in the mesenteric lymph node microbiome and
immune response during cirrhosis induction in rats. mSystems.
4:e00278–18. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Durand F and Valla D: Assessment of the
prognosis of cirrhosis: Child-Pugh versus MELD. J Hepatol. 42
(Suppl 1):S100–S107. 2005.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Tan S, Chen X, Xu M, Huang X, Liu H, Jiang
J, Lu Y, Peng X and Wu B: PGE2/EP4 receptor
attenuated mucosal injury via β-arrestin1/Src/EGFR-mediated
proliferation in portal hypertensive gastropathy. Br J Pharmacol.
174:848–866. 2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Tan S, Li L, Chen T, Chen X, Tao L, Lin X,
Tao J, Huang X, Jiang J, Liu H and Wu B: β-Arrestin-1 protects
against endoplasmic reticulum stress/p53-upregulated modulator of
apoptosis-mediated apoptosis via repressing p-p65/inducible nitric
oxide synthase in portal hypertensive gastropathy. Free Radic Biol
Med. 87:69–83. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Tan S, Lu Y, Xu M, Huang X, Liu H, Jiang J
and Wu B: β-Arrestin1 enhances liver fibrosis through
autophagy-mediated Snail signaling. FASEB J. 33:2000–2016.
2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Xiao Y, Liu X, Xie K, Luo J, Zhang Y,
Huang X, Luo J and Tan S: Mitochondrial dysfunction induced by
HIF-1α under hypoxia contributes to the development of gastric
mucosal lesions. Clin Transl Med. 14(e1653)2024.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Wang R, Li B, Huang B, Li Y, Liu Q, Lyu Z,
Chen R, Qian Q, Liang X, Pu X, et al: Gut microbiota-derived
butyrate induces epigenetic and metabolic reprogramming in
myeloid-derived suppressor cells to alleviate primary biliary
cholangitis. Gastroenterology. 167:733–749.e3. 2024.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Solé C, Guilly S, Da Silva K, Llopis M,
Le-Chatelier E, Huelin P, Carol M, Moreira R, Fabrellas N, De Prada
G, et al: Alterations in gut microbiome in cirrhosis as assessed by
quantitative metagenomics: relationship with acute-on-chronic liver
failure and prognosis. Gastroenterology. 160:206–218.e13.
2021.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Shen Y, Wu SD, Chen Y, Li XY, Zhu Q,
Nakayama K, Zhang WQ, Weng CZ, Zhang J, Wang HK, et al: Alterations
in gut microbiome and metabolomics in chronic hepatitis B
infection-associated liver disease and their impact on peripheral
immune response. Gut Microbes. 15(2155018)2023.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Hui W, Yu D, Cao Z and Zhao X: Butyrate
inhibit collagen-induced arthritis via Treg/IL-10/Th17 axis. Int
Immunopharmacol. 68:226–233. 2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Gevers D, Kugathasan S, Denson LA,
Vazquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song
SJ, Yassour M, et al: The treatment-naive microbiome in new-onset
Crohn's disease. Cell Host Microbe. 15:382–392. 2014.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Lepage P, Häsler R, Spehlmann ME, Rehman
A, Zvirbliene A, Begun A, Ott S, Kupcinskas L, Doré J, Raedler A
and Schreiber S: Twin study indicates loss of interaction between
microbiota and mucosa of patients with ulcerative colitis.
Gastroenterology. 141:227–236. 2011.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Ahn J, Sinha R, Pei Z, Dominianni C, Wu J,
Shi J, Goedert JJ, Hayes RB and Yang L: Human gut microbiome and
risk for colorectal cancer. J Natl Cancer Inst. 105:1907–1911.
2013.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei
D, Wang Y, Zhu B and Li L: Characterization of fecal microbial
communities in patients with liver cirrhosis. Hepatology.
54:562–572. 2011.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Sarangi AN, Goel A, Singh A, Sasi A and
Aggarwal R: Faecal bacterial microbiota in patients with cirrhosis
and the effect of lactulose administration. BMC Gastroenterol.
17(125)2017.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Qi P, Yang XX, Wang CK, Sang W, Zhang W
and Bai Y: Analysis of gut and circulating microbiota
characteristics in patients with liver cirrhosis and portal vein
thrombosis. Front Microbiol. 16(1597145)2025.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Jian Y, Zhang D, Liu M, Wang Y and Xu ZX:
The impact of gut microbiota on radiation-induced enteritis. Front
Cell Infect Microbiol. 11(586392)2021.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Wang Y, Wang Y, Ma J, Li Y, Cao L, Zhu T,
Hu H and Liu H: YuPingFengSan ameliorates LPS-induced acute lung
injury and gut barrier dysfunction in mice. J Ethnopharmacol.
312(116452)2023.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Jiang Y, Song J, Xu Y, Liu C, Qian W, Bai
T and Hou X: Piezo1 regulates intestinal epithelial function by
affecting the tight junction protein claudin-1 via the ROCK
pathway. Life Sci. 275(119254)2021.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Kumar A, Priyamvada S, Ge Y, Jayawardena
D, Singhal M, Anbazhagan AN, Chatterjee I, Dayal A, Patel M, Zadeh
K, et al: A novel role of SLC26A3 in the maintenance of intestinal
epithelial barrier integrity. Gastroenterology. 160:1240–1255.e3.
2021.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Jadhav K and Cohen TS: Can you trust your
gut? Implicating a disrupted intestinal microbiome in the
progression of NAFLD/NASH. Front Endocrinol (Lausanne).
11(592157)2020.PubMed/NCBI View Article : Google Scholar
|