Introduction
Meningiomas are the most common primary intracranial
tumors, originating from arachnoid cap cells (1) and classified into WHO Grades 1, 2 and
3, based on various histological and anaplastic features, number of
mitotic figures, invasive growth patterns and genetic
characteristics (2). While most
cases are benign Grade 1 tumors (80.5%), Grades 2 and 3 comprise
17.7 and 1.7%, respectively, and exhibit more aggressive behavior
(3). Distant metastasis occurs in
only 0.18% of cases, typically to the lungs, bones, spinal cord and
liver, and is associated after the primary tumor has been diagnosed
(93%). However, ~6% of metastatic meningiomas are found
simultaneously with the primary tumor at the time of diagnosis.
Metastatic meningiomas are associated with male sex, large tumor
size and WHO Grade 3(4). The
first-line treatment for all meningiomas is surgical resection
(5); however, surgery is not
entirely effective, as recurrences are reported more frequently in
patients with higher-grade tumors, even after extensive surgical
resection (6). Consequently, these
tumors are more likely to grow steadily and recur. Therefore,
treatments for high-grade meningiomas are still limited and are
being developed to better manage the morbidity and mortality of
patients with meningiomas (7).
One emerging area of interest in understanding the
aggressive behavior of such tumors is the study of sialylation, a
post-translational modification known to influence cancer
progression. Sialylation, the addition of sialic acids to proteins
and lipids, is a critical post-translational modification that
influences cell signaling, recognition and immune responses.
Sialylation occurs via three prominent linkages: α2,3, α2,6 and
α2,8, which are catalyzed by a group of sialyl-transferases (STs).
This process plays a crucial role in various biological functions
and cellular activities, such as cell-cell communication, cellular
recognition and cell adhesion, and affects the circulating
half-lives of numerous glycoproteins (8). Aberrant sialylation resulting from
altered expression of ST genes has been implicated in cancer
progression and development (9).
Alterations in this process can lead to changes in protein
structure or impair their function, thereby promoting oncogenic
activities, such as increased cell proliferation and survival. In
particular, aberrant sialylation, increased tumor growth, and
metastasis have been widely described (10). For instance, the addition of
α2,6-linked sialic acid to β1 integrin increases its affinity for
collagen I, thereby enhancing tumor cell migration and invasion
(11). Similarly, α2,3-sialylation
of CD44 enhances its interaction with hyaluronic acid (12), which in turn promote further cancer
cell migration and metastasis. While the role of sialylation in the
progression of various cancers has been extensively studied, its
specific impact on the progression and aggressiveness of
meningiomas remains poorly understood.
The presence of sialylated tumor-associated
carbohydrate antigens, such as sialyl Tn (STn), is frequently
linked to poor clinical outcomes due to their role in promoting
tumor invasiveness (13).
Additionally, a previous study by the authors found that patients
with meningiomas exhibit altered expression of glycosyl-transferase
enzymes (14). The present study
aims to investigate the expression patterns of STs specifically
β-galactoside α2,3 and α2,6 ST family members in different grades
of meningiomas and examine how altering sialylation affects the
migratory and invasive capabilities of meningioma cells.
Materials and methods
Gene expression analysis by Gene
Expression Omnibus (GEO) datasets
The gene expression levels of β-galactoside α2,3 and
α2,6 ST family members in meningiomas were retrieved from GEO
database (https://www.ncbi.nlm.nih.gov/), including entries
GSE16581, GSE74385, GSE136661 and GSE43290. Information on each GEO
entry is presented in Table I.
GEO2R is an interactive web tool that enables users to compare two
or more groups of Samples in a GEO series to identify genes that
are differentially expressed across experimental conditions. Thus,
all expression data from each GEO entry were used in GEO2R to
identify genes that were differentially expressed among the WHO
Grade 1, 2 and 3 meningiomas. All expression data were transformed
to log2 for analysis. The differential expression of ST genes
across WHO grades 1, 2 and 3 meningiomas was analyzed using
GraphPad Prism® (version 8.0; GraphPad Software, Inc.;
Dotmatics). Statistical significance was set at P<0.05.
 | Table IGEO entries used in the present
study. |
Table I
GEO entries used in the present
study.
Cell culture and 3Fax treatment
The human HKBMM cell line (Meningioma WHO grade 3)
was provided by Associate Professor Norie Araki (Kumamoto
University, Kumamoto, Japan). The cells were cultured in Dulbecco's
Modified Eagle's Medium (DMEM; cat. no. 121000-046; MilliporeSigma)
supplemented with 10% heat-inactivated fetal bovine serum (FBS;
cat. no. A5256701; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin (cat. no. 15140-122; Thermo Fisher
Scientific, Inc.). The cells were cultured in a humidified
incubator at 37˚C under 5% CO2. For 3Fax treatments, the
HKBMM cell line, at a cell density of 2x104 cells, was
seeded into a 24-well plate and cultured overnight in DMEM
containing 10% FBS. After overnight seeding, the culture
supernatant was replaced with a new medium containing 500 µM
3Fax-peracetyl-Neu5Ac (3Fax; cat. no. 566224; Merck KGaA) or DMSO
(cat. no. 04707516001; DMSO; Carlo Erba™) as a control, and the
cells were incubated for five days. The expression of sialic acids
using Maackia amurensis lectin II (MAL II) and Sambucus
nigra lectin (SNA; cat. no. PK-4000; Vector Laboratories Inc.)
staining was performed on both treated and control cells, and the
results were analyzed using an AttuneTM NxT Acoustic
Focusing Cytometer (Thermo Fisher Scientific, Inc.). FACS data were
analyzed using Flowing software version 2.5.1 (Turku Bioscience).
Sialic acid expression was measured before subsequent experiments.
The experiment was performed in three replicates.
Lectin staining and flow cytometry
analysis
3Fax-treated cells and control cells were
trypsinized using 0.25% trypsin-EDTA (cat. no. 25200-072; Gibco;
Thermo fisher Scientific, Inc.) and centrifuged at 400 x g for 3
min at 4˚C. The cell pellets were washed by resuspending in 1X PBS.
The cells were fixed using 4% paraformaldehyde in 1X PBS for 15 min
at room temperature. The fixed cells were then blocked for
non-specific binding with 2% BSA (HIMedia Laboratories, LLC) in 1X
PBS for 30 min on ice. Next, the cells were stained with SNA and
MAL II biotinylated lectin for 1 h on ice. After staining, the
stained cells were incubated with streptavidin-Alexa Fluor™ 488
conjugate (cat. no. S11223; Thermo Fisher Scientific, Inc.) for 30
min on ice. The cells were then resuspended in 2% FACS buffer and
analyzed using an AttuneTM NxT Acoustic Focusing
Cytometer. Forward and side scatter were used to select the
population. The BL1 channel was used to detect the expression of
SNA and MAL II positivity, and a histogram represented the positive
expression. The FACS data were analyzed using the Flowing software
version 2.5.1 (Turku Bioscience). The experiment was performed in
three replicates.
Cell proliferation by WST-8 viability
assay
3Fax-treated cells and control cells were
trypsinized, seeded at a density of 1,000 cells per well in a
96-well plate and incubated overnight. Cell proliferation was
measured daily using the Cell Counting Kit-8 (WST-8; cat. no.
ab228554; Abcam) at 10 µl per well for 7 days. Briefly, the cells
were incubated with the WST-8 solution for 3 h at 37˚C under 5%
CO2, and cell viability was assessed by measuring the
absorbance at 460 nm using a TECAN Infinite 200 Pro microplate
reader (Tecan Group, Ltd.). The experiment was performed in three
replicates.
Cell proliferation and survival by
colony formation assay
3Fax-treated cells and control cells were
trypsinized, seeded at a density of 500 cells in a 100-mm culture
dish and incubated overnight. The cells were cultured in complete
DMEM containing 10% FBS and incubated at 37˚C under 5%
CO2 for 14 days. The cells were then washed with 1X PBS,
fixed with absolute methanol for 30 min at -4˚C, and stained with
5% crystal violet for 30 min at room temperature (RT).
Subsequently, the stained cells were washed with tap water, and
images of colony formation were captured under a stereomicroscope.
Only colonies with >50 cells were included in the count. The
experiment was performed in three replicates.
Wound healing assay
The cells were seeded at a density of
2x104 cells in a 24-well plate and cultured overnight in
DMEM containing 10% FBS. After seeding for 24 h, the culture
supernatant was replaced with a new medium containing 500 µM of
3Fax or DMSO and incubated for 5 days to reach 90% confluence. The
3Fax-treated and control cells were scratched using 200-µl pipette
tips to create a cell-free area. The medium was then replaced with
serum-free medium. The cells were further incubated, and wound
closure in the cell-free area was assessed at 18 and 24 h. The
cell-free area was captured using an inverted light microscope, and
the images were analyzed using ImageJ software (version 1.54 g;
National Institutes of Health). Wound closure was calculated using
the following formula: (area of original wound-area of wound during
healing)/area of original wound. The experiment was performed in
three replicates.
Transwell migration and invasion
assay
The Transwell migration assay utilized 6.5 mm
inserts with 8.0-µM polycarbonate membranes (cat. no. 3422; Costar®
Transwell®; Corning Inc.) in 24-well plates. In the Transwell
invasion assay, Matrigel (cat. no. 354234; Corning Inc.) was
thawed, and 100 µl of a 6 mg/ml Matrigel matrix was added to 6.5-mm
inserts with 8.0-µM polycarbonate membranes. Matrigel was spread
evenly using a pipette tip and incubated at 37˚C for 2 h to allow
Matrigel to solidify. Subsequently, 3Fax-treated and control cells
at a density of 3x104 cells were suspended in 200 µl of
serum-free medium and seeded into the upper chamber. Meanwhile, 600
µl of DMEM containing 10% FBS was added to the lower chamber. The
cells were then incubated for 18 and 24 h. The cells that had
crossed the membrane were fixed with 10% trichloroacetic acid at
4˚C overnight and stained using the Sulforhodamine B colorimetric
assay according to a previous study (15) for visualization. Images were
captured using an inverted light microscope at x10 magnification.
The number of migratory cells was counted and analyzed using ImageJ
software (version 1.54 g; National Institutes of Health). The
experiment was performed in three replicates.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
3Fax-treated and control cells were extracted from
total RNA using TRIzol reagent (cat. no. 15596026; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
quantity and purity of RNA were measured using a NanoDrop 2000
spectrophotometer (Thermo Fisher Scientific, Inc.). RNA purity was
indicated via the A260/A280 nm ratio >1.8. RNA quality was
assessed using agarose gel electrophoresis. Total RNA was then
reverse transcribed into cDNA using the SensiFAST cDNA Synthesis
Kit (cat. no. 65054; Meridian Bioscience) according to the
manufacturer's protocol. qPCR was performed on the generated cDNA
to evaluate the expression levels of epithelial-mesenchymal
transition (EMT)-related transcription factors (TFs) and
EMT-related proteins using iTag Universal SYBR Green Supermix (cat.
no. 1725121; Bio-Rad Laboratories, Inc.). The thermocycling
conditions were performed by initial denaturation at 95˚C for 5
min, followed by 40 cycles of denaturation at 95˚C for 10 sec,
annealing at 55-63˚C for 10 sec and a final extension at 72˚C for
10 sec. The primers used and annealing temperatures for all
investigated genes in this experiment are shown in Table II. The beta-actin gene was used as
an internal control to normalize gene expression, and the relative
mRNA expression levels were calculated using the 2-ΔΔCq
method (16). The experiment was
performed in three replicates.
 | Table IIPrimer sequences used in expression
of EMT markers. |
Table II
Primer sequences used in expression
of EMT markers.
| Assay | Gene name | Primer sequence
(5'-3') | Annealing
temperature (˚C) |
|---|
| EMT-related
transcription factors | Slug | F: GCC TCC AAA AAG
CCA AAC TAC | 62 |
| | | R: GAG GAT CTC TGG
TTG TGG TAT GAC | |
| | ZEB1 | F: ACC TCT TCA CAG
GTT GCT CCT | 62 |
| | | R: AGT GCA GGA GCT
GAG AGT CA | |
| | Snail | F: TTC TCA CTG CCA
TGG AAT TCC | 62 |
| | | R: GCA GAG GAC ACA
GAA CCA GAA | |
| EMT-related
proteins | CDH1 | F: TAC GCC TGG GAC
TCC ACC TA | 62 |
| | | R: CCA GAA ACG GAG
GCC TGA T | |
| | CDH2 | F: ATC CTG CTT ATC
CTT GTG CTG | 60 |
| | | R: GTC CTG GTC TTC
TTC TCC TCC | |
| | MMP2 | F: CGT CTG TCC CAG
GAT GAC ATC | 63 |
| | | R: ATG TCA GGA GAG
GCC CCA TA | |
| | MMP7 | F:
CAGATGTGGAGTGCCAGATG | 60 |
| | | R:
TGTCAGCAGTTCCCCATACA | |
| | MMP9 | F: TTC CAG TAC CGA
GAG AAA GCC TAT | 62 |
| | | R: GGT CAC GTA GCC
CAC TTG GT | |
| Internal
control | ACTB | F: GAT CAG CAA GCA
GGA GTA TGA CG | 55-63 |
| | | R: AAG GGT GTA ACG
CAA CTA AGT CAT AG | |
SDS-PAGE and western blot
analysis
3Fax-treated cells and control cells were collected
for preparation of whole-cell lysate using protein lysis buffer
containing 10% TritonX, protease inhibitor cocktail (Roche
Diagnostics GmbH) and Tris-lysis buffer pH 7.5. The protein
concentrations of all samples were determined using the Pierce BCA
Protein Assay Kit (cat. no. 2322, Thermo Fisher Scientific, Inc.).
Total proteins (50 µg) were subjected to 10% SDS-polyacrylamide gel
electrophoresis and transferred to a nitrocellulose membrane (cat.
no. 1060003; MilliporeSigma). Non-specific binding was blocked
using 5% BSA at RT for 1 h. The membrane was probed with a specific
primary antibody overnight at 18˚C. The primary antibodies used
were matrix metalloproteinase 9 (MMP 9; cat. no. 3852),
Snail (cat. no. 3879), Akt (cat. no. 4685), phosphorylated
(p-)Akt (cat. no. 4060), Erk (cat. no. 4695) and p-Erk (cat. no.
9101); all antibodies were diluted at 1:1,000 and purchased from
Cell Signaling Technology, Inc. Simultaneously, β-actin (cat. no.
sc-47778) was obtained from Santa Cruz Biotechnology, Inc. After
incubation, secondary antibodies were added at a dilution of
1:2,000 and incubated for 1 h at RT. Anti-mouse IgG, HRP-linked
antibody (cat. no. 7074) and anti-rabbit IgG, HRP-linked antibody
(cat. no. 7076) were purchased from Cell Signaling Technology, Inc.
Immunodetection using a chemiluminescent HRP substrate was
performed to visualize the target protein signals using ImageQuant™
LAS 500 (Cytiva). The density bands of each target protein were
captured using ImageJ software (version 1.54 g; National Institutes
of Health). The experiment was performed in three replicates.
Statistical analysis
Data were analyzed using GraphPad Prism® (version
8.0; GraphPad Software, Inc.; Dotmatics). One-way ANOVA followed by
Bonferroni's multiple comparisons test was used to determine the
differential expression of ST genes across WHO Grade 1, 2 and 3
meningiomas. Differences between the two groups were further
analyzed using unpaired t-tests. P<0.05 was considered to
indicate a statistically significant difference.
Results
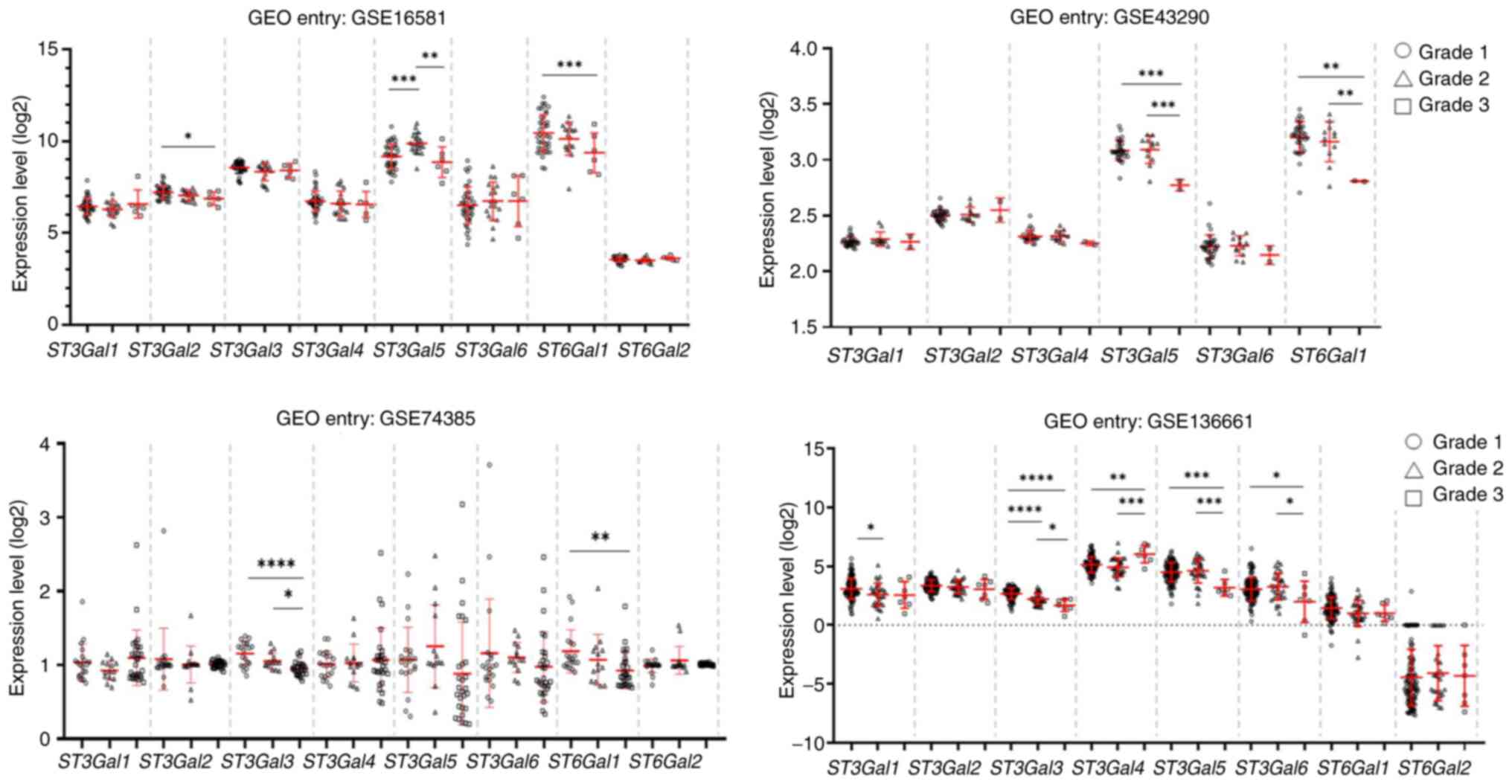
Downregulation of ST genes is
associated with the progression of meningiomas
To address the significance of ST in meningioma
development and progression, the gene expression of β-galactoside
α2,3 and α2,6 STs (ST3Gals and ST6Gals), including ST3Gal1, 2,
3, 4, and 5, and ST6Gal1 and 2, was obtained from four
GEO datasets: GSE16581, GSE43290, GSE74385 and GSE136661. The
results demonstrated that downregulation of the ST3Gal2,
ST3Gal3, ST3Gal4, ST3Gal5 and ST6Gal1 genes was observed
across four datasets and was associated with WHO Grade 3
meningiomas. Additionally, decreased expression of ST3Gal3 was
commonly observed in two datasets (GSE74385 and GSE136661), whereas
decreased expression of ST3Gal5 was observed in three
datasets, including GSE16581, GSE43290 and GSE136661. Moreover, the
expression of ST6Gal1 was significantly lower in WHO Grade 3
meningiomas than in WHO Grade 1 meningiomas in GSE16581, GSE43290
and GSE74385 (Fig. 1). These
findings suggested that the downregulation of these STs may play a
crucial role in meningioma progression.
Suppression of ST activity does not
affect meningioma cell proliferation
To investigate the role of ST in meningioma
development and progression, a malignant meningioma cell line,
HKBMM, was selected for functional studies. Talabnin et al
(14) demonstrated the high
expression of sialic acids and ST genes in HKBMM cells. HKBMM cells
were treated with a ST inhibitor (3Fax) or DMSO for 5 days. The
cell viability test demonstrated that 3Fax at 500 µM was not toxic
to HKBMM cells (Fig. 2A) and was
therefore selected for subsequent experiments. Surface sialic acid
expression was measured using MAL II to detect α2,3-sialylated
glycans and SNA to detect α2,6-sialylated glycans. The results
demonstrated that the staining intensities of both α2,3- and
α2,6-sialylated glycans were significantly reduced in 3Fax-treated
cells, indicating successful inhibition of sialylation (Fig. 2B). It was then investigated whether
the suppression of sialylation affects the proliferation of
meningioma cells. 3Fax-treated cells and control cells were used to
monitor cell proliferation via cell proliferation and colony
formation assays. The results showed no significant difference in
cell proliferation and survival (Fig.
2C and D) between the
3Fax-treated and control cells.
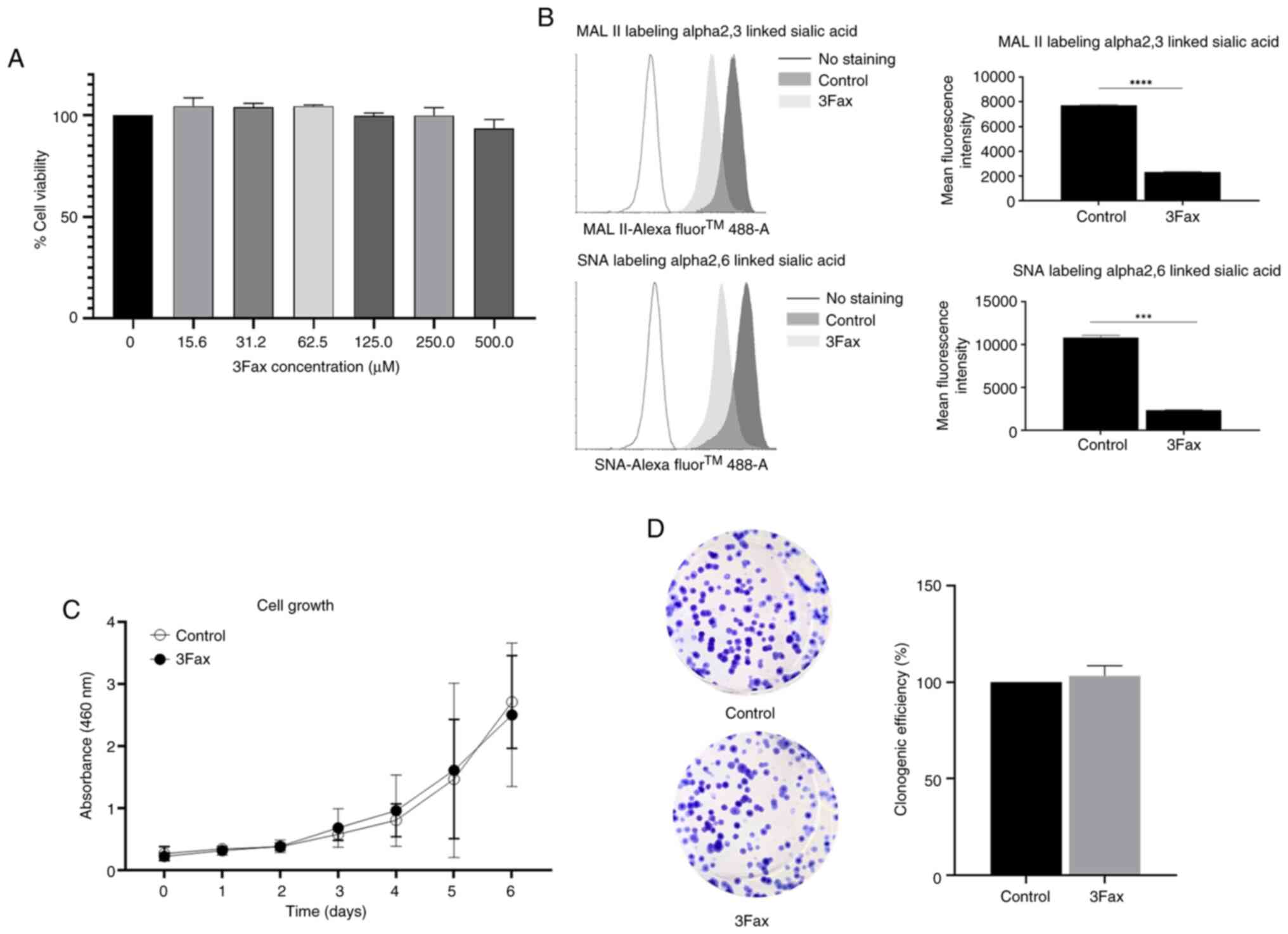 | Figure 2Suppression of sialylation does not
affect the proliferation of meningioma cells. HKBMM cells were
treated with various concentrations of 3Fax (0, 15.6, 31.2, 62.5,
125.0, 250.0, 500.0 µM) for 5 days. (A) Cytotoxic effect of 3Fax
was assessed using the WST-8 viability. (B) Histogram plots and
mean fluorescence intensity from lectin flow cytometry showed
decreased expression of α2,3-linked MAL II and α2,6-linked SNA
lectin sialic acids in 3Fax-treated cells (500 µM) compared with
DMSO-treated control cells. (C and D) Cell proliferation and
survival were assessed using the (C) WST-8 viability and (D) colony
formation assays. Values are expressed as the mean ± standard
deviation from three independent experiments. ***P≤0.001
and ****P≤0.0001 vs. control. MAL II, Maackia
amurensis lectin II; SNA, Sambucus nigra lectin. |
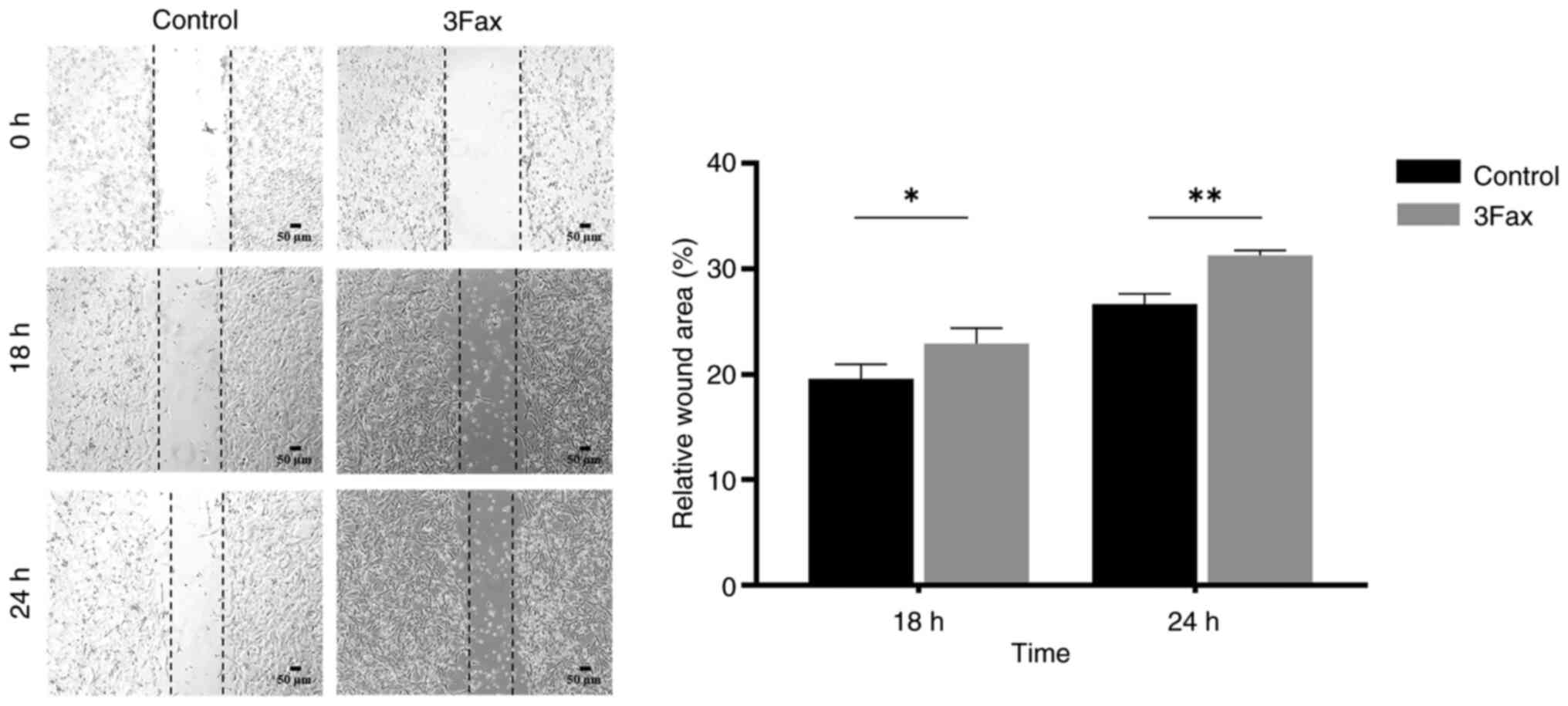
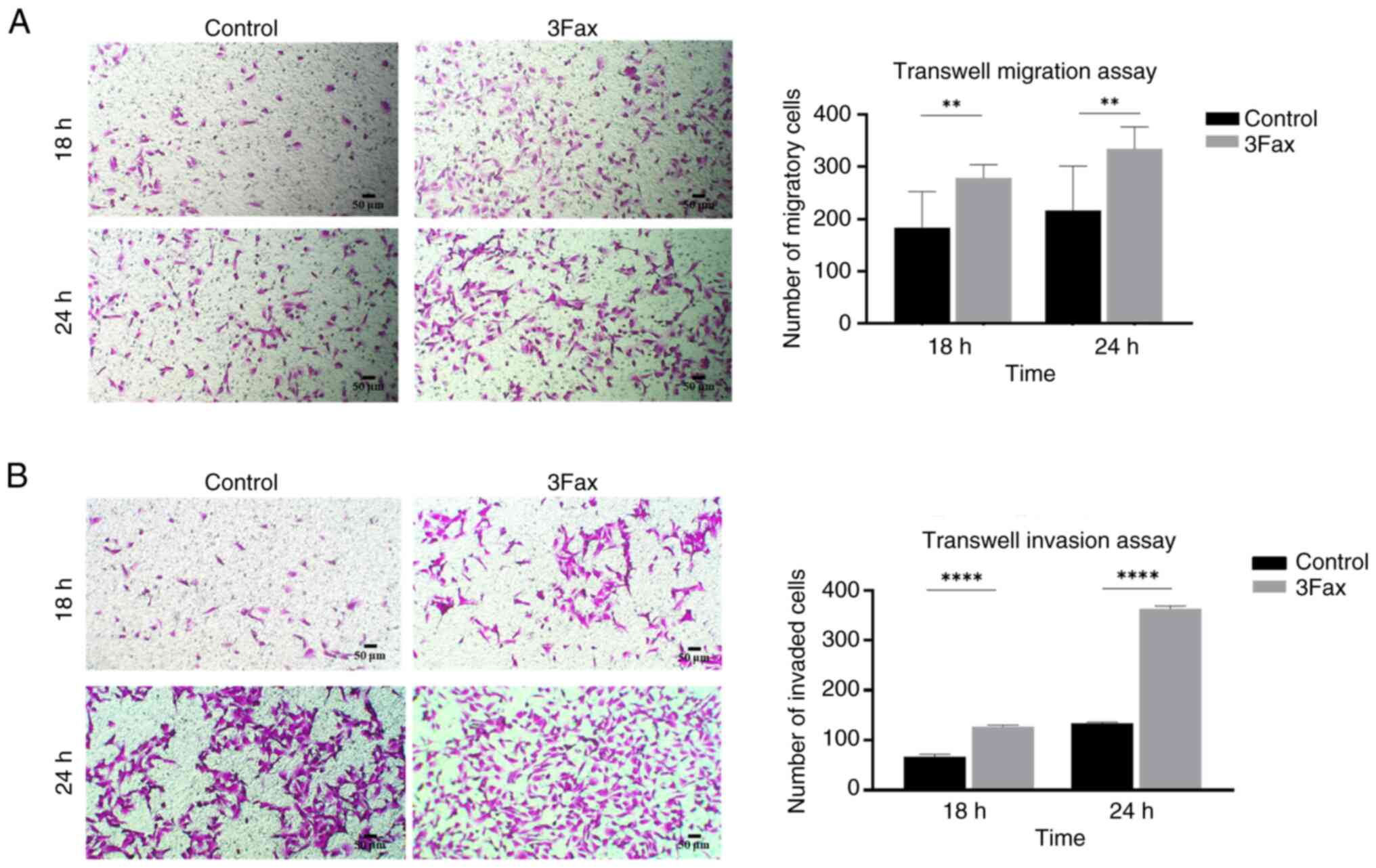
Suppression of ST activity promotes
migration and invasion abilities in the malignant meningioma
cells
To determine whether the suppression of sialylation
affects the migration and invasion of meningioma cells, wound
healing and Transwell migration or invasion assays were performed.
The wound healing assay demonstrated that the 3Fax-treated cells
had the ability to close the wound faster than the control cells at
both 18 h (% wound area: 19.6+1.0% vs. 22.9+1.2%, P<0.05) and 24
h (% wound area: 26.7+0.8% vs. 31.3+0.4%, P<0.01) (Fig. 3). These findings are consistent with
the results of the Transwell migration assay at 18 h (number of
migratory cells: 183+64 vs. 278+23, P<0.01) and 24 h (number of
migratory cells: 216+79 vs. 334+39, P<0.01) (Fig. 4A). Additionally, the Transwell
invasion assay showed that 3Fax-treated cells had a greater ability
to invade the extracellular matrix than control cells in a
time-dependent manner (number of invasive cells at 18 h: 66+4 vs.
127+2 and at 24 h: 134+1 vs. 363+4, P<0.0001) (Fig. 4B). These findings suggested that the
suppression of sialylation enhances the aggressive characteristics
of malignant meningioma cells.
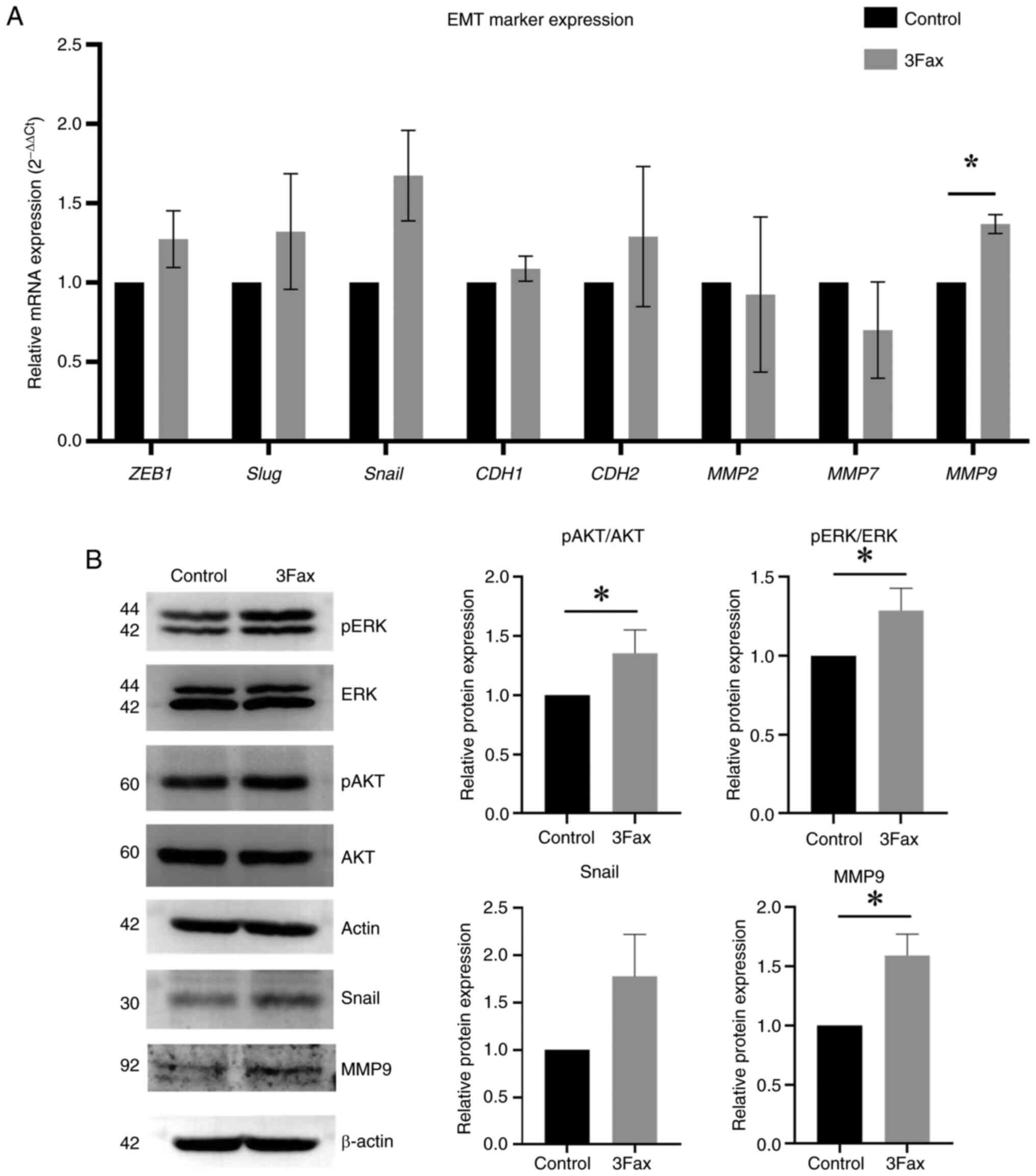
Suppression of ST activity promotes
the progression of meningiomas via EMT and the activation of
ERK/AKT pathways
The present study aimed to investigate whether the
suppression of sialylation enhances the aggressive characteristics
of malignant meningioma cells via EMT. The expression levels of
EMT-related TFs (ZEB1, Snail and Slug) and
EMT-related proteins (N-cadherin, E-cadherin and MMP2, 7 and
9 were determined in 3Fax-treated and control cells. The
results demonstrated that the expression of EMT-related TFs
(Snail: 1.7-fold change, P<0.05) and EMT-related proteins
(MMP9, 1.4-fold change) were increased in
3Fax-treated cells compared with the control (Fig. 5A). Western blot analysis
demonstrated an increase in the expression of MMP9 (1.6-fold
change, P<0,05), Snail (1.8-fold change), phosphorylation of AKT
(1.4-fold change, P<0.05) and ERK (1.3-fold change, P<0.05)
in 3Fax-treated cells (Fig. 5B).
These findings suggested that the suppression of ST activity
promotes cell migration and invasion by enhancing the EMT process
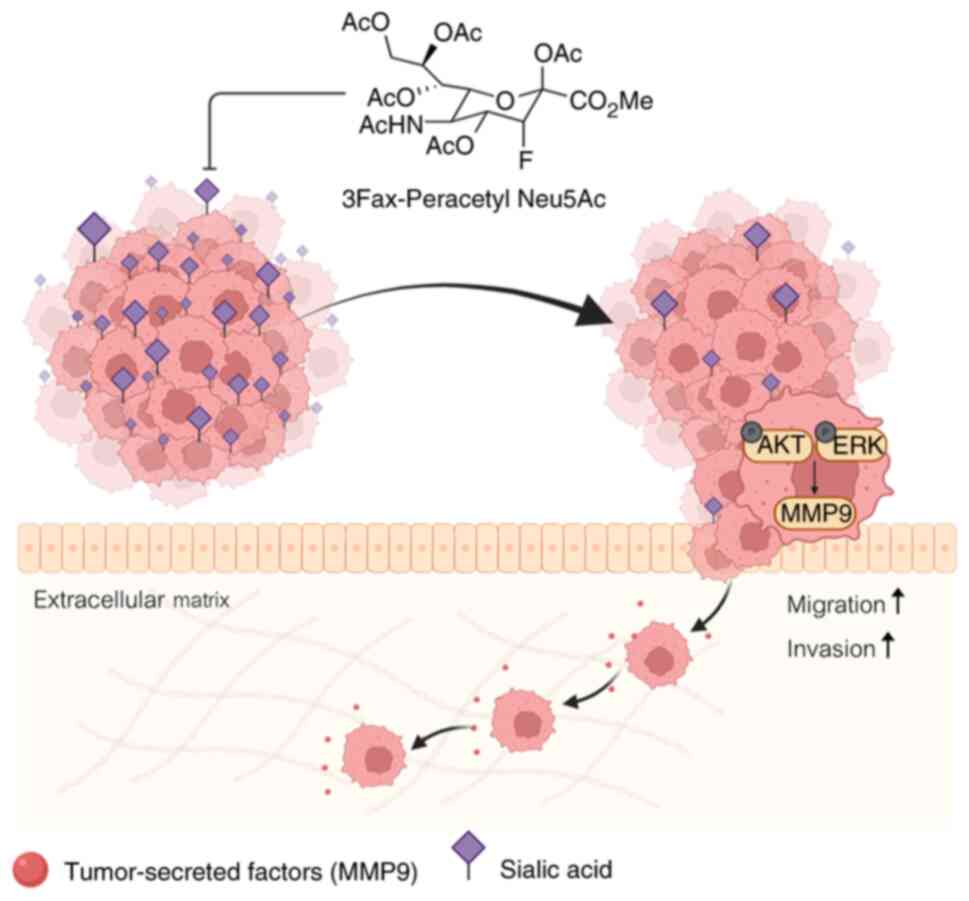
and activating the AKT/ERK pathways (Fig. 6).
Discussion
Aberrant sialylation is associated with cancer
progression by promoting tumor growth and metastasis (17), facilitating immune escape (17), conferring resistance to apoptosis
(18) and enhancing tumor
angiogenesis (19). In the present
study, the differential expression of β-galactoside α2,3 and α2,6
STs (ST3Gals and ST6Gals) was demonstrated in WHO Grade 1, 2 and 3
meningiomas using four different GEO datasets. The role of
sialylation in meningioma progression was further investigated by
chemically suppressing ST activity in malignant meningioma cells.
It was found that sialylation suppression plays a crucial role in
promoting the migration and invasion of malignant meningioma
cells.
In the GEO datasets (GSE16581, GSE43290, GSE74385
and GSE136661), the downregulation of ST3Gal5 and
ST6Gal1 was commonly observed across all four datasets and
was associated with high-grade meningiomas. These results are
consistent with studies in which downregulation of ST6Gal1
is associated with metastasis in liver cancer and
cholangiocarcinoma (20,21). Nevertheless, the upregulation of STs
is commonly observed in multiple cancer types and is associated
with tumor progression (22). The
role of ST in meningioma development and progression has not yet
been reported. In the present study, suppression of ST activity by
a pan-sialylation inhibitor (3Fax-peracetyl-Neu5Ac, 3Fax) did not
affect the proliferation or survival of malignant meningioma cells.
Several studies have demonstrated that aberrant ST expression is
primarily associated with invasion, metastasis and chemoresistance
(22). In breast cancer, ST3Gal3
has been found to induce invasiveness and metastasis by regulating
the integrinB1/MMP2/9 axis, and ST6Gal1 can promote the
invasiveness of breast cancer cells through TGF-β signaling
(23,24).
By contrast, suppression of sialylation promoted
meningioma cell migration and invasion. A similar observation was
made in colon cancer, where the depletion of ST6Gal1 enhanced the
migration and invasion of colon cancer cells (25). Additionally, ST6Gal1 silencing
promoted liver metastasis in vivo by reducing the cellular
pool of the metastasis suppressor KAI1(26). Furthermore, downregulation of
ST6Gal1 decreased α2,6 sialylation on MCAM, enhanced the
interaction between MCAM and galectin-3, leading to the
dimerization of MCAM on the cell surface and subsequently promoting
liver cancer metastasis (20).
Moreover, it was demonstrated that suppression of sialylation in
meningioma cells (HKBMM) induced EMT by increasing the expression
of Snail and MMP9. The present findings suggested that
sialylation by ST serves a protective function during meningioma
progression. However, this mechanism could be considered cell
type-specific because our explorations were performed using only
HKBMM cells. Therefore, further investigation is required in other
meningioma cell lines to confirm this.
EMT mechanisms are involved, including cytoskeletal
reorganization, altered expression of adhesion molecules, and
degradation of the basement membrane through the activation of MMP2
and MMP9(27). In multiple
carcinomas, activation of EMT results in a significant increase in
cancer cell proliferation and extravasation, enabling the expansion
of metastasis (28). In addition,
MMP9 has been observed in various malignancies, and overexpression
of MMP9 is positively correlated with poor prognosis and metastasis
in patients with cancer. MMP9 can interact with and proteolyze cell
surface proteins (CD44, E-cadherin, and α/β integrins) to promote
cell migration and invasion. Various downstream pathways, including
the PI3K/AKT and MAPK/ERK pathways, as well as various TFs, such as
NF-κB, SP1, AP1 and Snail, appear to play a significant role
in MMP9 expression and secretion (29,30).
In the present study, it was demonstrated that suppression of ST
activity markedly induced AKT and ERK phosphorylation. Taken
together, the current findings from in vitro experiments
suggest that the suppression of ST activity by a pan-sialylation
inhibitor (3Fax) enhances meningioma progression through the
AKT/ERK pathways. However, further in vivo studies are
required to establish the role of STs in meningioma progression.
Notably, the role of ST in meningiomas was demonstrated using a
pan-sialylation inhibitor. Therefore, further studies using
gene-specific knockdown or Clustered Regularly Interspaced Short
Palindromic Repeats techniques are required to identify which
specific ST plays a critical role in the progression of meningioma
and to investigate the underlying mechanism of the candidate ST
through the interaction with glycoprotein partners such as MCAM and
CD44.
In summary, differential expression of STs was
observed across the three grades of meningioma, suggesting
downregulation of ST gene expression in malignant meningiomas.
Additionally, suppression of sialylation using a ST inhibitor
significantly enhanced meningioma cell migration and invasion by
inducing the EMT process. These findings suggest that sialylation
plays an important role in meningioma progression. Therefore, the
sialylation status of meningioma tissues, as determined by MAL II
and SNA staining, could be a promising biomarker for assessing
meningioma progression.
Acknowledgements
The authors would like to thank Mr Bryan Roderick
Hamman (Publication Clinic, Research Affairs, Faculty of Medicine,
Khon Kaen University, Khon Kaen 40002, Thailand) for assistance
with the English language editing of the manuscript.
Funding
Funding: The present study was supported by Suranaree University
of Technology, Thailand Science Research and Innovation (TSRI),
National Science, Research, and Innovation Fund (NSRF) (grant no.
195616) and National Research Council of Thailand (NRCT) (grant no.
N41A670301).
Availability of data and materials
The data generated in the present study are included
in the figures and/or tables of this article.
Authors' contributions
CT and KT acquired funding, designed the study and
revised the manuscript. PJ and CT performed the biological
experiments and analyzed the data. PJ and CT confirm the
authenticity of all the raw data. KT, PA and PK analyzed and
interpretated the data. PJ drafted the manuscript. All authors read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Marosi C, Hassler M, Roessler K, Reni M,
Sant M, Mazza E and Vecht C: Meningioma. Crit Rev Oncol Hematol.
67:153–171. 2008.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 World Health Organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Ogasawara C, Philbrick BD and Adamson DC:
Meningioma: A review of epidemiology, pathology, diagnosis,
treatment, and future directions. Biomedicines.
9(319)2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Vuong HG, Ngo TNM and Dunn IF: Incidence,
risk factors, and prognosis of meningiomas with distant metastases
at presentation. Neurooncol Adv. 3(vdab084)2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Yamamoto J, Takahashi M, Idei M, Nakano Y,
Soejima Y, Akiba D, Kitagawa T, Ueta K, Miyaoka R and Nishizawa S:
Clinical features and surgical management of intracranial
meningiomas in the elderly. Oncol Lett. 14:909–917. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Yang CC, Tsai CC, Chen SJ, Chiang MF, Lin
JF, Hu CK, Chan YK, Lin HY and Cheng SY: Factors associated with
recurrence of intracranial meningiomas after surgical resection: A
retrospective single-center study. Int Journal of Gerontol.
12:57–61. 2018.
|
|
7
|
Hwang WL, Marciscano AE, Niemierko A, Kim
DW, Stemmer-Rachamimov AO, Curry WT, Barker FG II, Martuza RL,
Loeffler JS, Oh KS, et al: Imaging and extent of surgical resection
predict risk of meningioma recurrence better than WHO
histopathological grade. Neuro Oncol. 18:863–872. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Varki A: Biological roles of glycans.
Glycobiology. 27:3–49. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Dobie C and Skropeta D: Insights into the
role of sialylation in cancer progression and metastasis. Br J
Cancer. 124:76–90. 2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhou X, Yang G and Guan F: Biological
functions and analytical strategies of sialic acids in tumor.
Cells. 9(273)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Christie DR, Shaikh FM, Lucas JA IV, Lucas
JA III and Bellis SL: ST6Gal-I expression in ovarian cancer cells
promotes an invasive phenotype by altering integrin glycosylation
and function. J Ovarian Res. 1(3)2008.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhang X, Dou P, Akhtar ML, Liu F, Hu X,
Yang L, Yang D, Zhang X, Li Y, Qiao S, et al: NEU4 inhibits
motility of HCC cells by cleaving sialic acids on CD44. Oncogene.
40:5427–5440. 2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Cazet A, Julien S, Bobowski M,
Krzewinski-Recchi MA, Harduin-Lepers A, Groux-Degroote S and
Delannoy P: Consequences of the expression of sialylated antigens
in breast cancer. Carbohydr Res. 345:1377–1383. 2010.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Talabnin C, Trasaktaweesakul T,
Jaturutthaweechot P, Asavaritikrai P, Kongnawakun D, Silsirivanit
A, Araki N and Talabnin K: Altered O-linked glycosylation in benign
and malignant meningiomas. PeerJ. 12(e16785)2024.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Naradun N, Talabnin K, Ayuttha KIN and
Talabnin C: Piperlongumine and bortezomib synergically inhibit
cholangiocarcinoma via ER stress-induced cell death. Naunyn
Schmiedebergs Arch Pharmacol. 396:109–120. 2023.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Macauley MS, Crocker PR and Paulson JC:
Siglec-mediated regulation of immune cell function in disease. Nat
Rev Immunol. 14:653–666. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
18
|
Swindall AF and Bellis SL: Sialylation of
the Fas death receptor by ST6Gal-I provides protection against
Fas-mediated apoptosis in colon carcinoma cells. J Biol Chem.
286:22982–22990. 2011.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Chiodelli P, Urbinati C, Paiardi G, Monti
E and Rusnati M: Sialic acid as a target for the development of
novel antiangiogenic strategies. Future Med Chem. 10:2835–2854.
2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zou X, Lu J, Deng Y, Liu Q, Yan X, Cui Y,
Xiao X, Fang M, Yang F, Sawaki H, et al: ST6GAL1 inhibits
metastasis of hepatocellular carcinoma via modulating sialylation
of MCAM on cell surface. Oncogene. 42:516–529. 2023.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Park DD, Xu G, Park SS, Haigh NE, Phoomak
C, Wongkham S, Maverakis E and Lebrilla CB: Combined analysis of
secreted proteins and glycosylation identifies prognostic features
in cholangiocarcinoma. J Cell Physiol. 239(e31147)2024.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Al Saoud R, Hamrouni A, Idris A, Mousa WK
and Abu Izneid T: Recent advances in the development of
sialyltransferase inhibitors to control cancer metastasis: A
comprehensive review. Biomed Pharmacother.
165(115091)2023.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Cui HX, Wang H, Wang Y, Song J, Tian H,
Xia C and Shen Y: ST3Gal III modulates breast cancer cell adhesion
and invasion by altering the expression of invasion-related
molecules. Oncol Rep. 36:3317–3324. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Lu J, Isaji T, Im S, Fukuda T, Hashii N,
Takakura D, Kawasaki N and Gu J: β-Galactoside α2,
6-sialyltranferase 1 promotes transforming growth factor-β-mediated
epithelial-mesenchymal transition. J Biol Chem. 289:34627–34641.
2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhou L, Zhang S, Zou X, Lu J, Yang X, Xu
Z, Shan A, Jia W, Liu F, Yan X, et al: The β-galactoside
α2,6-sialyltranferase 1 (ST6GAL1) inhibits the colorectal cancer
metastasis by stabilizing intercellular adhesion molecule-1 via
sialylation. Cancer Manag Res. 11:6185–6199. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Jung YR, Park JJ, Jin YB, Cao YJ, Park MJ,
Kim EJ and Lee M: Silencing of ST6Gal I enhances colorectal cancer
metastasis by downregulating KAI1 via exosome-mediated exportation
and thereby rescues integrin signaling. Carcinogenesis.
37:1089–1097. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wick W, Platten M and Weller M: Glioma
cell invasion: Regulation of metalloproteinase activity by
TGF-beta. J Neurooncol. 53:177–185. 2001.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Shibue T, Brooks MW and Weinberg RA: An
integrin-linked machinery of cytoskeletal regulation that enables
experimental tumor initiation and metastatic colonization. Cancer
Cell. 24:481–498. 2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Augoff K, Hryniewicz-Jankowska A, Tabola R
and Stach K: MMP9: A tough target for targeted therapy for cancer.
Cancers (Basel). 14(1847)2022.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Hou S, Wang J, Li W, Hao X and Hang Q:
Roles of integrins in gastrointestinal cancer metastasis. Front Mol
Biosci. 8(708779)2021.PubMed/NCBI View Article : Google Scholar
|