Introduction
Polychlorinated biphenyls (PCBs) are synthetic
chlorinated compounds that were historically used in a numerous
industrial and commercial applications due to their chemical
stability, and insulating properties. Although the manufacture and
use of PCBs have been banned or severely restricted in many
countries, PCBs persist in the environment and continue to raise
significant public health concerns due to their bioaccumulative
nature and long biological half-life (1,2). Human
exposure occurs primarily through the consumption of contaminated
food, particularly animal fats, as well as through inhalation and
dermal contact (1,3). Among the various PCB congeners, PCB153
is one of the most abundant and persistent in human tissues
(2). Its strong lipophilicity and
resistance to metabolic degradation facilitate its accumulation in
the food chain, especially in lipid-rich food sources such as fish
and animal meat (4). The intestinal
epithelium, as the first site of dietary exposure and absorption,
represents a critical yet understudied target for PCB153
toxicity.
The intestinal epithelium plays a central role in
maintaining mucosal homeostasis, acting as both a physical barrier
and an active regulator of immune and metabolic processes.
Disruption of intestinal epithelial cell (IEC) function by
environmental toxicants has been linked to impaired barrier
integrity, chronic inflammation, metabolic dysregulation (5,6).
Previous studies have begun to elucidate the impact of PCB153 on
gut health. For example, Choi et al (7) demonstrated that PCBs disrupt
intestinal barrier function by inducing oxidative stress via NADPH
oxidase activation, which alters tight junction protein expression
and compromises epithelial integrity. Additionally, Phillips et
al (8) reported that intestinal
exposure to PCB153 activates the ATM/NEMO pathway, triggering
inflammation and epithelial injury. Together, these findings raise
important concerns about the impact of PCB153 on intestinal
homeostasis and underscore the need for deeper mechanistic studies
into how environmental toxicants drive intestinal dysfunction.
Significant knowledge gaps remain regarding the
broader transcriptional and cellular consequences of PCB153
exposure in normal human IECs. Prior studies have relied on colon
cancer cell lines or murine exposure models (7,8), while
may not accurately reflect the normal epithelial physiology.
To bridge these gaps, we utilized the
non-tumorigenic human IEC line NCM460D (9) to evaluate the dose-dependent effects
of PCB153 exposure on the profiling of whole transcriptome of IECs.
By integrating viability assays with RNA sequencing, we
systematically characterized how low and high concentrations of
PCB153 alter transcriptional programs and key biological pathways.
Our data reveal broad transcriptomic shifts involving inflammation,
cellular stress, metabolic disruption, impaired regenerative
signaling, and altered gut-brain communication. Notably, we found
the suppression of Wnt signaling and proliferation-associated
genes, suggesting mechanistic insights into how PCB153 compromises
epithelial renewal and barrier maintenance.
This study presents the first comprehensive,
dose-dependent transcriptomic analysis of PCB153 exposure in normal
human IECs, offering new perspectives on the molecular mechanisms
through which PCB153 compromises intestinal health.
Materials and methods
In vitro experiments
NCM460D cell line was purchased from INCELL
Corporation (San Antonio, TX). NCM460D is a non-transformed human
colonic epithelial cell line originally derived from normal colon
tissue of a 68-year-old male donor. The cells were selected for
stable in vitro growth and were not infected, immortalized,
or genetically modified, preserving a normal genotype.
Phenotypically, NCM460D cells express key intestinal epithelial
markers, including cytokeratins, villin, colonic epithelial
antigens, and a subset of cells exhibit mucin production,
consistent with differentiated epithelial features (9). NCM460D cells are non-tumorigenic and
grown in INCELL's enriched M3:10™ medium (INCELL Corp.)
to maintain their physiological phenotype, supplemented with 10%
fetal bovine serum (Gibco, Thermo Fisher Scientific, Waltham, MA),
100 U/ml penicillin and 100 µg/ml streptomycin (Gibco), at 37˚C in
a humidified atmosphere containing 5% CO2.
PCB153 (2,2',4,4',5,5'-hexachlorobiphenyl) was
purchased from Sigma-Aldrich (St. Louis, MO) and dissolved in
dimethyl sulfoxide (DMSO) to generate a 10 mM stock solution.
Working concentrations were freshly prepared by diluting the stock
in M3:10™ complete medium, ensuring the final DMSO
concentration did not exceed 0.1% (v/v) in any treatment. Cells
were seeded at 2x104 cells/well in 96-well plates for
viability assays or 2x105 cells/well in 6-well plates
for RNA extraction. For the cell viability assay, cells were
treated with 0.5, 5, 50, or 200 µM PCB153, or an equivalent volume
of DMSO (vehicle control), for 6, 24, or 48 h. For transcriptomic
analysis and RT-qPCR validation, cells were exposed to PCB153 (0,
5, or 50 µM) or DMSO for 24 h under identical culture conditions in
a 37˚C humidified atmosphere containing 5% CO2.
Cell viability assay
The genotoxicity of PCB153 on intestinal epithelial
cells was determined by PrestoBlueTM HS Reagent
(Invitrogen, Thermo Fisher Scientific, Waltham, MA). Briefly,
NCM460D cells (2x104 cells/well) were seeded into a
96-well plate and treated with PCB153 at final concentrations of 0,
0.5, 5, 50 and 200 µM, with DMSO serving as the vehicle control.
Treatments were performed for 6, 24, and 48 h. After treatment,
PrestoBlue reagent was added directly to the culture medium as
1/10th of the total well volume and incubated for 1 h in a cell
culture incubator at 37˚C, protected from direct light. Absorbance
was measured at 570 nm, using 600 nm as a reference wavelength.
Cell viability was calculated as the ratio of OD570/600.
Reverse transcription-quantitative PCR
(RT-qPCR)
NCM460D cells (2 x 105 cells/well in
6-well plate) were treated with PCB153 (0, 5, or 50 µM) for 24 h,
with DMSO serving as the vehicle control. Total RNA was extracted
from the cells using TRIzol reagent (Invitrogen, Thermo Fisher
Scientific, Waltham, MA) according to the manufactory protocol. RNA
purity and concentration were assessed using the NanoDrop
spectrophotometer (Agilent Technologies, Santa Clara, CA). Total 1
µg of RNA was reverse-transcribed to cDNA using high-capacity cDNA
reverse transcription kit (Applied Biosystems, Thermo Fisher
Scientific, Waltham, MA). Reverse transcription was performed at
25˚C for 10 min, followed by 37˚C for 120 min, 85˚C for 5 min, and
then held at 4˚C. Quantitative PCR was performed with SYBR Green
master mix (Applied Biosystems) on a QuantStudio 3 Real-Time PCR
System (Thermo Fisher Scientific). The thermocycling program
consisted of an initial denaturation at 95˚C for 10 min, followed
by 40 cycles of 95˚C for 15 sec and 60˚C for 60 sec. The relative
expression of target gene was analyzed by the
2-ΔΔCq method (10), normalizing each target to
GAPDH and expressing data as fold-change relative to the
solvent control. Primer sequences of MT1G, MT2A,
LGR5, DACT1, LEF1 and GAPDH are
provided in Table SI.
RNA sequencing
NCM460D cells (2x105 cells/well in 6-well
plate) were treated with PCB153 (5, or 50 µM) for 24 h, with DMSO
serving as the vehicle control. Total RNA was extracted from the
cells using TRIzol reagent (Invitrogen, Thermo Fisher Scientific,
Waltham, MA) as previously described and purified by RNA Clean
& Concentrator kit (Zymo Research, Irvine, CA) according to the
manufactory protocol. RNA integrity and quantity were further
assessed using Agilent Bioanalyzer (Agilent Technologies, Santa
Clara, CA) before the library construction for RNA sequencing.
Total RNA was processed for Poly(A) mRNA isolation and preparation
of cDNA libraries using NEBNext Ultra II RNA Library Prep Kit (New
England Biolabs, Ipswich, MA) and barcoded with NEBNext Multiplex
Oligos for Illumina (NEB), all according to the manufacturer's
manuals. Library concentration was measured by Agilent Bioanalyzer
(Agilent Technologies) and 2.6x10-10 moles desired
loading of final library were subjected to 100 bp pair-end
sequenced (PE100) on the Illumina NovaSeqX platform at the
University of Chicago Genomics Core Facility (Chicago, IL). RNA-Seq
raw data has been uploaded to the database at the National Center
for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/bioproject/) under
BioProject accession number PRJNA1285092.
RNA sequencing data analysis
For transcriptomic analysis, we performed two
independent biological experiments, each containing duplicate
cultures per treatment condition (0, 5, and 50 µM PCB153). To
minimize technical variation and maximize consistency across
experiments, the duplicate cultures within each treatment group
were pooled prior to RNA extraction, resulting in two biological
replicates per treatment group used for RNA-seq. The output FASTQ
sequences were aligned to reference human genome annotation
(GENCODE version 48) using STAR software (version 2.7) (11). The aligned sequences were further
sorted with Samtools (version 1.18) (12). Counting reads in features with
htseq-count (version 2.0.2) (13).
The count matrix was then normalized and differentially expressed
were identified (adjusted P-value <0.05) with the DESeq2 package
(version 1.46) (14) in R software
(version 4.4). Gene Ontology (GO) was identified by clusterProfiler
(version 4.14) (15) to discover
the functional enrichment of pathways comparing experimental
groups. ShinyGO 0.82 was used to visualize the pathway networks
(16).
Statistical analysis
All statistical analyses were performed using
GraphPad Prism (version 10.5). For cell viability assays (n=6) and
RT-qPCR (n=3) experiments, differences among treatment groups were
analyzed using one-way ANOVA, followed by Tukey's post hoc test for
multiple comparisons. Data are presented as mean ± standard
deviation (SD). A P-value of P<0.05 was considered statistically
significant. Significance levels are indicated as
*P<0.05, **P<0.01,
***P<0.001, ****P<0.0001.
Results
Establishing RNA-seq compatible PCB153
exposure conditions
To evaluate the cellular consequences of PCB153
exposure in a physiologically relevant intestinal model, we first
established an in vitro system using the non-tumorigenic
human intestinal epithelial cell line NCM460D. This line is derived
from normal colonic mucosa and retains key features of
differentiated intestinal epithelial cells, including expression of
cytokeratins, villin, and other colonic epithelial markers, without
exogenous genetic modifications (9). Due to these characteristics, NCM460D
cells provide an appropriate model for examining transcriptomic
disruptions in normal IECs.
To determine suitable exposure conditions for
transcriptomic profiling, we next assessed the effects of PCB153 on
NCM460D cell viability across a range of concentrations (0.5, 5,
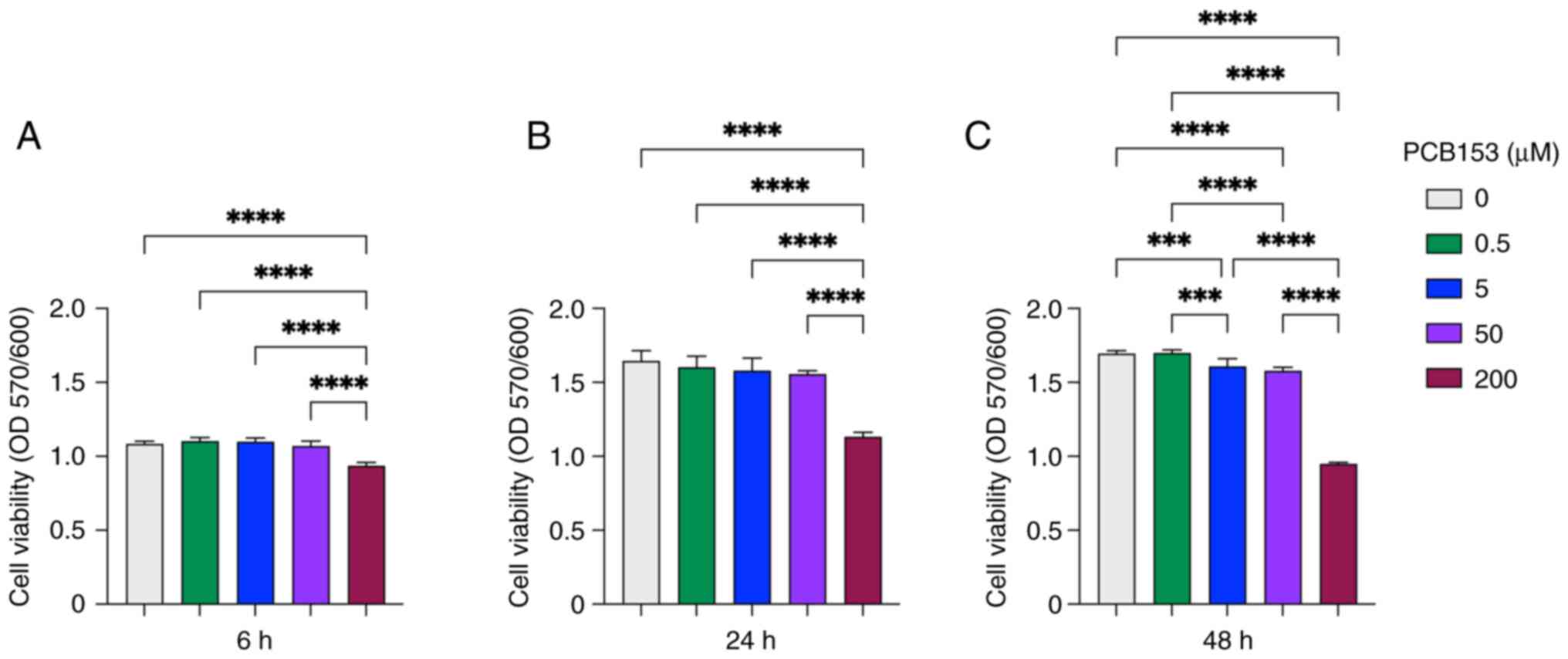
50, and 200 µM) and exposure times (6, 24, and 48 h) (Fig. 1). This analysis was conducted to
identify non-cytotoxic conditions appropriate for detection of
meaningful alterations by RNA-seq, rather than to define a full
cytotoxicity curve. At 6 h, cells exposed to 0.5-50 µM PCB153
showed little change in viability, indicating that this short
exposure primarily captures immediate early-response genes and is
not ideal for broader transcriptional profiling (Fig. 1A). In contrast, 200 µM PCB153 caused
acute viability loss at this early time point, making it unsuitable
for mechanistic studies (Fig. 1A).
Cells were maintained >90% viability when exposed with both 5
and 50 µM PCB153 at 24 h, providing sufficient time for primary
transcriptomic responses to develop while avoiding overt
cytotoxicity (Fig. 1B). By
comparison, 0.5 µM PCB153 produced minimal biological impact on
NCM460D cells and was unlikely to yield detectable transcriptional
changes (Fig. 1A-C). However,
subtle but statistically significant reductions in cell viability
emerged at 48 h, even at 5 and 50 µM intermediate doses, indicating
that longer exposures may introduce nonspecific cytotoxic effects
(Fig. 1C). Together, these findings
identify 5 and 50 µM PCB153 for 24 h as optimal exposure conditions
that balance IEC cellular integrity with meaningful transcriptional
alterations while minimizing confounding cytotoxic effects and thus
were selected for subsequent RNA-seq experiments.
Transcriptome changes in IECs exposed
to PCB153
Although recent studies have identified various
adverse effects of PCB153 exposure, significant knowledge gaps
remain regarding its impact and underlying mechanisms of
cytotoxicity in normal intestinal epithelial cells. In addition, no
studies have systematically investigated the dose-dependent effects
of PCB153 exposure on normal IECs. To address these gaps, this
study aimed to characterize the transcriptional responses and
signaling pathways affected by PCB153 in human normal IECs using
RNA-seq analysis. Principal component analysis (PCA) revealed a
significant transcriptomic shift between control and PCB153-treated
cells. More importantly, the transcriptomic changes followed a
dose-dependent pattern in IECs exposed to PCB153 (Fig. 2A). Further analysis of
differentially expressed genes (DEGs), using an adjusted P-value
cutoff of <0.05, identified 329 upregulated and 246
downregulated genes in IECs treated with 5 µM of PCB153 compared to
controls (Fig. 2B). Moreover,
exposure to 50 µM of PCB153 resulted in 4,006 upregulated and 3,070
downregulated genes (Fig. 2C).
Among these significantly differentially expressed genes, 136 were
commonly downregulated, and 181 were commonly upregulated in both
the 5 and 50 µM PCB153 treatment groups compared to controls
(Fig. 2D). These results suggest
that acute PCB153 exposure significantly alters the transcriptome
of IECs, with higher doses exerting more pronounced effects than
lower doses. The higher PCB153 exposure resulted in thousands of
gene expression changes, indicating a dose-dependent impact on the
IEC transcriptome. To validate the RNA-seq results, RT-qPCR was
performed for several representative differentially expressed
genes, including MT1G, MT2A, LGR5,
DACT1, and LEF1. The RT-qPCR results confirmed a
strong induction of MT1G and MT2A, and a marked
suppression of LGR5, DACT1, and LEF1 following
PCB153 exposure, consistent with the RNA-seq data (Fig. 2E). Taken together, these findings
suggest that PCB153 exposure may profoundly influence IEC cellular
functions and biological processes.
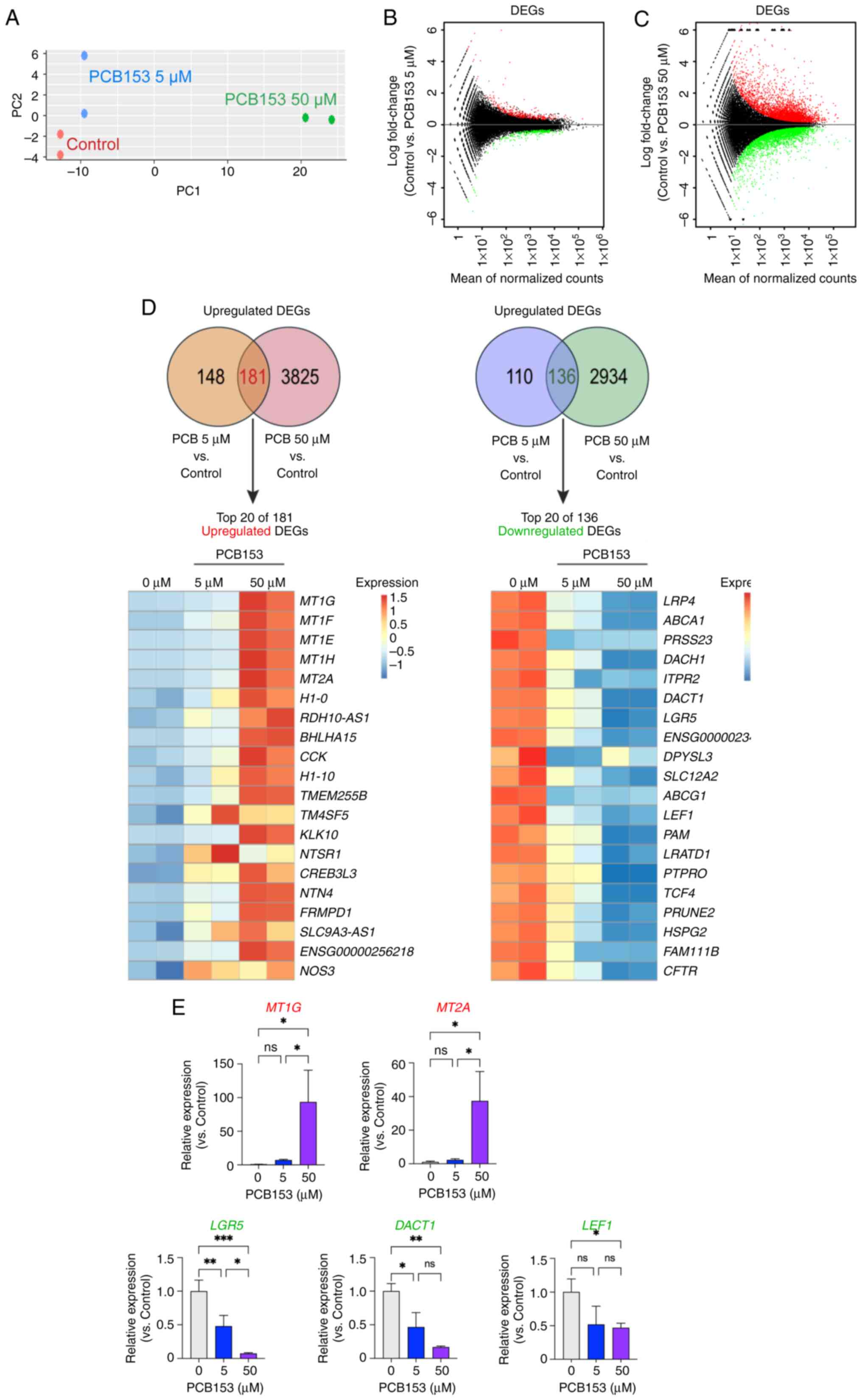 | Figure 2Dose-dependent transcriptomic
alterations in human IECs following PCB153 exposure. (A) PC
analysis of RNA-seq data showing distinct clustering of control and
PCB153-treated IECs, with a clear dose-dependent separation between
the 5 and 50 µM exposure groups. (B) MA plot of DEGs in IECs
treated with 5 µM PCB153 compared with the control group,
highlighting 329 upregulated and 246 downregulated genes (adjusted
P<0.05). (C) MA plot of DEGs in IECs treated with 50 µM PCB153
compared with the control group, showing 4,006 upregulated and
3,070 downregulated genes (adjusted P<0.05). (D) Venn diagram
illustrating the overlap of DEGs between the 5 and 50 µM PCB153
treatment groups, identifying 181 commonly upregulated and 136
commonly downregulated genes, with the top 20 DEGs (ranked by
P-value) visualized in a heatmap. RNA-seq data from two independent
biological experiments, each performed in duplicate are presented.
(E) Reverse transcription-quantitative PCR validation of selected
representative DEGs (MT1G, MT2A, LGR5,
DACT1 and LEF1) confirming induction of
metallothionein genes and suppression of Wnt-related targets,
consistent with RNA-seq results. n=3 for each group. Statistical
significance was determined using one-way ANOVA.
*P<0.05, **P<0.01,
***P<0.001. DEG, differentially expressed gene; IEC,
intestinal epithelial cell; ns, not significant; PC, principal
component; PCB153, polychlorinated biphenyl 153; RNA-seq, RNA
sequencing; MA plot, minus-average plot. |
PCB153 exposure leads to intestinal
epithelial cell dysfunction, metabolic disruption, and tumorigenic
potential
Through DEG analysis, we identified a core set of
317 genes (136 downregulated and 181 upregulated) shared between
two doses of PCB153 exposure. Pathway analysis of this core set
revealed significant alterations in several key biological
processes. GO terms analysis of the 181 upregulated genes indicated
significant enrichment in pathways related to mineral absorption,
ribosome function, coronavirus disease, and axon guidance (Fig. 3A and B). The upregulation of mineral absorption
pathways suggests that PCB153 disrupts metal homeostasis,
particularly affecting zinc (Zn) and copper (Cu) transport.
Metallothionein (MT) family genes (MT1E, MT1F,
MT1G and MT2A), regulate essential metal ions
(17,18), dramatically induced, suggesting an
adaptive response to PCB153-induced metal transporter disruption,
potentially leading to increased cellular stress due to metal
accumulation or dysregulation of Zn/Cu levels. Additionally, the
increased ribosome activity observed following PCB153 exposure
suggests enhanced protein synthesis, which may burden cellular
machinery, triggering ER stress, misfolded proteins, and conditions
favoring tumorigenesis (19,20).
The upregulation of coronavirus disease pathway (Path:hsa05171,
aggregates 232 pathway genes) likely reflects the activation of a
general ‘viral response-related pathways’ or inflammatory stress
response, suggesting PCB153 exposure alters immune responses in the
IECs, potentially increasing susceptibility to viral infections and
chronic inflammation. Taken together, network analysis further
revealed a potential connection between ribosome and coronavirus
disease pathways in response to PCB153, suggesting that ribosome
dysregulation may exacerbate gut inflammation during infections
(Fig. 3B). Interestingly, PCB153
exposure also disrupted axon guidance signaling, suggesting
potential alterations in gut-brain axis communication. Axon
guidance genes, such as NTN4 [a member of the netrin family
(21)], regulate neuronal
connectivity, and their dysregulation may impair gut-brain
signaling, potentially affecting gut motility and neurological
function.
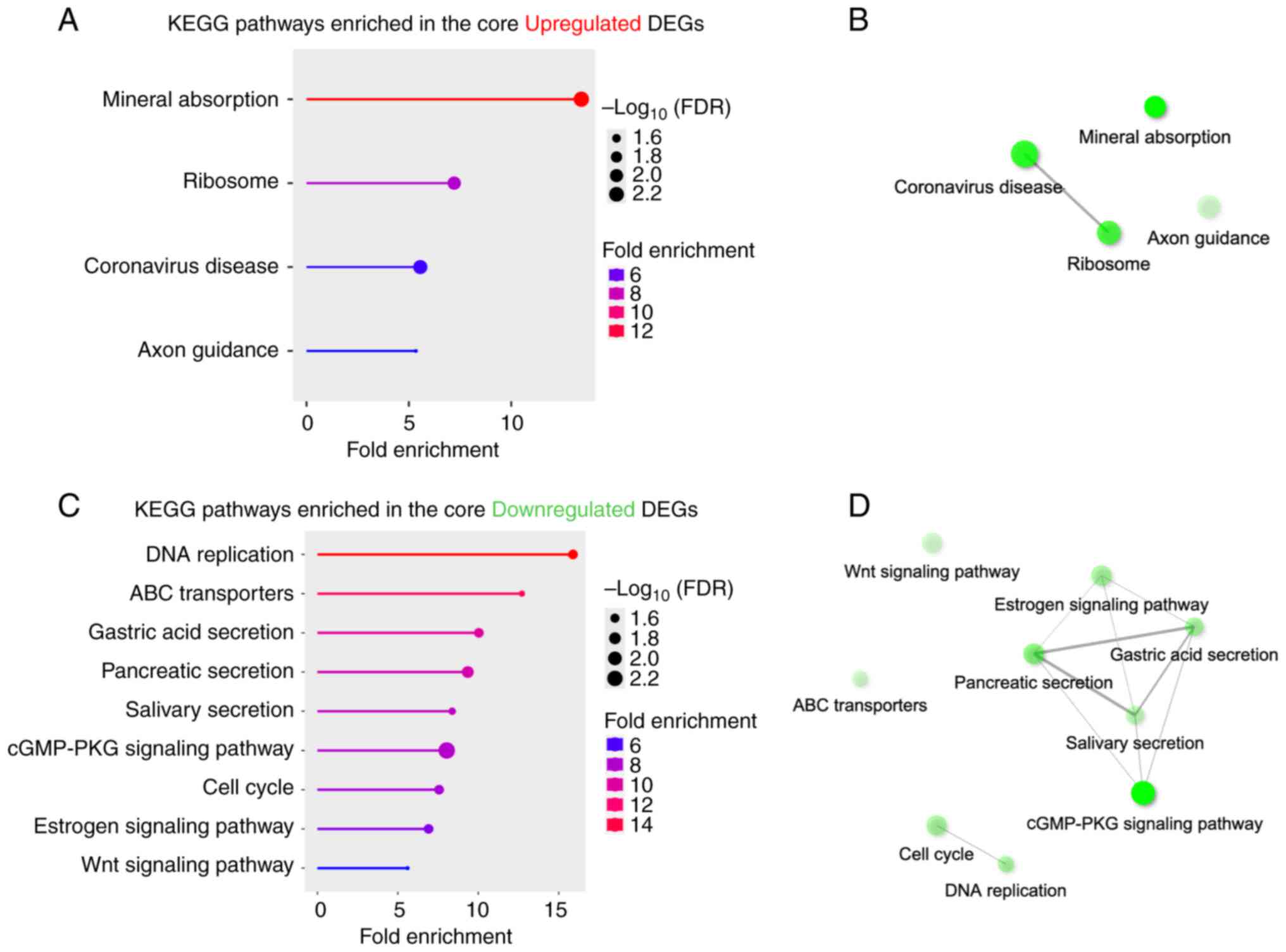 | Figure 3Pathway enrichment analysis of
commonly dysregulated genes in IECs following PCB153 exposure. (A)
KEGG pathway enrichment analysis of 181 commonly upregulated genes
revealed significant enrichment in pathways related to ‘mineral
absorption’, ‘ribosome’, ‘coronavirus disease’ and ‘axon guidance’.
(B) Network analysis of upregulated pathways highlighted potential
interactions between ‘ribosome’ and viral response-related pathways
(‘coronavirus disease’). (C) KEGG pathway enrichment analysis of
136 commonly downregulated genes identified significant suppression
of pathways involved in ‘DNA replication’, ‘ABC transporters’,
digestive secretions (‘gastric acid secretion’, ‘pancreatic
secretion’ and ‘salivary secretion’), ‘cGMP-PKG signaling’, ‘cell
cycle’, ‘estrogen signaling’ and ‘Wnt signaling’. (D) Network
analysis of downregulated pathways showed interconnected
suppression of digestive secretion pathways and the ‘cGMP-PKG
signaling pathway’, indicating impaired intestinal epithelial
physiology. (B and D) Interactive plot shows the relationship
between enriched pathways. Two pathways (nodes) are connected if
they share ≥20% of genes. Darker nodes are more significantly
enriched gene sets. Bigger nodes represent larger gene sets.
Thicker edges represent more overlapped genes. DEG, differentially
expressed gene; FDR, false discovery rate; IEC, intestinal
epithelial cell; KEGG, Kyoto Encyclopedia of Genes and Genomes;
PCB153, polychlorinated biphenyl 153. |
On the other hand, PCB153 exposure led to the
downregulation of 136 core genes, which were significantly enriched
in pathways related to DNA replication, ABC transporters, digestive
secretions (gastric acid, pancreatic, and salivary), cGMP-PKG
signaling, cell cycle, estrogen signaling, and Wnt signaling
(Fig. 3C and D). Among these, the suppression of DNA
replication and cell cycle genes suggests reduced IEC
proliferation. Network analysis highlighted a core connection
between gastric acid secretion, pancreatic secretion, and salivary
secretion, indicating that PCB153 may significantly disrupt
digestive processes (Fig. 3D). The
downregulation of these pathways may impair protein, fat, and
carbohydrate digestion, leading to malabsorption disorders
(22). Furthermore, the cGMP-PKG
signaling pathway is also connected to the digestive secretion
function, which plays a role in intestinal motility and cellular
homeostasis (23,24), was also downregulated (Fig. 3C and D). The suppression of ABC transporters
(e.g., ABCA1, ABCG1) impairs cholesterol efflux,
leading to lipid accumulation and oxidative stress (25). Similarly, downregulation of estrogen
signaling in IECs may contribute to the disruptions in intestinal
lipid homeostasis. Since PCBs are lipophilic and accumulate in
lipid membranes, the inhibition of ABC transporters may increase
intracellular PCB retention, exacerbating cellular toxicity and
bioaccumulation. Most importantly, PCB153 exposure significantly
disrupted the Wnt signaling pathway, which is critical for
intestinal stem cell renewal and epithelial integrity (26,27)
(Fig. 3C). Notably, due to the
downregulation of WNT signaling, LGR5, a stemness-associated
molecule, was significantly downregulated (Fig. 2D), suggesting impaired intestinal
regeneration and homeostasis. Additionally, TCF4 and
LEF1, two major Wnt transcription factors essential for IEC
proliferation, ranked among the top 20 downregulated genes
(Fig. 2D). Their suppression
directly contributes the disrupts of WNT signaling linking to IECs
renewal and regeneration, highlighting the detrimental impact of
PCB153 on intestinal epithelial integrity.
Taken together, PCB153 exposure profoundly alters
multiple essential signaling pathways in normal IECs, disrupting
cell proliferation, lipid metabolism, immune responses, and
gut-brain axis communication. These findings suggest that PCB153
exposure not only impairs gut homeostasis but may also contribute
to tumorigenic potential and long-term metabolic dysfunction.
IECs displays a dose-dependent
toxicant response to PCB153 exposure
In this study, we further evaluated the
dose-dependent effects of PCB153 on cell fate and signaling in
IECs. We found that the 5 µM of PCB153 exposure resulted in the
upregulation of the same cellular pathways as identified with the
common core upregulated gene set, suggesting the lower dose of
PCB153 exposure can initiate the cellular changes of mineral
absorption, ribosome function, coronavirus disease, and axon
guidance (Fig. 4A). We found that
relatively low PCB153 exposure in IECs led to the downregulation in
DNA replication, mismatch repair, homologous recombination, ABC
transporters, cell cycle regulation, and cGMP-PKG signaling,
effects that would be predicted to result in severe gut dysfunction
(Fig. 4B). Impaired DNA repair and
cell cycle arrest increase genomic instability, raising the risk of
mutations and colorectal cancer (28,29).
Downregulated ABC transporters reduce toxin efflux, suggesting that
PCB153 exposure could augment further cellular PCB153 retention.
Additionally, weakened intestinal regeneration can impair the
barrier integrity and contribute to chronic inflammation and
disrupted homeostasis. These changes collectively are predicted to
compromise gut health, increasing susceptibility to disease,
oxidative stress, and metabolic imbalances.
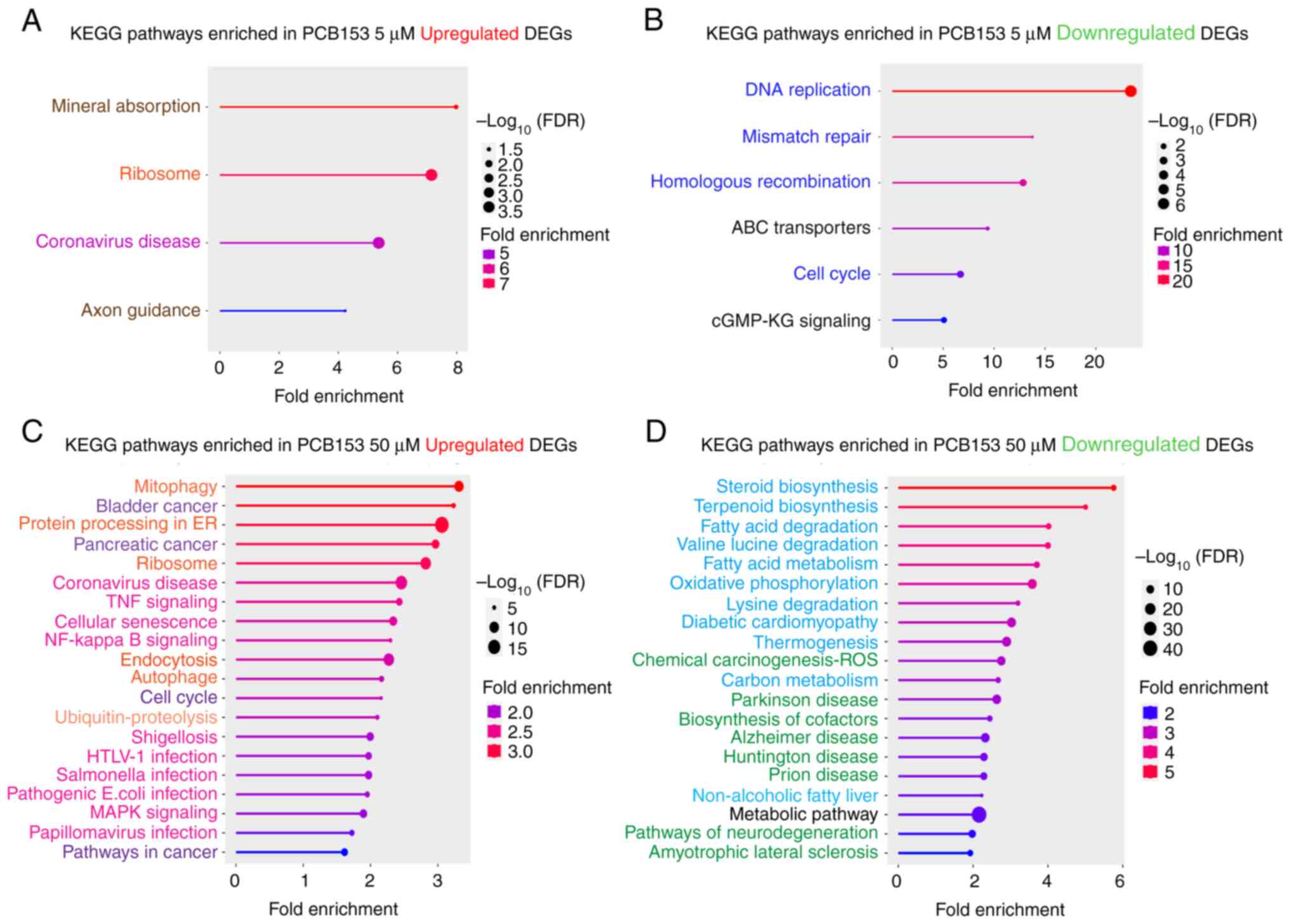 | Figure 4Dose-dependent pathway alterations in
IECs following PCB153 exposure. KEGG pathway enrichment analysis of
(A) upregulated genes and (B) downregulated genes in IECs treated
with 5 µM PCB compared with the control group. (C and D) The top 20
enriched pathways in IECs treated with 50 µM PCB153 were ranked by
fold enrichment and reorganized into functional modules (indicated
by the font color) to improve clarity and interpretability. (A-D)
Upregulated pathways were grouped into ‘Immune & Host-Defense’
(magenta), ‘Proteostasis & Translation’ (orange),
‘Proliferation/Oncogenic’ (violet) and ‘Others’ (brown) categories,
whereas downregulated pathways were grouped into ‘Core Energy &
Lipid Metabolism’ (light blue), ‘Genome Maintenance &
Proliferation’ (dark blue), ‘Neurodegeneration & Proteostasis
Stress’ (green) and ‘Transport/Signaling & Global’ (black)
categories. The significance of pathway enrichment is shown as
-log10(FDR), with higher values indicating stronger
statistical enrichment. DEG, differentially expressed gene; FDR,
false discovery rate; IEC, intestinal epithelial cell; KEGG, Kyoto
Encyclopedia of Genes and Genomes; PCB153, polychlorinated biphenyl
153. |
On the other hand, 50 µM of PCB153 exposure in IECs
resulted in dramatical impairments of multiple signaling pathways.
Using an FDR <0.05 as the significance cutoff, a total of 111
pathways were significantly upregulated and 68 were downregulated.
The top 20 pathways are shown in Fig.
4C and D. To enhance
interpretability, the enriched pathways were consolidated into
functional modules based on shared biological relevance.
Upregulated pathways were grouped into four major categories:
‘Immune & Host-Defense’, ‘Proteostasis & Translation’,
‘Proliferation/Oncogenic’, and ‘Others’. Downregulated pathways
were classified into ‘Core Energy & Lipid Metabolism’, ‘Genome
Maintenance & Proliferation’, ‘Neurodegeneration &
Proteostasis Stress’, and ‘Transport/Signaling & Global’
modules. This modular grouping highlights the extensive remodeling
of immune, metabolic, and stress-related processes in intestinal
epithelial cells following high-dose PCB153 exposure.
To provide deeper biological context, we next
analyzed the pathways driving these functional modules to delineate
the specific cellular processes affected by PCB153 exposure.
Exposure to 50 µM PCB153 in IECs induces significant cellular
stress, inflammation, and cancer-related responses. Upregulation of
mitophagy, autophagy, and protein processing in the ER suggests
heightened stress adaptation mechanisms, while activation of TNF
and NF-kB signaling points to chronic inflammation and immune
dysregulation (30). Increased
expression of cancer-related pathways, including bladder and
pancreatic cancer, as well as cell cycle dysregulation and
senescence, implies a potential pro-tumorigenic environment.
Additionally, pathways linked to bacterial and viral infections
(Shigellosis, Salmonella, E. coli, and
Papillomavirus) are upregulated, suggesting gut barrier
dysfunction and increased pathogen susceptibility (31). Thus, these alterations indicate that
PCB153 compromises intestinal homeostasis, promotes inflammatory
and oncogenic signaling, and weakens immune defenses, increasing
the risk of gut-related diseases and malignancies. Conversely, we
found that exposure to 50 µM of PCB153 in IECs intensively
downregulates key metabolic, mitochondrial, and detoxification
pathways, effects likely leading to impaired cellular function.
Suppression of steroid and terpenoid biosynthesis, fatty acid
metabolism, and amino acid degradation (valine, leucine, lysine)
suggests disrupted lipid and energy homeostasis, which may
contribute to metabolic disorders. Downregulation of oxidative
phosphorylation and thermogenesis points to mitochondrial
dysfunction and reduced energy production, potentially compromising
cellular survival (32). Inhibition
of chemical carcinogenesis (ROS detoxification) and carbon
metabolism indicates reduced detoxification capacity, likely
rendering PCB153-exposed cells more vulnerable to oxidative stress
and secondary toxic insults. Furthermore, repression of pathways
related to neurodegenerative diseases (Alzheimer's, Parkinson's,
Huntington's, and ALS) raises concerns about systemic neurotoxic
effects of PCB153 exposure. Collectively, these alterations suggest
that high dose of PCB153 disrupts cellular metabolism,
mitochondrial function, and detoxification mechanisms, potentially
contributing to epithelial cell dysfunction, metabolic disorders,
and increased disease susceptibility.
Taken together, both concentrations of PCB153
disrupt IEC homeostasis. The exposure to PCB153 exhibits
dose-dependent transcriptomic shifts in IECs. The higher dose of
PCB153 (50 µM) leads to a broader range of cellular effects,
exacerbating cellular stress, metabolic disruption, and
cancer-related signaling compared to lower doses (5 µM). These
findings highlight the potential role of PCB153 in promoting
epithelial cell dysfunction, immune dysregulation, and disease
susceptibility.
Discussion
This study provides new insights into the cellular
and transcriptomic responses of normal human intestinal epithelial
cells to PCB153 exposure, advancing our understanding of how
environmental toxicants disrupt intestinal epithelial cell
homeostasis. Our data demonstrate that PCB153 exposure leads to
broad transcriptional reprogramming that impacts key biological
pathways related to epithelial renewal, detoxification, metabolism,
immune regulation, and gut homeostasis.
The non-tumorigenic NCM460D cells express key
intestinal epithelial markers with differentiated epithelial
features, which is widely regarded as a physiologically relevant
model for studying normal intestinal epithelial cell biology,
though not a model of intestinal stem cells. Therefore, the
reductions in stemness-associated transcripts (e.g., LGR5)
observed after PCB153 exposure should be interpreted as suppression
of stemness-related gene programs, rather than loss of bona
fide intestinal stem cells. Several pathways altered in this
study align with previously reported mechanisms of PCB-induced
intestinal toxicity. Choi et al (7) demonstrated that PCBs compromise
intestinal barrier integrity by inducing oxidative stress via NADPH
oxidase activation, resulting in altered expression of tight
junction proteins. While our study did not directly assess tight
junction components, we identified the downregulation of genes
involved in cell adhesion and epithelial barrier regulation,
including key metabolic and signaling pathways that support
epithelial integrity, such as cGMP-PKG signaling and Wnt signaling.
The repression of Wnt pathway components (e.g., LEF1, DACT1, and
LGR5) in particular points to impaired epithelial renewal and
homeostatic maintenance, effects predicted to contribute to barrier
dysfunction as reported in Choi's study. Our data support the
hypothesis that PCB153 compromises intestinal barrier function
through multiple molecular mechanisms, including both oxidative
stress and suppression of regenerative signaling. Similarly,
Phillips et al (8)
previously reported that PCB153 activates the ATM/NEMO inflammatory
axis leading to intestinal inflammation in mouse models. In our
transcriptomic analysis, we observed upregulation of immune-related
pathways, including TNF signaling, NF-κB signaling, and genes
associated with viral response and inflammation. These changes
suggest that PCB153 elicits an epithelial inflammatory response,
even in the absence of immune cell stimulation, which may act as a
priming signal for more robust inflammation in vivo. The
upregulation of coronavirus disease-related genes and ribosomal
genes may also reflect a heightened stress and immune surveillance
state, consistent with the immune-activating effects described by
Phillips et al (8). Thus,
our findings in human IECs support and expand upon the
inflammation-related mechanisms impaired by PCB153 exposure.
Importantly, our study also revealed novel
transcriptomic responses to PCB153 exposure, particularly in
pathways related to axon guidance, metallothionein expression, and
lipid metabolism, which have not been widely reported in the
context of intestinal toxicology. These alterations suggest that
PCB153 may affect not only local gut epithelial function but also
broader intercellular communication processes, including gut-brain
signaling and systemic metabolic regulation. In particular,
enrichment of the axon guidance pathway points to potential
modulation of neuro-epithelial communication, as several axon
guidance molecules (e.g., semaphorins, netrins) are increasingly
recognized for their roles in regulating epithelial integrity,
inflammation, and neuronal-immune interactions along the gut-brain
axis (33,34), which represents an intriguing
direction for future research. Metallothioneins, MT1G and MT2A are
regulators of intracellular divalent metal ions, with their highest
binding affinity for zinc (Zn²+) and copper
(Cu+/Cu²+). MT1 and MT2 isoforms also
participate in buffering cadmium (Cd²+), mercury
(Hg²+), and other heavy metals when present. In the
context of intestinal epithelial cells, the PCB-induced
upregulation of MT1G and MT2A most likely reflects
the dysregulation of zinc homeostasis, with potential secondary
effects on copper handling (17,18).
Additionally, the downregulation of ABC transporters and estrogen
signaling pathways points to impaired lipid handling and
detoxification, mechanisms with potential implications for both
epithelial toxicity and systemic exposure burden (25).
In this study, we performed two independent
biological experiments for the RNA-seq transcriptomic profiling.
Although this replicate number is at the lower end of standard
RNA-seq designs, several factors support that it is sufficient for
robust bioinformatic analysis and downstream DEG identification.
First, the two biological replicates showed excellent concordance,
as demonstrated by the tight clustering in the PCA plot (Fig. 2A), which indicates low within-group
variability and strong separation among treatment conditions.
Second, despite the modest replicate number, the RNA-seq dataset
yielded the coherent sets of differentially expressed genes and
enriched pathways, including metallothionein genes, Wnt signaling,
metabolism, and inflammatory pathways. Third, we independently
validated several representative DEGs (MT1G, MT2A,
LGR5, DACT1, LEF1) by RT-qPCR in a separate
experiment with biological triplicates, all of which confirmed the
direction and magnitude of RNA-seq changes.
While the findings of this study significantly
enhance our understanding of PCB153-induced transcriptomic changes
in human IECs, several limitations should be acknowledged. The aim
of the cell viability assay in our study was not to establish a
full pharmacological dose response relationship for PCB153, but
rather to identify treatment conditions that allow detectable
transcriptomic changes without inducing overt cytotoxicity. We
found the sharp viability decline observed at 200 µM compared with
0-50 µM does not imply a mechanistically meaningful PCB153 dose
response inflection point. Instead, it reflects that 200 µM PCB153
exceeds the cytotoxic threshold for the NCM460D intestinal
epithelial cells. This abrupt reduction may correlate with the
physicochemical properties of PCB153, such as its hydrophobicity
and membrane-associated toxicity. In addition, although the
concentrations used in this study (5 and 50 µM) exceed typical
human serum PCB153 levels reported in epidemiological studies
(approximately 0.01-0.1 µM) (35-38),
these exposures remain relevant for in vitro mechanistic
investigation. PCB153 exhibits strong lipophilicity and binds
extensively to serum proteins, lipids, and culture plastics,
substantially reducing the freely bioavailable fraction in cell
culture systems. Therefore, higher concentrations in cell culture
system are often required to achieve intracellular levels
comparable to chronic low-dose exposures in vivo, but does
not fully reflect the chronic, low-level exposures experienced by
most human populations. Future studies incorporating multiple time
points or repeated lower-dose exposure models will be important for
capturing more physiologically relevant outcomes. Furthermore,
although our results align with previous studies in animal models
and provide new mechanistic insights, the use of a single in
vitro cell culture system inherently limits biological
complexity and does not fully capture the multicellular complexity
of the intestinal environment, including stromal, immune, and
microbial components. In whole organisms, compensatory mechanisms
such as epithelial regeneration and systemic detoxification
processes may partially counterbalance or adapt to such cellular
perturbations, potentially attenuating the magnitude of
transcriptomic changes observed in vitro. Validation in
intestinal organoids or in vivo models will be critical to
determine the functional relevance of the pathways identified here.
In addition, while RNA-sequencing offers a powerful tool for global
transcriptomic profiling, it is limited to changes at the mRNA
level. Because transcriptional responses do not always correlate
with protein expression or cellular function, additional validation
is warranted. Future work will focus on confirming major
differentially expressed genes at the protein level, assessing key
signaling activity (i.e. Wnt/cGMP-PKG) and performing functional
assays such as transepithelial electrical resistance (TEER),
oxidative stress measurements, and inflammatory cytokine release,
to determine whether the observed molecular changes translate into
physiological effects and strengthen the mechanistic interpretation
of the RNA-seq findings. These future directions will allow us to
build upon the transcriptomic signatures identified in this study
and more fully delineate the molecular consequences of PCB153
exposure in the intestinal epithelium. Nevertheless, the findings
from this study have important implications for populations exposed
to PCB153 through dietary or environmental sources. By uncovering
specific gene networks and signaling pathways disrupted by PCB153
in normal intestinal epithelial cells, this research offers a
molecular framework that may aid in the development of early
biomarkers for exposure-related gut dysfunction. Pathways such as
Wnt signaling, ABC transporter activity, and metallothionein
regulation could serve as candidate targets for monitoring
epithelial health or assessing individual susceptibility to
PCB-related toxicity. Furthermore, understanding how even low-dose
PCB exposure can impair epithelial regeneration, metabolism, and
immune regulation supports the need for health surveillance in
exposed populations, particularly those with high dietary intake of
contaminated fish or those residing in regions with persistent PCB
contamination. In this aspect, the in vivo toxicokinetic
studies of PCB153 bioaccumulation, including absorption,
distribution, tissue persistence, and dose-time relationships in
animal models, would indeed be highly valuable for future work.
Such studies would help define physiologically relevant exposure
ranges in the intestine and provide quantitative context for
interpreting the transcriptomic responses observed in vitro.
While toxicokinetic modeling and in vivo bioaccumulation
analyses are beyond the scope of the present study, we believe that
integrating these approaches in future investigations would
strengthen translational interpretation and help bridge in
vitro responses with real world exposure scenarios. These
mechanistic insights may also inform future strategies to mitigate
toxicant effects, such as dietary or pharmacological interventions
that restore epithelial integrity or enhance detoxification
capacity, eventually offering a strategy toward personalized
approaches to environmental health protection.
In conclusion, our findings support and extend prior
research indicating that PCB153 exposure adversely affects
intestinal epithelial function. We demonstrate that PCB153 disrupts
multiple pathways essential for epithelial renewal, immune
regulation, and metabolic balance. These findings emphasize the gut
epithelium as a critical and sensitive target of environmental
toxicants and underscore the need for further research using
physiologically relevant exposure models and mechanistic validation
to better assess the health risks associated with PCB exposure.
Supplementary Material
Primers for reverse
transcription-quantitative PCR.
Acknowledgements
Not applicable.
Funding
Funding: This work was supported by the Department of
Pediatrics, University of Illinois at Chicago and US National
Institutes of Health (grant no. P30ES027792).
Availability of data and materials
The RNA sequencing data generated in the present
study may be found in the National Center for Biotechnology
Information database under accession number PRJNA1285092 or at the
following URL: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1285092.
The other data generated in the present study may be requested from
the corresponding author.
Authors' contributions
HH and PS conceived the study, analyzed data and
wrote the manuscript. SCT analyzed data and edited the manuscript.
RMS and GSP acquired funding, interpreted data, and reviewed and
edited the manuscript. HG supervised the study, designed the
methodology, interpreted data, and wrote, reviewed and edited the
manuscript. HH and HG confirm the authenticity of all the raw data.
All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Carpenter DO: Polychlorinated biphenyls
(PCBs): Routes of exposure and effects on human health. Rev Environ
Health. 21:1–23. 2006.PubMed/NCBI View Article : Google Scholar
|
|
2
|
McLachlan MS, Undeman E, Zhao F and
MacLeod M: Predicting global scale exposure of humans to PCB 153
from historical emissions. Environ Sci Process Impacts. 20:747–756.
2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wood SA, Armitage JM, Binnington MJ and
Wania F: Deterministic modeling of the exposure of individual
participants in the National Health and Nutrition Examination
Survey (NHANES) to polychlorinated biphenyls. Environ Sci Process
Impacts. 18:1157–1168. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Desvignes V, Volatier JL, de Bels F,
Zeghnoun A, Favrot MC, Marchand P, Le Bizec B, Riviere G, Leblanc
JC and Merlo M: Study on polychlorobiphenyl serum levels in French
consumers of freshwater fish. Sci Total Environ. 505:623–632.
2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Chelakkot C, Ghim J and Ryu SH: Mechanisms
regulating intestinal barrier integrity and its pathological
implications. Exp Mol Med. 50:1–9. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Peterson LW and Artis D: Intestinal
epithelial cells: Regulators of barrier function and immune
homeostasis. Nat Rev Immunol. 14:141–153. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Choi YJ, Seelbach MJ, Pu H, Eum SY, Chen
L, Zhang B, Hennig B and Toborek M: Polychlorinated biphenyls
disrupt intestinal integrity via NADPH oxidase-induced alterations
of tight junction protein expression. Environ Health Perspect.
118:976–981. 2010.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Phillips MC, Dheer R, Santaolalla R,
Davies JM, Burgueño J, Lang JK, Toborek M and Abreu MT: Intestinal
exposure to PCB 153 induces inflammation via the ATM/NEMO pathway.
Toxicol Appl Pharmacol. 339:24–33. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Moyer MP, Manzano LA, Merriman RL,
Stauffer JS and Tanzer LR: NCM460, a normal human colon mucosal
epithelial cell line. In Vitro Cell Dev Biol Anim. 32:315–317.
1996.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Dobin A, Davis CA, Schlesinger F, Drenkow
J, Zaleski C, Jha S, Batut P, Chaisson M and Gingeras TR: STAR:
Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21.
2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Li H, Handsaker B, Wysoker A, Fennell T,
Ruan J, Homer N, Marth G, Abecasis G and Durbin R: 1000 Genome
Project Data Processing Subgroup. The sequence alignment/Map format
and SAMtools. Bioinformatics. 25:2078–2079. 2009.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Anders S, Pyl PT and Huber W: HTSeq-a
Python framework to work with high-throughput sequencing data.
Bioinformatics. 31:166–169. 2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15(550)2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ge SX, Jung D and Yao R: ShinyGO: A
graphical gene-set enrichment tool for animals and plants.
Bioinformatics. 36:2628–2629. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Socha-Banasiak A, Sputa-Grzegrzółka P,
Grzegrzółka J, Pacześ K, Dzięgiel P, Sordyl B, Romanowicz H and
Czkwianianc E: Metallothioneins in inflammatory bowel diseases:
Importance in pathogenesis and potential therapy target. Can J
Gastroenterol Hepatol. 2021(6665697)2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Waeytens A, De Vos M and Laukens D:
Evidence for a potential role of metallothioneins in inflammatory
bowel diseases. Mediators Inflamm. 2009(729172)2009.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Nait Slimane S, Marcel V, Fenouil T, Catez
F, Saurin JC, Bouvet P, Diaz JJ and Mertani HC: Ribosome biogenesis
alterations in colorectal cancer. Cells. 9(2361)2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Silva J, Alkan F, Ramalho S, Snieckute G,
Prekovic S, Garcia AK, Hernández-Pérez S, van der Kammen R, Barnum
D, Hoekman L, et al: Ribosome impairment regulates intestinal stem
cell identity via ZAKa activation. Nat Commun.
13(4492)2022.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhang H, Vreeken D, Leuning DG, Bruikman
CS, Junaid A, Stam W, de Bruin RG, Sol WMPJ, Rabelink TJ, van den
Berg BM, et al: Netrin-4 expression by human endothelial cells
inhibits endothelial inflammation and senescence. Int J Biochem
Cell Biol. 134(105960)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Sensoy I: A review on the food digestion
in the digestive tract and the used in vitro models. Curr Res Food
Sci. 4:308–319. 2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Fleckenstein JM and Bitoun JP: Changing
the locks on intestinal signaling. Cell Host Microbe. 29:1335–1337.
2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Rappaport JA and Waldman SA: The guanylate
cyclase C-cGMP signaling axis opposes intestinal epithelial injury
and neoplasia. Front Oncol. 8(299)2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Dietrich CG, Geier A and Oude Elferink
RPJ: ABC of oral bioavailability: Transporters as gatekeepers in
the gut. Gut. 52:1788–1795. 2003.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Flanagan DJ, Austin CR, Vincan E and
Phesse TJ: Wnt signalling in gastrointestinal epithelial stem
cells. Genes (Basel). 9(178)2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Mah AT, Yan KS and Kuo CJ: Wnt pathway
regulation of intestinal stem cells. J Physiol. 594:4837–4847.
2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Lazarova D and Bordonaro M: Multifactorial
causation of early onset colorectal cancer. J Cancer. 12:6825–6834.
2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Li J, Ma X, Chakravarti D, Shalapour S and
DePinho RA: Genetic and biological hallmarks of colorectal cancer.
Genes Dev. 35:787–820. 2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ullman TA and Itzkowitz SH: Intestinal
inflammation and cancer. Gastroenterology. 140:1807–1816.
2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Stolfi C, Maresca C, Monteleone G and
Laudisi F: Implication of intestinal barrier dysfunction in gut
dysbiosis and diseases. Biomedicines. 10(289)2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zong Y, Li H, Liao P, Chen L, Pan Y, Zheng
Y, Zhang C, Liu D, Zheng M and Gao J: Mitochondrial dysfunction:
Mechanisms and advances in therapy. Signal Transduct Target Ther.
9(124)2024.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Jacobson A, Yang D, Vella M and Chiu IM:
The intestinal neuro-immune axis: Crosstalk between neurons, immune
cells, and microbes. Mucosal Immunol. 14:555–565. 2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Treps L, Le Guelte A and Gavard J:
Emerging roles of Semaphorins in the regulation of epithelial and
endothelial junctions. Tissue Barriers. 1(e23272)2013.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Agudo A, Goñi F, Etxeandia A, Vives A,
Millán E, López R, Amiano P, Ardanaz E, Barricarte A, Chirlaque MD,
et al: Polychlorinated biphenyls in Spanish adults: Determinants of
serum concentrations. Environ Res. 109:620–628. 2009.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Cerná M, Malý M, Grabic R, Batáriová A,
Smíd J and Benes B: Serum concentrations of indicator PCB congeners
in the Czech adult population. Chemosphere. 72:1124–1131.
2008.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Leong JY, Blachman-Braun R, Patel AS,
Patel P and Ramasamy R: Association between polychlorinated
biphenyl 153 exposure and serum testosterone levels: Analysis of
the national health and nutrition examination survey. Transl Androl
Urol. 8:666–672. 2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Longnecker MP, Wolff MS, Gladen BC, Brock
JW, Grandjean P, Jacobson JL, Korrick SA, Rogan WJ,
Weisglas-Kuperus N, Hertz-Picciotto I, et al: Comparison of
polychlorinated biphenyl levels across studies of human
neurodevelopment. Environ Health Perspect. 111:65–70.
2003.PubMed/NCBI View Article : Google Scholar
|