Introduction
Bone remodeling is an ongoing and intricate
physiological process essential for preserving bone integrity and
functionality. It depends on the balance between
osteoclast-mediated bone resorption and osteoblast-driven bone
formation (1). Osteoporosis is
defined by a disruption in the equilibrium between bone resorption
and formation, resulting in diminished bone density and a
heightened susceptibility to fractures (2). The compromised ability of bone marrow
mesenchymal stem cells (BMSCs) to undergo osteogenic
differentiation is a critical factor underlying the reduced
capacity for bone formation (3).
BMSCs are pivotal in bone formation during remodeling and repair.
Research into conditions such as bone defects and osteoporosis
focuses on guiding bone remodeling toward formation by modulating
BMSCs (4). In the field of oral
regenerative repair, BMSCs have demonstrated tremendous potential
in promoting alveolar bone repair and regeneration. Numerous
studies have shown that BMSCs significantly enhance the
regeneration of alveolar bone defects whether used alone or in
combination with materials such as fibrin glue or scaffolds
(5,6). The use of BMSCs in periodontal bone
tissue regeneration and repair has significant potential to enhance
the scope of tooth movement and extend the applicability to
orthodontic and dental interventions, ultimately delivering safer
and more effective treatment alternatives for patients.
Autophagy plays an essential role in osteogenesis
and periodontal bone tissue regeneration. Autophagy is a cellular
self-degradation and recycling process in which damaged or excess
organelles, proteins, and other macromolecules are enclosed in
autophagosomes and eventually degraded within lysosomes (7). This mechanism supports cellular
homeostasis and allows cells to manage stress conditions such as
nutrient deprivation or oxidative stress, significantly impacting
numerous physiological and pathological processes (8). Autophagy is integral to the
development and progression of human diseases, with numerous
studies highlighting its close association with various conditions
(9); it suppresses early
tumorigenesis but promotes survival in advanced cancer (10), contributes to neurodegeneration (for
example Parkinson s disease) via defective protein/organelle
clearance (11), modulates
cardiovascular injury outcomes (12), exacerbates metabolic disorders such
as type 2 diabetes when impaired (13), and participates in host defense
against pathogens (14). Research
on cellular autophagy is crucial for understanding cell biology and
disease mechanisms, potentially leading to novel therapeutic
discoveries. Studies have shown that autophagy levels increase in a
time-dependent manner during the osteogenic differentiation of
BMSCs, highlighting its essential role in promoting osteogenesis
(15,16). Specifically, autophagy activation
promotes the differentiation of BMSCs into osteoblasts. Moreover,
under conditions of nutrient deprivation or oxidative stress,
autophagy can sustain osteoblast activity by clearing damaged
mitochondria and proteins (17).
Research has shown that hypoxic conditions in the local periodontal
microenvironment caused by orthodontic treatments can induce
autophagy, thereby affecting the remodeling of periodontal bone
tissue (18). Therefore,
understanding the mechanisms underlying autophagy in BMSCs holds
substantial clinical and research value.
Autophagy progresses from initiation to
autophagosome-lysosome fusion and the eventual degradation of
cellular contents (19). The
mammalian target of rapamycin (mTOR) pathway is a central regulator
of cellular autophagy, orchestrating the autophagy process through
diverse molecular mechanisms and signaling pathways. mTOR responds
to various signals such as cellular nutritional status, energy
levels, and growth factors to adjust the metabolism and growth of
the cell (20). mTOR affects
autophagy through downstream proteins such as neural precursor
cell-expressed developmentally down-regulated protein 4 (NEDD4).
NEDD4 consists of an N-terminal C2 domain, a C-terminal HECT
domain, and two to four WW domains (21). Studies have shown that NEDD4 is
essential for the survival of cranial neural crest cells and the
development of craniofacial bones (22). Mice with Nedd4 gene deletion
were shown to exhibit notable craniofacial defects (23). Furthermore, osteoprogenitor cells
lacking NEDD4 exhibited impaired osteogenic differentiation
(24). Our preliminary research has
also shown that NEDD4 knockout promotes osteogenic differentiation
in BMSCs (25). Notably, the role
that NEDD4 plays in autophagy has also attracted attention. NEDD4,
an E3 ubiquitin ligase, facilitates protein degradation through
ubiquitination. NEDD4 influences the stability and function of
autophagy-related proteins, including light chain 3 (LC3) and
Beclin-1, via direct or indirect ubiquitination. These proteins are
recognized autophagy markers and exhibit increased expression
levels during autophagic activity (26-29).
Tumor cell research has shown that NEDD4 promotes autophagy and is
associated with the mTOR signaling pathway (30). Meanwhile, our preliminary research
revealed that NEDD4 knockout increased the expression of
mTORC1(25). mTORC1, a key element
of the mTOR signaling pathway, was essential for autophagy
regulation.
Based on these observations, it was hypothesized
that NEDD4 expression in BMSCs promotes autophagy via the mTOR
pathway. To test this hypothesis, autophagy was induced in BMSCs
under starvation conditions and NEDD4 expression was modulated by
lentiviral knockdown. The changes were then examined in autophagy
(LC3 and Beclin-1 levels) and mTOR pathway activity in response to
NEDD4 knockdown. Through this experimental design, the effect of
NEDD4 on starvation-induced autophagy in BMSCs via mTOR was sought
to be elucidated, and the potential of NEDD4 as a novel therapeutic
target for conditions such as periodontal bone defects and
osteoporosis was aimed to be determined.
Materials and methods
Animals
Male, 4-week-old Sprague-Dawley rats (n=8; body
weight, 80-100 g) were obtained from the Experimental Animal Center
of Southwest Medical University (Luzhou, China). The animals were
housed under standard laboratory conditions with a controlled
temperature of 20-26˚C, a relative humidity of 40-70%, a 12-h
light/dark cycle, and ad libitum access to standard rodent
chow and sterile drinking water. After euthanasia by carbon dioxide
asphyxiation, femurs were dissected for BMSC extraction. For
euthanasia, carbon dioxide (CO2) was introduced into an
uncharged chamber at a displacement rate of 30-70% of the chamber
volume per minute, in accordance with the 2020 AVMA Guidelines.
Death was confirmed by cessation of respiration and heartbeat, lack
of corneal reflexes, and a secondary physical method (cervical
dislocation). Ethical approval for the animal experiments was
granted by the Ethics Committee of Southwest Medical University in
Luzhou, Sichuan, China (approval no. 20221206-005).
Cell culture and identification
The rat femurs were dissected and rinsed with
sterile phosphate-buffered saline (PBS; cat. no. 1001002; Gibco;
Thermo Fisher Scientific, Inc. 3), and the soft tissues attached to
the femurs were removed in a sterile biosafety cabinet. α-Minimum
essential medium (α-MEM; cat no. 12561056; Gibco, Thermo Fisher
Scientific, Inc.) containing 10% fetal bovine serum (FBS; cat. no.
10099141; Gibco;Thermo Fisher Scientific, Inc.) was used for cell
culture.
The BMSC cell suspension was centrifuged at 1,000 x
g at 4˚C for 5 min and then resuspended in α-MEM supplemented with
10% FBS, 100 µg/ml penicillin and 100 µg/ml streptomycin (cat. no.
15140122; Gibco; Thermo Fisher Scientific, Inc.), and incubated in
a humidified atmosphere containing 5% CO2 at 37˚C. After
48 h, the medium was replaced with fresh medium to remove
non-adherent cells. When cell confluency reached 80%, the cells
were passaged, and the passaged cells were designated as passage 1
(P1). Subsequent experiments were performed using third-passage
cells (P3).
Flow cytometry was performed to detect the surface
markers of P1 BMSCs (CD29, CD90, CD45, and CD34). The following
primary antibodies were used to label the corresponding markers:
FITC-conjugated anti-rat CD29 antibody (cat. no. 102206, dilution,
1:100; BioLegend, Inc.), PE-Cy7-conjugated anti-rat CD90 antibody
(cat. no. 202517; dilution, 1:100; BioLegend, Inc.), PE-conjugated
anti-rat CD34 antibody (cat no. 551387; dilution, 1:100; BD
Pharmingen; BD Biosciences), and AF647-conjugated anti-rat CD45
antibody (cat. no. 160303; dilution, 1:100; BioLegend, Inc.). All
primary antibodies were directly conjugated with fluorochromes
(FITC, PE-Cy7, PE, and AF647 as reporter moieties); therefore, no
secondary antibodies were required. Detection was performed using a
direct immunofluorescence assay with a flow cytometer, and the
fluorochrome-conjugated primary antibodies were excited at their
respective optimal wavelengths to detect fluorescence signals.
After incubation for 30 min at 4˚C in the dark, the cells were
analyzed using a fluorescence-activated cell sorter (FACSCalibur;
BD Biosciences). Data acquisition and analysis were performed using
FlowJo software (version 10.8.1; FlowJo LLC). The protocol for
isolating and identifying BMSCs was consistent with previously
described methods (31).
Multilineage differentiation induction
and starvation-induced autophagy
P3 BMSCs were cultured in 6-well plates until cell
confluence exceeded 80%, after which the medium was switched to
either adipogenic or osteogenic induction medium, with the medium
changed every three days. The adipogenic induction medium consisted
of α-MEM supplemented with 10% FBS, 1 µM dexamethasone (cat. no.
D8040), 0.5 mM 3-isobutyl-1-methylxanthine (IBMX; cat. no.
II00101), 10 µg/ml insulin (cat. no. I8040), and 100 µM
indomethacin (cat. no. II0100; all from Beijing Solarbio Science
& Technology Co., Ltd.). The osteogenic induction medium
consisted of α-MEM with 10% FBS, 10 mM β-glycerophosphate (cat. no.
G9140), 50 µg/ml L-ascorbic acid-2-phosphate sesquimagnesium salt
hydrate (cat. no. A8960-5G; Sigma-Aldrich; Merck KGaA), and 100 nM
dexamethasone (cat. no. D8040; all from Beijing Solarbio Science
& Technology Co., Ltd.). Cells were washed three times with PBS
and fixed in 4% paraformaldehyde for 30 min at room temperature on
the 14th day of adipogenic induction and the 21st day of osteogenic
induction. Alizarin Red S (cat. no. G1452) and Oil Red O (cat. no.
G1262; both from Beijing Solarbio Science & Technology Co.,
Ltd.) were used to stain osteogenically and adipogenically induced
cells, respectively, according to the manufacturer s
instructions.
Autophagy was induced by nutrient starvation.
Nutrient deprivation using Earle s balanced salt solution
(EBSS; cat. no. H2045) is a standard approach to trigger autophagy
by acutely limiting amino acids and growth factors; this method is
widely used to assess upstream regulators of autophagy. EBSS was
therefore selected to induce autophagy in BMSCs (32,33).
Briefly, P3 BMSCs in a logarithmic growth condition were switched
to EBSS once confluence reached ~70%, marking time point 0 as the
start of induction.
Lentiviral infection
Lentiviruses expressing short hairpin RNA (shRNA)
targeting NEDD4 (sh-NEDD4) and a non-targeting control shRNA
(sh-NC) were constructed using the GV493 lentiviral vector
(hU6-MCS-CBh-gcGFP-IRES-puromycin; Shanghai GeneChem Co., Ltd.).
Lentiviral packaging was performed using 293T cells (cat. no.
SNL-015; Wuhan SunnCell Biotech Co., Ltd.) as the packaging cell
line. For each 6-well plate transfection, 4 µg of lentiviral
plasmids (sh-NEDD4 or sh-NC) were utilized, with lentiviral,
packaging (psPAX2) and envelope (pMD2.G) plasmids mixed at a strict
mass ratio of 4:3:1. The transfection was carried out in at 37˚C
incubator supplemented with 5% CO2 for 48 h
consecutively. Culture supernatants harboring lentiviral particles
were harvested at 48 and 72 h post-transfection, subjected to
centrifugation at 1,000 x g for 20 min at 4˚C and filtration
through a 0.45-µm filter membrane to eliminate cell debris, thus
obtaining purified lentiviral particles. Following lentiviral
transduction of BMSCs, the cells were cultured for an additional 72
h to ensure sufficient viral integration prior to initiating
subsequent nutrient deprivation experiments. Puromycin (cat. no.
P8230; Beijing Solarbio Science & Technology Co., Ltd.) was
utilized for positive selection of stably transduced cell clones,
with a selection concentration of 3.5 µg/ml and a maintenance
concentration of 1.5 µg/ml during long-term subsequent culture. The
specific target sequences of the shRNAs are detailed in Table I. For the transduction procedure,
passage 3 (P3) BMSCs in the logarithmic growth phase were detached
with trypsin and adjusted to a cell density of 5x104
cells/ml. Each well of a 6-well plate was seeded with 1 ml of the
cell suspension, after which the cells were co-incubated with
lentivirus in the presence of HitransG P transduction enhancer for
16 h at 37˚C. Fresh growth medium was replenished at 24 h
post-transduction. The multiplicity of infection (MOI) was
determined using the formula: (virus titer x volume of virus
added)/number of cells, and an optimal MOI of 50 was selected and
applied in all experiments of this study.
 | Table IshRNA target sequences used in this
study. |
Table I
shRNA target sequences used in this
study.
| shRNA ID | Accession | Target sequence
(5 →3 ) |
|---|
| sh-NC | NM_012986 |
TTCTCCGAACGTGTCACGT |
| sh1 | NM_012986 |
AGCCACAAATCAAGAGTTAAA |
| sh2 | NM_012986 |
TTGGAAGGACCTACTACGTAA |
| sh3 | NM_012986 |
CTGGATTGAGTTTGATGGTGA |
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cells using the TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, inc.) according to
the manufacturer s instructions. cDNA synthesis was carried
out with the PrimeScript Reverse Transcription Kit (Takara Bio,
Inc.). RT-qPCR with SYBR Premix Ex Taq (Takara Bio, Inc.) was used
to quantify the gene expression levels of NEDD4, LC3, and Beclin-1
(Becn1) with glyceraldehyde-3-phosphate dehydrogenase
(GAPDH) as the reference gene. The thermocycling conditions were as
follows: Initial denaturation at 95˚C for 30 sec, followed by 40
cycles at 95˚C for 5 sec and 60˚C for 30 sec; a melt-curve analysis
was performed to confirm specificity. Relative expression was
calculated using the 2-ΔΔCq method (34). Primer sequences are provided in
Table II.
 | Table IIPrimer sequences of the measured
genes. |
Table II
Primer sequences of the measured
genes.
| Genes | Sequence of primers
(5 →3 ) |
|---|
| GAPDH | R:
TAGCCCAGGATGCCCTTTAGT |
| | F:
CCCCCAATGTATCCGTTGTG |
| LC3B | R:
GCCGAAGGTTTCTTGGGAGG |
| | F:
TTGGTCAAGATCATCCGGCG |
|
Beclin-1 | R:
AATTGTCCGCTGTGCCAGATATG |
| | F:
GCTCAAGAGTGTAGAGAACCAGA |
| NEDD4 | R:
GCCAGACCTATGCCAGCTAT |
| | F:
GCTTTCGGAGGACGAGGTAT |
Western blotting
Cellular protein was extracted with RIPA lysis
buffer containing 1 mM phenylmethylsulfonyl fluoride (Beyotime
Institute of Biotechnology) according to the manufacturer s
instructions. The protein concentration was measured using a BCA
assay kit (Beyotime Institute of Biotechnology). Western blotting
was performed according to standard protocols (35). Specifically, equal amounts of total
protein (20-30 µg per lane) were separated by 12% SDS-PAGE and
transferred to PVDF membranes (0.45 µm). Membranes were blocked
with 5% bovine serum albumin (BSA; cat. no. SW3015; Beijing
Solarbio Science & Technology Co., Ltd.) in TBST containing
0.1% Tween-20 for 1 h at room temperature, then incubated overnight
at 4˚C with primary antibodies: GAPDH [cat. no. 2118;
1:3,000-1:5,000; Cell Signaling Technology (CST)], LC3B (cat. no.
ab192890; 1:1,000; Abcam), Beclin-1 (cat. no. 3495; 1:1,000; CST),
NEDD4 (cat. no. 2740; 1:1,000; CST), total mTOR (cat. no. 2983;
1:1,000; CST), and phospho-mTOR (Ser2448; cat. no. 5536; 1:1,000;
CST). After washing, membranes were incubated with HRP-conjugated
secondary antibodies (anti-rabbit IgG, cat. no. 7074; anti-mouse
IgG, 7076; each at 1:5,000; CST) for 1 h at room temperature. Bands
were visualized using an enhanced chemiluminescent substrate
(SuperSignal West Pico PLUS; cat. no. 34580; Thermo Fisher
Scientific, Inc.) and imaged on a digital chemiluminescence system.
Within each composite figure panel, proteins were probed on the
same membrane (using membrane cutting or strip-and-reprobe) unless
otherwise indicated. Densitometric analysis of the blots was
performed using ImageJ software (version 1.8.0_172; National
Institutes of Health).
Immunofluorescence
Cells were seeded into 24-well plates at a density
of 1x104 cells per well. After adherence, cells were
fixed with 4% paraformaldehyde for 15 min at room temperature,
permeabilized with 0.5% Triton X-100 for 15 min at room
temperature, and blocked with 5% BSA for 60 min at room
temperature. Cells were then incubated overnight at 4˚C with an
anti-LC3 primary antibody (cat. no. ab192890; 1:1,000; Abcam). The
following day, cells were washed three times with PBS and incubated
for 1 h at room temperature with an Alexa Fluor 594-conjugated goat
anti-rabbit secondary antibody (cat. no. 31460; Thermo Fisher
Scientific, Inc.) at a dilution of 1:1,000 in 5% BSA/PBS for 1 h at
room temperature. Nuclei were counterstained with
4 ,6-diamidino-2-phenylindole (DAPI; cat. no. C0065; Beijing
Solarbio Science & Technology Co., Ltd.) at a dilution of
1:5,000 in PBS for 10 min at room temperature. After washing with
PBS, the cells were observed under a fluorescence microscope
(Olympus Corporation). These immunocytochemical procedures followed
previously published protocols (35).
Transmission electron microscopy
(TEM)
Autophagic vesicles (AVs) were visualized using TEM.
Cells were collected by gentle scraping and initially fixed in 3%
glutaraldehyde (cat. no. G5882; Sigma-Aldrich; Merck KGaA) in 0.1 M
phosphate buffer (pH 7.4) at 4˚C for 2 h, followed by post-fixation
with 1% osmium tetroxide (cat. no. 201030; Sigma-Aldrich; Merck
KGaA) at 4˚C for 30 min. Cells were dehydrated through a graded
ethanol series, embedded in Epon resin (cat. no. 45345;
Sigma-Aldrich; Merck KGaA), and ultrathin sections with a thickness
of 50 nm were prepared. The sections were stained with 2% uranyl
acetate (cat. no. S9062-5; RuiTaibio) at room temperature for 15
min, followed by lead citrate (cat. no. 17800; Electron Microscopy
Sciences) at room temperature for 10 min. Subcellular structures
were examined using a JEM-2000EX transmission electron microscope
(JEOL, Ltd.).
Statistical analysis
Data processing and visualization were performed
using GraphPad Prism (version 9.0; GraphPad Software; Dotmatics).
All experiments were conducted in triplicate (n=3). Results are
presented as the mean ± standard deviation (SD). One-way ANOVA was
used to evaluate group differences. Following one-way ANOVA,
Tukey s multiple-comparison post hoc test was applied for
pairwise comparisons. A two-sided α value of 0.05 was considered
the threshold for statistical significance; P-values <0.05 were
considered to indicate a statistically significant difference.
Results
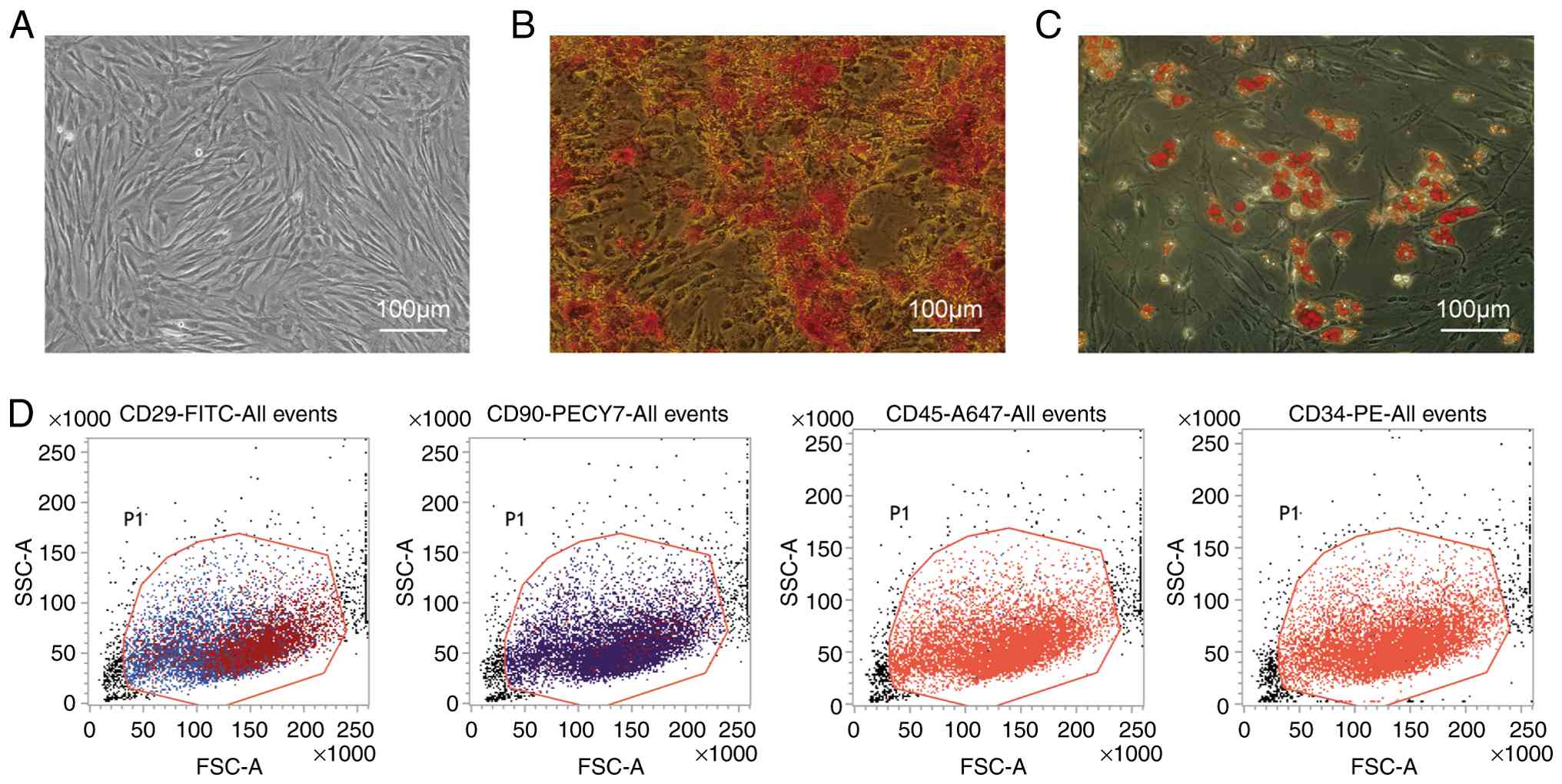
Culture and identification of
BMSCs
Cells were extracted from rat femurs using whole
bone marrow adherent culture techniques. After passaging, the cells
exhibited a uniform fibroblast-like morphology and were arranged in
a vortex pattern (Fig. 1A).
Multilineage differentiation potential was observed. On day 21 of
osteogenic induction, Alizarin Red S staining revealed numerous
orange-red calcium nodules (Fig.
1B). Oil Red O staining after 14 days of adipogenic induction
showed the presence of red lipid droplets, demonstrating the
adipogenic potential of the cultured cells (Fig. 1C). Flow cytometric analysis
indicated high expression of mesenchymal stem cell markers CD90
(98.73%) and CD29 (95.99%), whereas hematopoietic stem cell markers
CD34 and CD45 were minimally expressed (0.3 and 0.05%,
respectively), consistent with mesenchymal stem cell
characteristics (Fig. 1D). These
findings verify the successful isolation and culture of rat
BMSCs.
Autophagy induction and expression of
autophagy-related genes
Autophagy in BMSCs was induced by EBSS starvation,
and cells were collected at specific time points. RT-qPCR analysis
showed that the mRNA expression of NEDD4 and autophagy-related
genes LC3 and Beclin-1 gradually increased, peaking at 30 min
post-induction, and then decreased by 1 h (Fig. 2A). Western blot analysis showed that
NEDD4 protein levels, along with autophagy markers LC3-II and
Beclin-1, were elevated in BMSCs following EBSS starvation (with a
maximum expression at ~30 min; Fig.
2B and C). Immunofluorescent
staining further confirmed autophagy induction in BMSCs (Fig. 2D). LC3 was mainly expressed in the
cytoplasm, and the fluorescence intensity of LC3 increased during
the first 30 min of starvation. After 1 and 2 h, LC3 fluorescence
remained strong, but the number of cells in the field of view
decreased significantly and cell morphology changed. By 2 h,
numerous cells exhibited pronounced shrinkage.
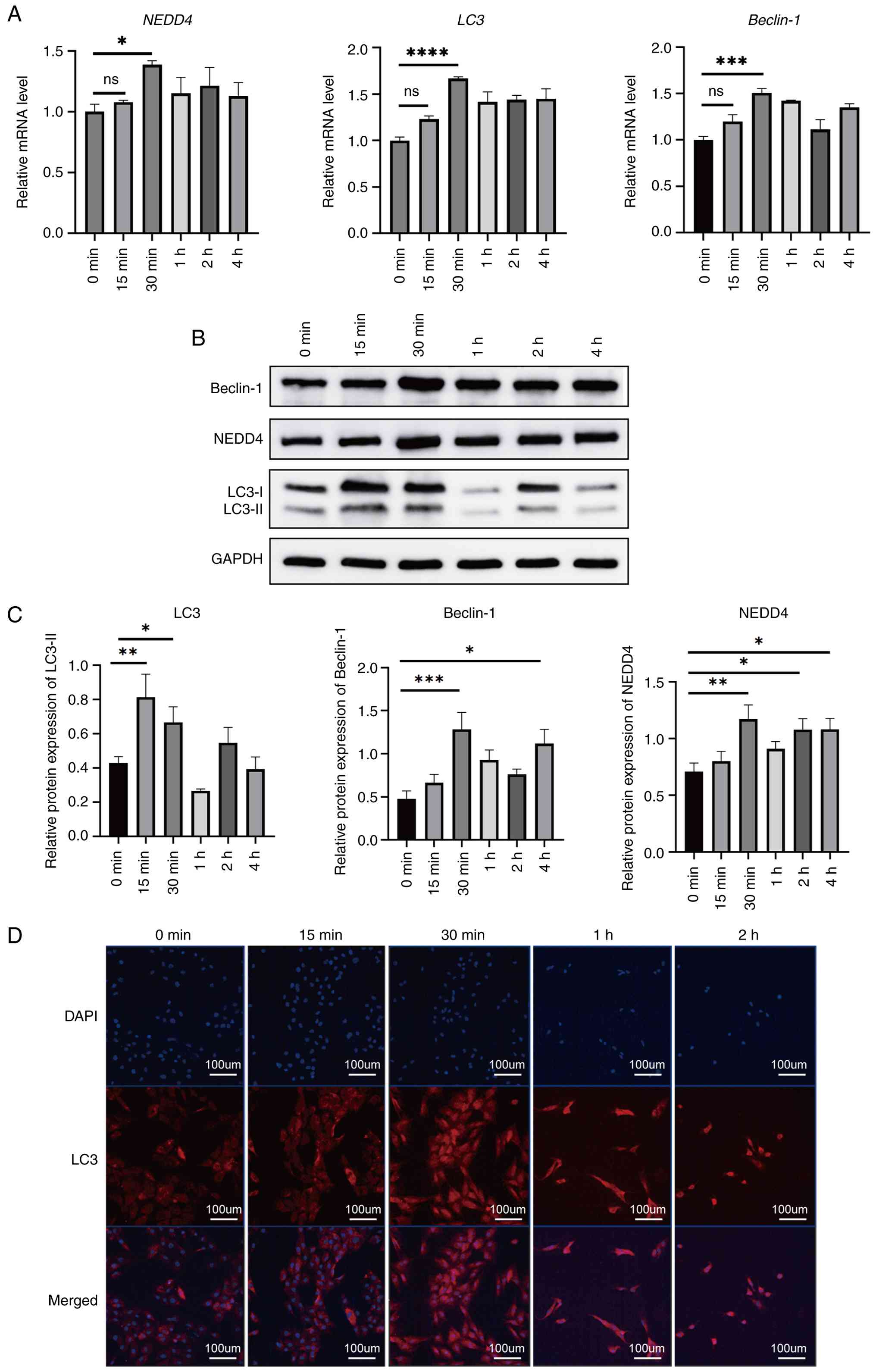 | Figure 2Time-course of autophagy induction in
BMSCs following EBSS starvation. (A) RT-qPCR analysis of NEDD4,
LC3, and Beclin-1 mRNA levels at 0, 15, 30 min, and 1, 2, 4 h
post-starvation. (B and C) A representative western blot image and
quantification of autophagy-related proteins NEDD4, LC3-I and -II,
and Beclin-1 over time. (D) Representative immunofluorescent images
showing LC3 (red) localization in the cytoplasm of BMSCs after
starvation. Nuclei were counterstained with DAPI (blue). Scale bar,
100 µm. Data are presented as the mean ± SD (n=3).
*P≤0.05, **P≤0.01, ***P≤0.001 and
****P≤0.0001. BMSCs, bone marrow mesenchymal stem cells;
EBSS, Earle s balanced salt solution; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; NEDD4, neural
precursor cell-expressed developmentally down-regulated protein 4;
LC3, light chain 3; DAPI, 4 ,6-diamidino-2-phenylindole; ns,
not significant. |
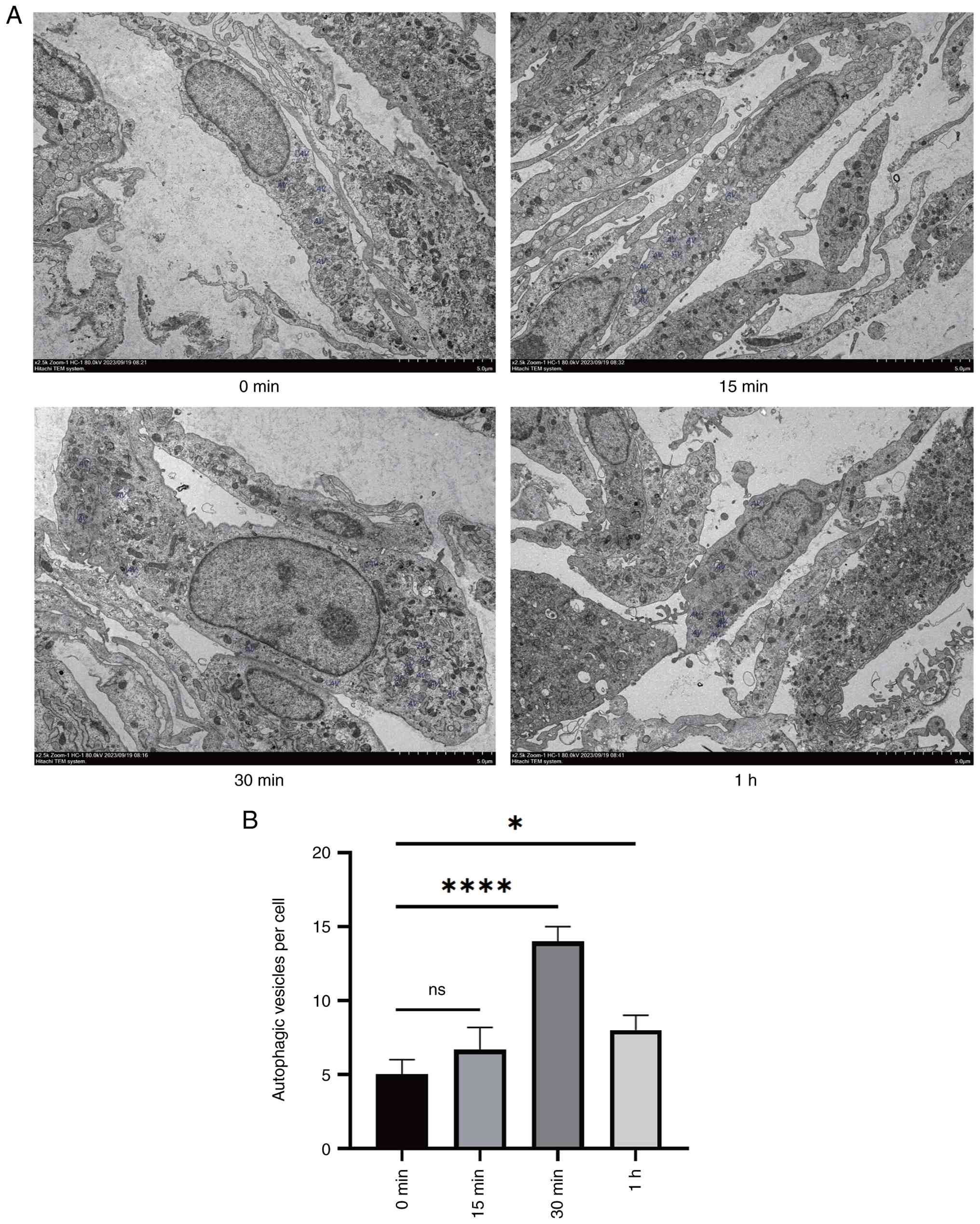
TEM was employed to evaluate the formation of AVs
among different groups. The results showed that there was a
tendency for AV accumulation after starvation induction, reflecting
autophagy induction, and the 30-min starvation group exhibited
significant accumulation (Fig. 3).
In addition, morphological changes such as nuclear shrinkage,
fragmentation, and irregular edges were observed in cells 1 h after
starvation, indicating the progression to adverse cellular
conditions or apoptosis (consistent with excessive stress).
Collectively, these results indicated that BMSCs undergo
significant autophagy after 30 min of EBSS induction, which may be
related to increased NEDD4 expression.
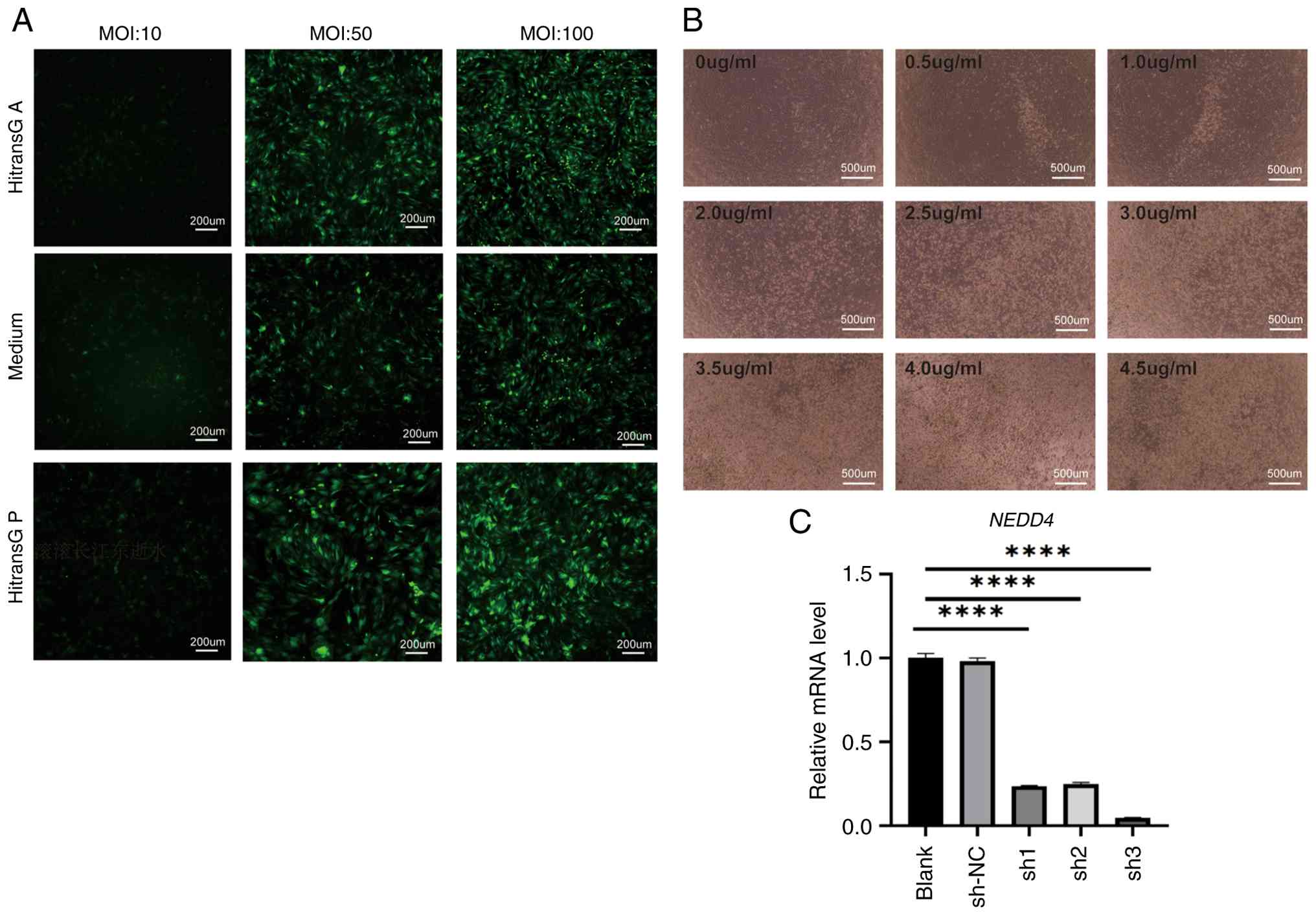
Knockdown of NEDD4 inhibits autophagy
in BMSCs
Lentivirus-mediated NEDD4 knockdown was performed in
BMSCs to investigate the role of NEDD4 in autophagy. The optimal
infection condition was achieved using HitransG P with an MOI of 50
(Fig. 4A). Successful infection was
further confirmed by puromycin selection, with optimal selection
efficiency observed at a puromycin concentration of 3.5 µg/ml
(Fig. 4B). RT-qPCR analysis
revealed that among three shRNA sequences assessed, the sh3
sequence produced the most effective silencing of the NEDD4 gene.
Therefore, the sh3 virus was selected for use in subsequent
experiments (Fig. 4C).
Upon EBSS starvation, NEDD4-knockdown BMSCs
exhibited a marked reduction in autophagy compared with controls.
NEDD4 knockdown significantly blunted the starvation-induced
increases in LC3 and Beclin-1 expression, keeping them below
baseline levels (Fig. 5A).
Immunofluorescent staining of LC3 (Fig.
5B) further demonstrated that autophagy was markedly suppressed
in NEDD4-deficient cells: LC3 puncta formation and intensity were
significantly lower in the sh-NEDD4 + EBSS group than in the EBSS
(starvation only) group. These results indicated that NEDD4 is
necessary for the full autophagic response in BMSCs under
starvation, suggesting that NEDD4 mediates or positively regulates
autophagy in BMSCs.
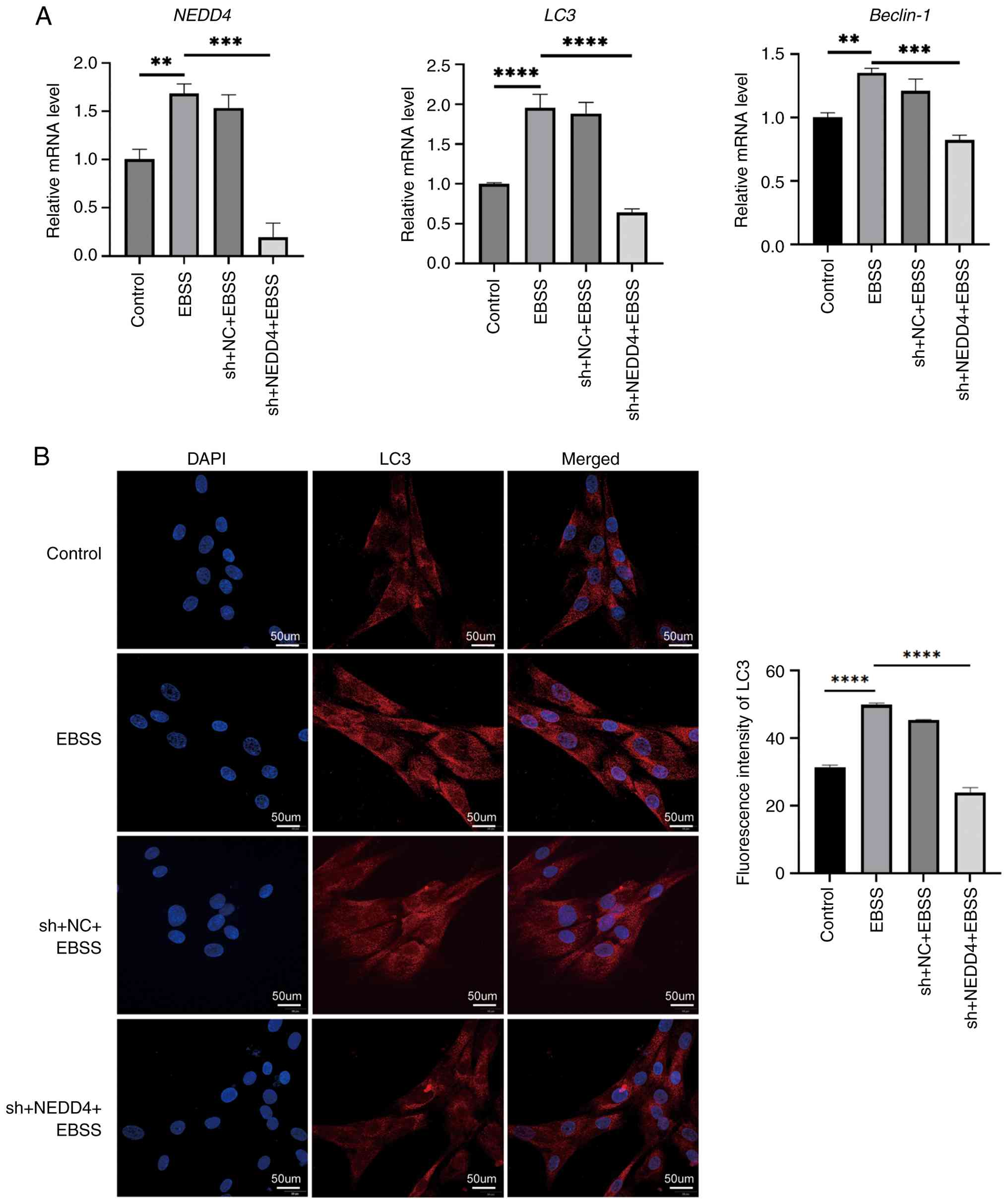 | Figure 5NEDD4 knockdown impairs
starvation-induced autophagy in BMSCs. (A) RT-qPCR analysis of
NEDD4, LC3 and Beclin-1 mRNA expression under control, EBSS, sh-NC
+ EBSS, and sh-NEDD4 + EBSS conditions. (B) Representative LC3
immunofluorescent images and quantification. LC3 puncta are
markedly reduced in the NEDD4 knockdown group. Scale bar, 50 µm.
Data are expressed as mean ± SD (n=3). **P≤0.01,
***P≤0.005 and ****P≤0.0001. NEDD4, neural
precursor cell-expressed developmentally down-regulated protein 4;
BMSCs, bone marrow mesenchymal stem cells; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; LC3, light
chain 3; EBSS, Earle s balanced salt solution; sh-, short
hairpin; NC, negative control. |
Possible association between NEDD4 and
the mTOR pathway in the regulation of autophagy
The role of NEDD4 in autophagy in BMSCs was
investigated by examining the mTOR pathway. Western blot analyses
were used to assess mTOR activation status (p-mTOR) as well as
autophagy markers, with and without NEDD4 knockdown (Fig. 6). The results showed that starvation
(EBSS) led to increased protein levels of NEDD4, LC3-II, and
Beclin-1, accompanied by a notable reduction in the level of
p-mTOR, indicating suppression of mTOR activity during autophagy
induction. When NEDD4 was knocked down, this trend was reversed.
p-mTOR levels were restored (reaching higher levels than in starved
control cells), while the levels of LC3-II and Beclin-1 were lower
(consistent with the inhibition of autophagy). These results
suggest that NEDD4 may activate autophagy at least in part by
inhibiting the mTOR signaling pathway. In other words, NEDD4 acts
upstream to negatively regulate mTOR during autophagy. This finding
aligns with previous findings on the role of mTOR as a negative
regulator of autophagy (30). In
our prior research, it was observed that an increase in osteogenic
differentiation markers in BMSCs was associated with improved
bone-forming capability when autophagy was inhibited (25), suggesting a possible link between
autophagy and osteogenesis. The present study identified NEDD4 as
an upstream regulator of mTOR in BMSCs, offering novel insights
into the molecular mechanisms governing BMSC autophagy and its
potential impact on bone remodeling.
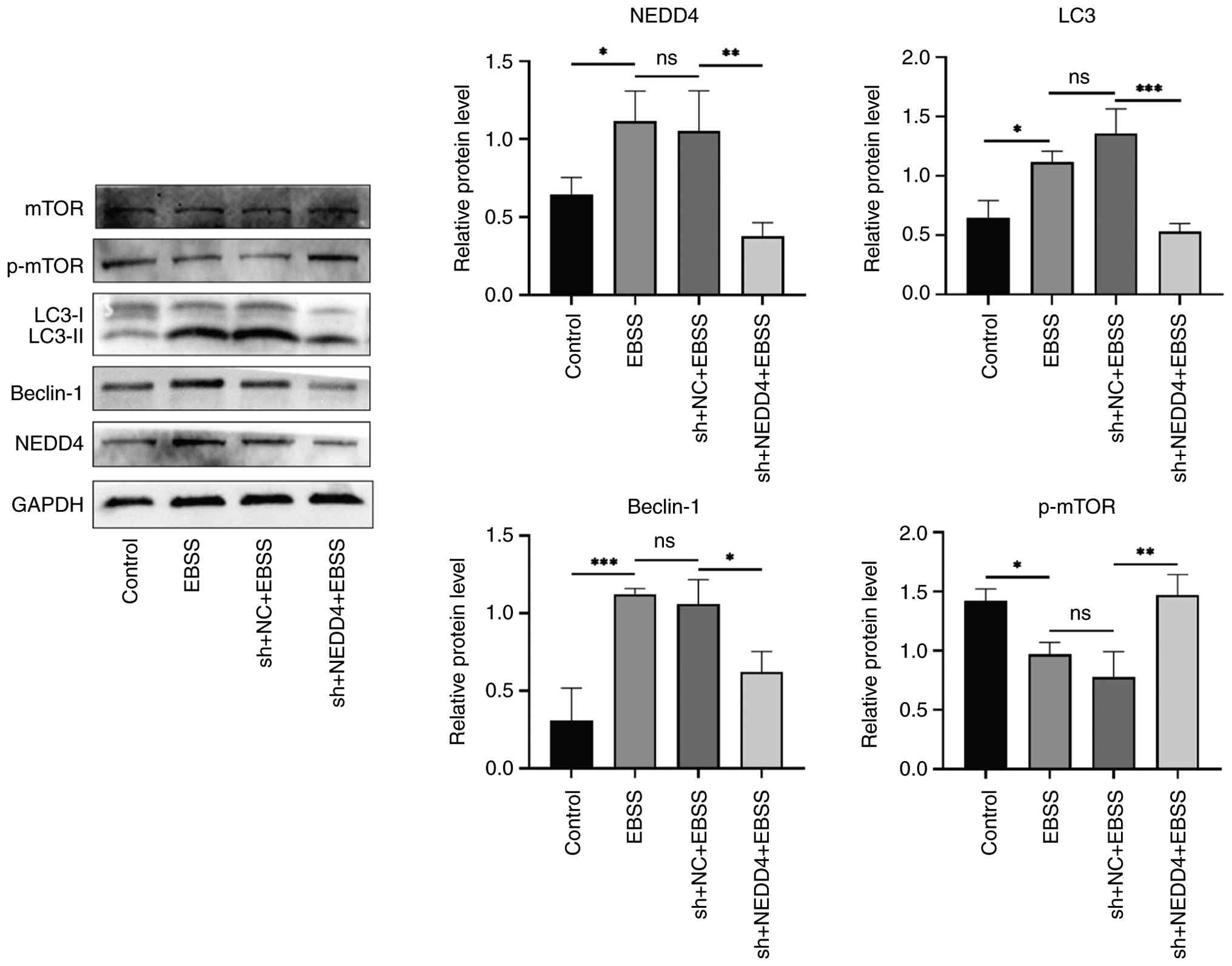 | Figure 6Effects of NEDD4 knockdown on
autophagy markers and mTOR signaling. A representative western blot
image and quantification of LC3-II, Beclin-1, total mTOR, and
p-mTOR in BMSCs under EBSS treatment, with and without NEDD4
knockdown. NEDD4 knockdown reverses EBSS-induced reduction in
p-mTOR, indicating mTOR reactivation. Data are shown as mean ± SD
(n=3). *P≤0.01, **P≤0.005,
***P≤0.0001. Proteins shown within the panel were
detected on the same membrane (cut by molecular weight) unless
otherwise indicated. NEDD4, neural precursor cell-expressed
developmentally down-regulated protein 4; mTOR, mammalian target of
rapamycin; p-, phosphorylated; BMSCs, bone marrow mesenchymal stem
cells; EBSS, Earle s balanced salt solution; sh-, short
hairpin; NC, negative control. |
Discussion
The present study investigated the involvement of
NEDD4 in BMSC autophagy. Our experimental findings demonstrated
that NEDD4 is essential for the initiation and maintenance of
autophagy in BMSCs. Through culturing of rat BMSCs, autophagy
induction via starvation, and NEDD4 knockdown experiments, it was
found that NEDD4 significantly influences the expression of
autophagy-related genes and regulates this process through
interactions with the mTOR signaling pathway. The findings offer
novel insights into how NEDD4 and autophagy regulate BMSC
function.
In the present study, autophagy was induced by EBSS
starvation. RT-qPCR and western blot analyses indicated elevated
Beclin-1 and LC3 expression following starvation induction,
suggesting enhanced autophagic activity due to their essential
roles in autophagy (36). Beclin-1
and LC3 expression are typically associated with autophagic
activity. However, an increase in these proteins alone is
insufficient to confirm autophagy, as such changes may reflect the
initiation of autophagy rather than its completion. LC3, Beclin-1,
and p62 are among the most well-known core autophagy proteins
(37). To fully evaluate autophagy,
measuring p62 levels, which typically decrease when autophagic flux
increases, would be informative and p62 will be assessed in future
experiments.
RT-qPCR and western blot analyses also revealed a
marked increase in NEDD4 expression in BMSCs after 30 min of EBSS
starvation-induced autophagy. NEDD4 expression was revealed to be
positively associated with autophagy-related genes LC3 and
Beclin-1, suggesting its role as a positive regulator of autophagy
in BMSCs. NEDD4 is suggested to promote cell survival during stress
(such as nutrient deprivation) by supporting autophagy.
Regarding the starvation duration, various time
points (0-4 h) were assessed and it was found that BMSCs maintained
acceptable viability up to ~30 min of starvation. After 1 h of
starvation, cells exhibited contraction, rounding, and some cell
detachment, with reduced cell density. TEM at 1 h revealed nuclear
shrinkage, fragmentation, and irregular nuclear edges, indicating
that prolonged starvation could initiate apoptotic changes.
Starvation is known to activate autophagy as an adaptive response
to help cells maintain energy balance by degrading and recycling
intracellular components. However, prolonged starvation can trigger
apoptosis. Autophagy and apoptosis, while distinct, have complex
interrelationships that can be synergistic, mutually inhibitory, or
even interconvertible under different conditions (38,39).
Some studies suggest cells can undergo apoptosis and autophagy
simultaneously (40,41). In our experiments, after 15 min of
starvation, autophagy markers had begun to increase (as determined
by RT-qPCR and western blotting) but not to the extent observed at
30 min, indicating that 15 min may have been insufficient for
robust autophagy induction. Based on our comprehensive autophagy
assessments (molecular and ultrastructural), 30 min of EBSS
starvation was selected as the optimal induction duration for
subsequent experiments.
The experiments in the present study showed that
knocking down NEDD4 significantly reduced the expression of LC3 and
Beclin-1 in starved BMSCs. This finding aligns with that of Sun
et al (27), who
demonstrated that stable NEDD4 knockdown in A549 lung cancer cells
inhibited starvation- and rapamycin-induced autophagy, thereby
reducing LC3-II levels. In addition, NEDD4 directly interacted with
LC3. These results indicated that NEDD4 is not only a positive
regulator of autophagy but is also essential for proper execution
of the autophagic process.
A potential mechanism underlying NEDD4-mediated
autophagy, was further investigated. mTOR is a well-established
negative regulator of autophagy, suppressing autophagy under
nutrient-rich conditions (42,43).
Moreover, mTOR is a crucial regulator of osteogenic differentiation
in mesenchymal stem cells such as stem cells from apical papilla,
BMSCs, and periodontal ligament stem cells (44-47).
In the experiments of the present study, EBSS-induced autophagy led
to increased expression of NEDD4 and autophagy-related proteins
(LC3-II and Beclin-1), along with a notable reduction in p-mTOR,
indicating suppressed mTOR activity and enhanced autophagy. When
NEDD4 was knocked down, p-mTOR levels were restored (signifying
reactivation of mTOR), and autophagy (LC3-II and Beclin-1 levels)
was inhibited. This suggests that NEDD4 may activate the autophagy
program by inhibiting the mTOR signaling pathway. This
interpretation is consistent with the well-known role of mTOR in
autophagy regulation (30). In our
prior research, it was observed that increased expression of
osteogenic differentiation markers in BMSCs was associated with
improved bone formation capacity (25). Other research indicates a potential
link between suppressed autophagy and enhanced osteogenesis,
although the exact mechanism remains to be explored (48). Thus, the present study identified
NEDD4 as an upstream regulator of mTOR, offering novel insights
into the molecular mechanisms connecting BMSC autophagy and bone
remodeling.
In addition, it is acknowledged that further
mechanistic validation is needed. Only nutrient starvation was
employed to induce autophagy. Using specific autophagy modulators
(such as rapamycin to inhibit mTOR or chloroquine to block
autophagy flux) could strengthen the conclusions. In future
studies, such modulators are planned to be used to further verify
the role of NEDD4. Moreover, the present study design did not
include a ‘rescue’ experiment (restoring NEDD4 expression after
knockdown) to fulfill molecular causation criteria, nor were assays
such as co-immunoprecipitation performed to test for direct
interactions (for example between NEDD4 and components of the mTOR
pathway) or ubiquitination assays conducted to confirm the E3
ligase activity of NEDD4 on autophagy-related substrates. These
experiments are beyond the scope of the present study and will be
addressed in future research. Nevertheless, by combining our
results with previous findings that NEDD4 promotes autophagy in
other cell types (27), compelling
evidence that NEDD4 acts as an upstream regulator of autophagy in
BMSCs is provided. As a further step, ALP staining and
mineralization assays (such as Alizarin Red S staining) could be
employed in future work to directly evaluate how changes in NEDD4
expression or autophagy levels affect the osteogenic
differentiation of BMSCs. These follow-up studies will help clarify
the relationship between autophagy induced by NEDD4 and
osteogenesis.
In summary, the present study revealed an important
role for NEDD4 in regulating autophagy in BMSCs. By establishing
in vitro BMSC models with NEDD4 knockdown, it was
demonstrated that NEDD4 depletion impairs autophagy. The limited
osteogenic differentiation of BMSCs is a key factor in reduced bone
formation; the results of the present study suggest that NEDD4 (via
autophagy activation through mTOR inhibition) could potentially
enhance the osteogenic capacity of BMSCs. This positions NEDD4 as a
promising therapeutic target for conditions such as osteoporosis.
Future studies will further investigate the interplay between
NEDD4-regulated autophagy and BMSC osteogenesis in vivo.
Acknowledgements
Not applicable.
Funding
Funding: The present study was funded by a grant from the Luzhou
Science and Technology Plan Project (grant no. 2020-JYJ-40), the
Natural Science Foundation of Southwest Medical University (grant
no. 2021ZKMS020), the Tutor Group Capacity Improvement Project of
The Affiliated Stomatological Hospital of Southwest Medical
University (grant no. 2022DS16), and the Sichuan College Students
Innovation and Entrepreneurship Training Program (grant no.
S202210632170).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors contributions
CL and FL conceived and designed the study. CL and
XY carried out the methodology design and performed data analysis
using software. SZ and XX conducted the formal analysis. BL was
responsible for data curation and investigation. CL and XY wrote
the original draft. FL and XX reviewed and edited the manuscript.
CL and FL confirm the authenticity of all the raw data. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Ethical approval for the animal experiments was
granted by the Ethics Committee of Southwest Medical University,
Luzhou, Sichuan, China (approval no. 20221206-005). All animal
procedures were conducted in accordance with institutional
guidelines, the National Research Council s Guide for the Care
and Use of Laboratory Animals (8th ed.), and the AVMA Guidelines
for the Euthanasia of Animals (2020).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wang L, You X, Zhang L, Zhang C and Zou W:
Mechanical regulation of bone remodeling. Bone Res.
10(16)2022.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zhang G, Kang Y, Dong J, Shi D, Ziang Y,
Gao H, Lin Z, Wei X, Ding R, Fan B, et al: Fluffy hybrid
nanoadjuvants for reversing the imbalance of osteoclastic and
osteogenic niches in osteoporosis. Bioact Mater. 39:354–374.
2024.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Liu F, Yuan L, Li L, Yang J, Liu J, Chen
Y, Zhang J, Lu Y, Yuan Y and Cheng J: S-sulfhydration of SIRT3
combats BMSC senescence and ameliorates osteoporosis via
stabilizing heterochromatic and mitochondrial homeostasis.
Pharmacol Res. 192(106788)2023.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Trompet D, Melis S, Chagin AS and Maes C:
Skeletal stem and progenitor cells in bone development and repair.
J Bone Miner Res. 39:633–654. 2024.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Shahnaser S, Sheikhi M, Hashemibeni B,
Mousavi AS and Soltani P: Comparison of autogenous bone graft and
tissue-engineered bone graft in alveolar cleft defects in canine
animal models using digital radiography. Indian J Dent Res.
31:118–123. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zhang L, Wang P, Mei S, Li C, Cai C and
Ding Y: In vivo alveolar bone regeneration by bone marrow stem
cells/fibrin glue composition. Arch Oral Biol. 57:238–244.
2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Mizushima N and Komatsu M: Autophagy:
Renovation of cells and tissues. Cell. 147:728–741. 2011.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Mizushima N and Levine B: Autophagy in
human diseases. N Engl J Med. 383:1564–1576. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Klionsky DJ, Petroni G, Amaravadi RK,
Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cadwell K,
Cecconi F, Choi AMK, et al: Autophagy in major human diseases. EMBO
J. 40(e108863)2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Amaravadi RK, Kimmelman AC and White E:
Recent insights into the function of autophagy in cancer. Genes
Dev. 30:1913–1930. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Menzies FM, Fleming A and Rubinsztein DC:
Compromised autophagy and neurodegenerative diseases. Nat Rev
Neurosci. 16:345–357. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
Lavandero S, Chiong M, Rothermel BA and
Hill JA: Autophagy in cardiovascular biology. J Clin Invest.
125:55–64. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
13
|
Jung HS, Chung KW, Won Kim J, Kim J,
Komatsu M, Tanaka K, Nguyen YH, Kang TM, Yoon KH, Kim JW, et al:
Loss of autophagy diminishes pancreatic beta cell mass and function
with resultant hyperglycemia. Cell Metab. 8:318–324.
2008.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Huang J and Brumell JH: Autophagy in
immunity against intracellular bacteria. Curr Top Microbiol
Immunol. 335:189–215. 2009.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Su Z, Chen D, Huang J, Liang Z, Ren W,
Zhang Z, Jiang Q, Luo T and Guo L: Isoliquiritin treatment of
osteoporosis by promoting osteogenic differentiation and autophagy
of bone marrow mesenchymal stem cells. Phytother Res. 38:214–230.
2024.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Zeng C, Wang S, Chen F, Wang Z, Li J, Xie
Z, Ma M, Wang P, Shen H and Wu Y: Alpinetin alleviates osteoporosis
by promoting osteogenic differentiation in BMSCs by triggering
autophagy via PKA/mTOR/ULK1 signaling. Phytother Res. 37:252–270.
2023.PubMed/NCBI View
Article : Google Scholar
|
|
17
|
Liu Y, Lin S, Xu Z, Wu Y, Wang G, Yang G,
Cao L, Chang H, Zhou M and Jiang X: High-performance
hydrogel-encapsulated engineered exosomes for supporting
endoplasmic reticulum homeostasis and boosting diabetic bone
regeneration. Adv Sci (Weinh). 11(e2309491)2024.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang M, Zhang L, Lin F, Zheng Q, Xu X and
Mei L: Dynamic study into autophagy and apoptosis during
orthodontic tooth movement. Exp Ther Med. 21(430)2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Xu HM and Hu F: The role of autophagy and
mitophagy in cancers. Arch Physiol Biochem. 128:281–289.
2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ceccariglia S, Cargnoni A, Silini AR and
Parolini O: Autophagy: A potential key contributor to the
therapeutic action of mesenchymal stem cells. Autophagy. 16:28–37.
2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Ingham RJ, Gish G and Pawson T: The Nedd4
family of E3 ubiquitin ligases: Functional diversity within a
common modular architecture. Oncogene. 23:1972–1984.
2004.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wiszniak S, Harvey N and Schwarz Q: Cell
autonomous roles of Nedd4 in craniofacial bone formation. Dev Biol.
410:98–107. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wiszniak S, Kabbara S, Lumb R, Scherer M,
Secker G, Harvey N, Kumar S and Schwarz Q: The ubiquitin ligase
Nedd4 regulates craniofacial development by promoting cranial
neural crest cell survival and stem-cell like properties. Dev Biol.
383:186–200. 2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Jeon SA, Lee JH, Kim DW and Cho JY:
E3-ubiquitin ligase NEDD4 enhances bone formation by removing
TGFβ1-induced pSMAD1 in immature osteoblast. Bone. 116:248–258.
2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Li B, Zhang S, Yun X, Liu C, Xiao R, Lu M,
Xu X and Lin F: NEDD4 s effect on osteoblastogenesis potential
of bone mesenchymal stem cells in rats concerned with PI3K/Akt
pathway. Differentiation. 141(100830)2025.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Luo M, Ye L, Chang R, Ye Y, Zhang Z, Liu
C, Li S, Jing Y, Ruan H, Zhang G, et al: Multi-omics
characterization of autophagy-related molecular features for
therapeutic targeting of autophagy. Nat Commun.
13(6345)2022.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Sun A, Wei J, Childress C, Shaw JH IV,
Peng K, Shao G, Yang W and Lin Q: The E3 ubiquitin ligase NEDD4 is
an LC3-interactive protein and regulates autophagy. Autophagy.
13:522–537. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Xie W, Jin S and Cui J: The NEDD4-USP13
axis facilitates autophagy via deubiquitinating PIK3C3. Autophagy.
16:1150–1151. 2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Xie W, Jin S, Wu Y, Xian H, Tian S, Lie
DA, Guo Z and Cui J: Auto-ubiquitination of NEDD4-1 Recruits USP13
to facilitate autophagy through deubiquitinating VPS34. Cell Rep.
30:2807–2819.e4. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Li Y, Zhang L, Zhou J, Luo S, Huang R,
Zhao C and Diao A: Nedd4 E3 ubiquitin ligase promotes cell
proliferation and autophagy. Cell Prolif. 48:338–347.
2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Farahzadi R, Fathi E, Mesbah-Namin SA and
Vietor I: Granulocyte differentiation of rat bone marrow resident
C-kit+ hematopoietic stem cells induced by mesenchymal
stem cells could be considered as new option in cell-based therapy.
Regen Ther. 23:94–101. 2023.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Mizushima N, Yoshimori T and Levine B:
Methods in mammalian autophagy research. Cell. 140:313–326.
2010.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Klionsky DJ, Abdel-Aziz AK, Abdelfatah S,
Abdellatif M, Abdoli A, Abel S, Abeliovich H, Abildgaard MH, Abudu
YP, Acevedo-Arozena A, et al: Guidelines for the use and
interpretation of assays for monitoring autophagy (4th
edition)1. Autophagy. 17:1–382. 2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Farahzadi R, Valipour B, Anakok OF, Fathi
E and Montazersaheb S: The effects of encapsulation on NK cell
differentiation potency of C-kit-hematopoietic stem cells via
identifying cytokine profiles. Transpl Immunol.
77(101797)2023.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Vicente GP, Della Salda L and Strefezzi
RF: Beclin-1 and LC3B expression in canine mast cell tumours: An
immuno-ultrastructural and immunohistochemical study of autophagy.
Vet Q. 44:1–15. 2024.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Su S, Wu Y, Wang D and Hai J: Inhibition
of excessive autophagy and mitophagy mediates neuroprotective
effects of URB597 against chronic cerebral hypoperfusion. Cell
Death Dis. 9(733)2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Biswas U, Roy R, Ghosh S and Chakrabarti
G: The interplay between autophagy and apoptosis: Its implication
in lung cancer and therapeutics. Cancer Lett.
585(216662)2024.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Remadevi V, Jaikumar VS, Vini R,
Krishnendhu B, Azeez JM, Sundaram S and Sreeja S: Urolithin A,
induces apoptosis and autophagy crosstalk in oral squamous cell
carcinoma via mTOR/AKT/ERK1/2 pathway. Phytomedicine.
130(155721)2024.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zhu Y, Wang H, Wang J, Han S, Zhang Y, Ma
M, Zhu Q, Zhang K and Yin H: Zearalenone Induces apoptosis and
cytoprotective autophagy in chicken granulosa cells by
PI3K-AKT-mTOR and MAPK signaling pathways. Toxins (Basel).
13(199)2021.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Li S, Xu B, Luo Y, Luo J, Huang S and Guo
X: Autophagy and apoptosis in rabies virus replication. Cells.
13(183)2024.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Kim J, Kundu M, Viollet B and Guan KL:
AMPK and mTOR regulate autophagy through direct phosphorylation of
Ulk1. Nat Cell Biol. 13:132–141. 2011.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhang Y, Vasheghani F, Li YH, Blati M,
Simeone K, Fahmi H, Lussier B, Roughley P, Lagares D, Pelletier JP,
et al: Cartilage-specific deletion of mTOR upregulates autophagy
and protects mice from osteoarthritis. Ann Rheum Dis. 74:1432–1440.
2015.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Chen M, Jing D, Ye R, Yi J and Zhao Z:
PPARβ/δ accelerates bone regeneration in diabetic mellitus by
enhancing AMPK/mTOR pathway-mediated autophagy. Stem Cell Res Ther.
12(566)2021.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Tanaka Y, Sonoda S, Yamaza H, Murata S,
Nishida K, Hama S, Kyumoto-Nakamura Y, Uehara N, Nonaka K, Kukita T
and Yamaza T: Suppression of AKT-mTOR signal pathway enhances
osteogenic/dentinogenic capacity of stem cells from apical papilla.
Stem Cell Res Ther. 9(334)2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Zhao X, Sun W, Guo B and Cui L: Circular
RNA BIRC6 depletion promotes osteogenic differentiation of
periodontal ligament stem cells via the miR-543/PTEN/PI3K/AKT/mTOR
signaling pathway in the inflammatory microenvironment. Stem Cell
Res Ther. 13(417)2022.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Jiang Z, Huang H, Luo L and Jiang B: The
role of autophagy on osteogenesis of dental follicle cells under
inflammatory microenvironment. Oral Dis. 31:928–940.
2025.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Zheng J, Gao Y, Lin H, Yuan C and Zhi K:
Corrigendum to ‘Enhanced autophagy suppresses inflammation-mediated
bone loss through ROCK1 signaling in bone marrow mesenchymal stem
cells’ [Cells Dev 167 (2021) 203687]. Cells Dev.
176(203867)2023.PubMed/NCBI View Article : Google Scholar
|