Introduction
Giant congenital melanocytic nevi (GCMN) are rare
pigmented skin lesions present at birth, often enlarging to 20 cm
in adulthood (1,2). Their development results from an
abnormal migration and proliferation of neural crest-derived
melanoblasts, which may involve not only the skin but also deeper
structures such as leptomeninges (1). Individuals with GCMN face a higher
lifetime risk of malignant melanoma and neurocutaneous melanosis
(1,2). Modern surveillance combines clinical
examination with total body mapping and artificial intelligence
(AI)-assisted dermoscopy. These technologies allow sensitive
detection of morphological changes and objective comparison over
time, thus facilitating early recognition of malignant
transformation (3,4). Alongside imaging, genetic
predisposition strongly influences melanoma risk. The melanocortin
1 receptor (MC1R) gene is central in regulating pigmentation, and
its high-risk variants are well documented as low- to
moderate-penetrance melanoma susceptibility alleles. Identifying
such variants may explain individual nevus burden and guide
tailored follow-up. The aim of the present case report was to
present a patient with a bathing-trunk-type GCMN who underwent
integrative follow-up with AI-enhanced dermoscopy, histopathology,
and germline genetic analysis. The case illustrates how combining
clinical and molecular tools supports personalized surveillance in
high-risk patients (5,6).
Case report
Patient
A 55-year-old woman was referred for dermatologic
evaluation due to a bathing-trunk type GCMN and multiple additional
nevi distributed over the trunk and extremities. The present study
was approved (approval no.755/19.12.2023) by the Ethics Commission
of the Medical University-Pleven, (Pleven Bulgaria). Written
informed consent for participation and publication was obtained
from the patient in the present study.
Dermatological еvaluation
Cutaneous examination was performed with total body
mapping using an AI-assisted dermoscopy system (MoleAnalyzer Pro
version 3.5.6.0; FotoFinder Systems GmbH). The software applies
convolutional neural networks trained on large annotated image
datasets to identify and score melanocytic lesions according to
asymmetry, border, color, and structure. Sequential imaging allows
objective detection of new or evolving lesions.
Germline pathogenic variant
detection
After informed consent, peripheral blood was
collected in EDTA tubes. DNA extraction was performed with the
MagCore® Genomic DNA Whole Blood Kit (cat. no.
MGB400-04; RBC Bioscience Corp.). Genomic DNA integrity was
assessed using an Agilent 2200 TapeStation system with Genomic DNA
ScreenTape (Agilent Technologies, Inc.), following the
manufacturer's instructions. Concentration and purity were
evaluated using the Qubit 4 Fluorometer and NanoDrop One
spectrophotometer (both from Thermo Fisher Scientific, Inc.).
Genetic analysis was performed with massive parallel
next-generation sequencing (NGS). Library preparation was carried
out using the TruSight One Expanded Panel (6699 genes; Illumina,
Inc.), followed by paired-end sequencing (2x150 bp) on the Illumina
NextSeq 550 High Output Kit v2.5 (300 cycles; cat. no. 20024907;
Illumina Inc.). Bioinformatic processing included alignment to hg19
and generation of gVCF files. The mean sequencing depth across all
targeted regions was 256. Coverage analysis demonstrated that ≥99%
of target regions were covered at ≥20x. Variants were filtered
using custom thresholds: Minimum depth ≥20x, variant allele
frequency (VAF) ≥0.20, exclusion of synonymous variants, and
prioritization of nonsynonymous, splicing, and frameshift changes.
Variant annotation used ClinVar, dbSNP, and Ensembl databases.
Classification followed the ACMG five-tier framework (7).
Histopathology
Excised lesions were fixed in 10% neutral buffered
formalin (Sigma-Aldrich; Merck KGaA) for 12-24 h at room
temperature, paraffin-embedded, and sectioned at 3-4 µm. Sections
were stained with hematoxylin and eosin following standard
histopathological procedures. Hematoxylin staining was performed at
room temperature for 4-6 min, followed by eosin staining at room
temperature for 1-2 min. Hematoxylin (Mayer's hematoxylin; Merck
KGaA) and eosin (Eosin Y; Merck KGaA) were used according to the
manufacturer's protocols. Formalin-fixed, paraffin-embedded tissue
sections (4 µm) were deparaffinized in xylene, rehydrated through a
descending ethanol series, and subjected to heat-induced antigen
retrieval in citrate buffer (pH 6.0) at 95-98˚C for 20 min.
Endogenous peroxidase activity was quenched using 3% hydrogen
peroxide for 10 min, followed by blocking with 5% normal goat serum
(Vector Laboratories, Inc.) for 20 min at room temperature. Primary
antibodies were applied under the following conditions: S100
(1:400; overnight at 4˚C; cat. no. IR504; Dako; Agilent
Technologies, Inc.); SOX10 (1:200; overnight at 4˚C; cat. no.
A-1028, Cell Marque/Merck KGaA); Melan-A (1:100; 1 h at room
temperature; code no. M7196; Dako; Agilent Technologies, Inc.)
Sections were incubated with the EnVision+ System HRP secondary
antibody (cat. no. K4001; Dako; Agilent Technologies, Inc.) for 30
min at room temperature, and chromogenic detection was performed
using DAB+ (code no. K3468; Dako; Agilent Technologies, Inc.).
Slides were counterstained with Mayer's hematoxylin (Merck KGaA) at
room temperature (20-25˚C) for 1-2 min, dehydrated through graded
alcohols, cleared in xylene, and mounted. Immunoreactivity was
evaluated using a bright-field microscope (Olympus BX43; Olympus
Corporation) at a magnification of x100-x400.
Clinical presentation and
histopathological findings
The female patient presented with a large
bathing-trunk-type GCMN, accompanied by over 30 satellite nevi
(0.5-7 cm) across the trunk and limbs. Neurological examinations,
performed due to prior seizure episodes, revealed no structural
abnormalities. In 2023 (at the age of 54 years), three pigmented
tumors (total diameter 28 cm) in the lumbar region were surgically
excised. Histology revealed a multinodular plexiform melanocytic
schwannoma composed of spindle-shaped melanocytes, extending into
subcutaneous tissue, with low mitotic activity.
Immunohistochemistry showed diffuse S100 and SOX10 positivity,
focal Melan A expression, and a low Ki-67 index. These markers
supported schwannoma with melanocytic differentiation rather than
malignant melanoma.
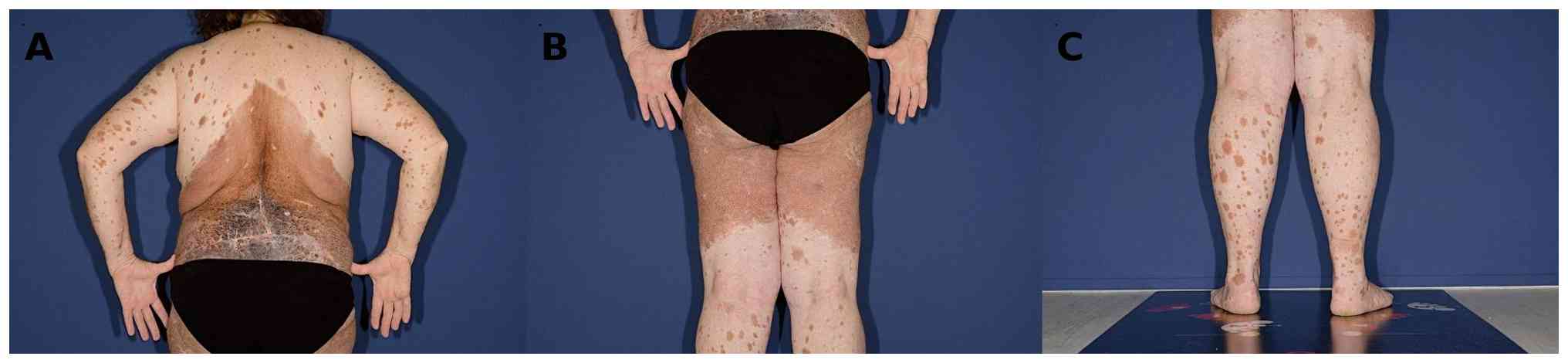
Dermatological status
Cutaneous examination revealed extensive pigmentary
changes involving the face, trunk, and both upper and lower
extremities. The findings included a symmetrically distributed
bathing-trunk-type GCMN, accompanied by multiple satellite nevi
ranging from 0.5 to 7 cm in diameter. In the sacral region, a
linear, cross-shaped atrophic scar was noted, consistent with the
site of prior surgical excision. The scar was surrounded by
numerous dark brown to black colored macules. (Fig. 1A-C) The dermatological examination
with a Total body mapping and AI dermoscopy device (MoleAnalyzer
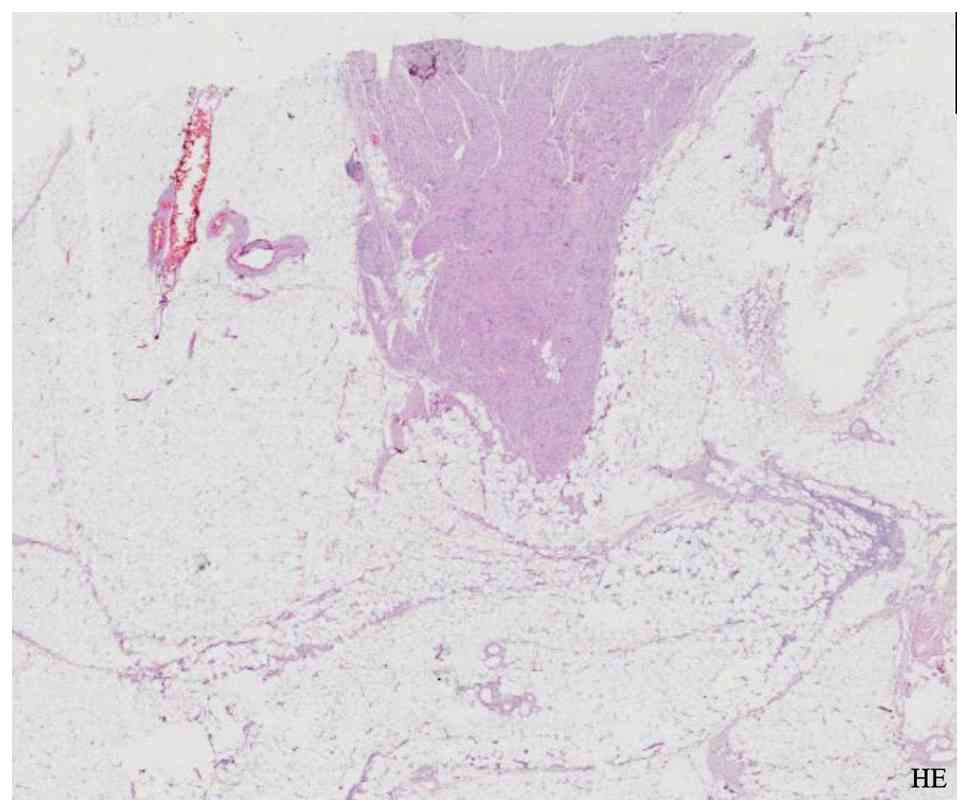
Pro) did not identify melanoma-like evolution. Histopathological
analysis of the excised multinodular, plexiform, non-encapsulated
lesion demonstrated a proliferation of oval and spindle-shaped
cells extending from the epidermis through the dermis and into the
deep subcutaneous adipose tissue, as visualized with hematoxylin
and eosin staining (Fig. 2).
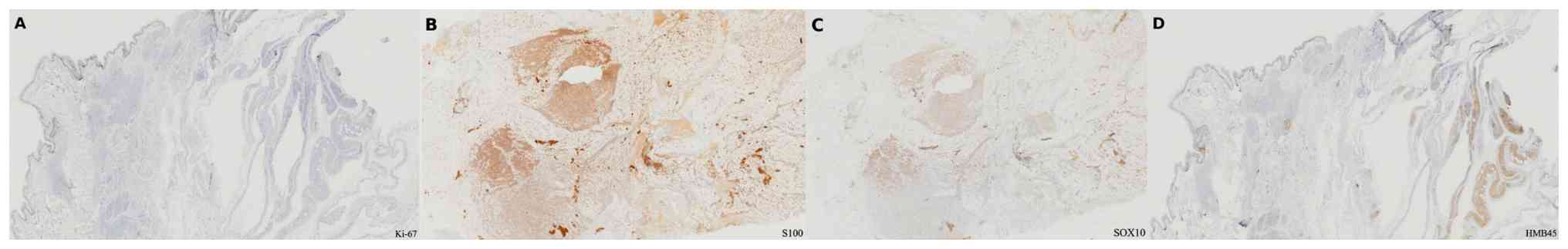
Immunohistochemistry showed diffuse S100 and SOX10 positivity,
focal Melan A expression, and a low Ki-67 index (Fig. 3A-D). These markers supported
schwannoma with melanocytic differentiation rather than malignant
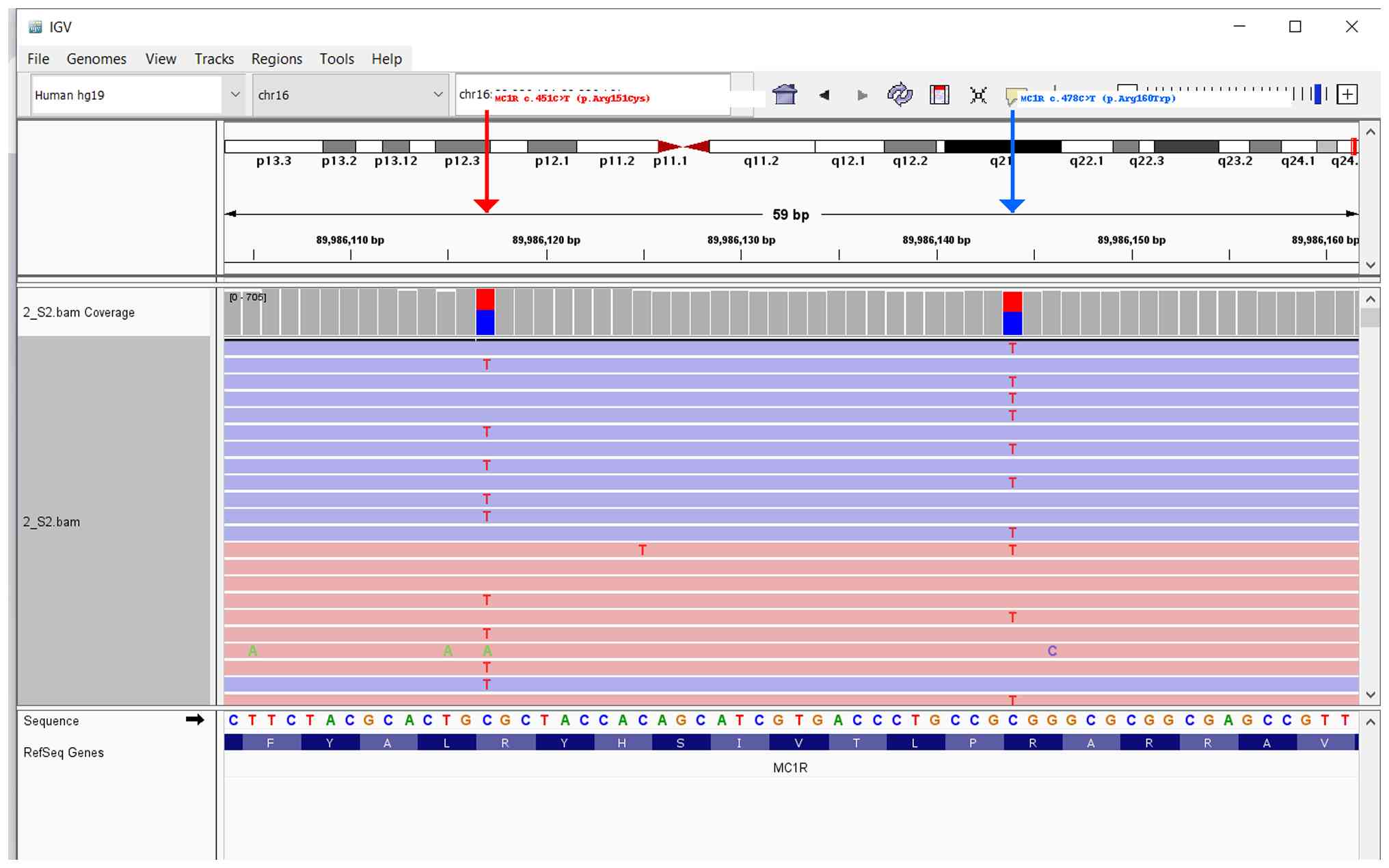
melanoma. NGS analysis identified two pathogenic MC1R variants:
c.451C>T (p.Arg151Cys) and c.478C>T (p.Arg160Trp), both in
compound heterozygous state (Fig.
4). No pathogenic variants were detected in NRAS or BRAF. No
additional high-risk variants were identified across the panel.
Discussion
GCMN represent rare developmental anomalies of
melanoblast migration and proliferation, with a reported melanoma
risk of ~5% over a lifetime (8,9). Early
recognition of malignant transformation is essential. These
developmental abnormalities are frequently linked to somatic
mutations in genes such as NRAS (particularly Q61 variants) and
BRAF (notably V600 mutations), as well as dysregulation of pathways
involving hepatocyte growth factor/scatter factor (10).
Various classification systems exist for CMN, with
the most widely accepted being that of Kopf et al (11), which categorizes lesions by maximum
diameter: Small (<1.5 cm), medium (1.5-19.9 cm), and large or
giant CMN (>20 cm) . The overall incidence of CMN in neonates
ranges from 0.2 to 2.1%, with a slight female predominance reported
(male-to-female ratios between 1:1.17 and 1:1.4) (12). Data from the Giant Congenital
Melanocytic Nevus Registry at the Federal University of Minas
Gerais, based on patient records between 1999 and 2011, estimated
the lifetime risk of melanoma in individuals with GCMN to be ~5%
(13). Anatomical distribution
patterns showed a predominance on the trunk (68.4%), followed by
the head and neck (17.5%), and the extremities (14.1%) (13).
Total body mapping involves systematic photographic
documentation of the entire skin surface, followed by digital
dermoscopic analysis of selected melanocytic lesions. This approach
facilitates longitudinal comparison, enabling the detection of
subtle morphological changes or the emergence of new lesions over
time (14). In the present case,
total body mapping combined with AI-enhanced dermoscopy was
employed, offering a sensitive and objective method for
surveillance. Regular imaging at 6 to 12-month intervals is
especially beneficial in individuals with GCMN, where early
recognition of malignant transformation is critical.
Surgical management remains a topic of ongoing
discussion. A systematic review by Krengel et al (12), analyzing 49 reported cases of
CMN-associated melanoma, found that in 67% of patients the
malignant transformation occurred within the congenital lesion
itself . Based on such findings, prophylactic surgical excision is
sometimes advocated, particularly in large or atypical nevi. While
the impact of surgery on melanoma risk reduction remains
inconclusive, the procedure often yields aesthetic and functional
improvements, which are important considerations for both patients
and families when deciding on treatment.
The histological diagnosis of plexiform melanocytic
schwannoma required careful differentiation from melanoma. Positive
staining for S100 and SOX10 confirmed Schwann cell origin, while
focal Melan A expression reflected melanocytic differentiation.
Similar immunoprofiles have been reported in rare schwannoma
variants (15). Although an
association between MC1R and schwannomas was not established, the
findings of the present study raise the question whether
pigmentation genes may modulate tumor phenotype, a hypothesis
warranting further investigation. Ranson et al (16) described a case involving a
66-year-old man with a tumor sharing similar histological and
immunophenotypic characteristics, including co-expression of S100,
SOX10, and HMB45, consistent with the findings in the present
case.
Pigmentation traits such as red hair, fair skin,
freckling, and poor tanning capacity have long been recognized as
genetically driven risk factors for both melanoma and
non-melanocytic skin cancers particularly when combined with
environmental exposure to ultraviolet radiation (17). MC1R, a G protein-coupled receptor
expressed in cutaneous melanocytes located in the basal epidermal
layer, plays a central role in regulating pigment production
(18,19). Genetic variation within the MC1R
gene contributes substantially to interindividual differences in
skin phototype and pigmentation phenotype.
MC1R variants are typically categorized into two
groups: Low-penetrance alleles (denoted as ‘r’) and high-penetrance
alleles (‘R’) in relation to their association with red hair color
and melanoma risk (20,21). Common ‘r’ alleles include Val60Leu,
Val92Met, and Arg163Gln, while ‘R’ alleles, such as Asp84Glu,
Arg142His, Arg151Cys, Ile155Thr, Arg160Trp, and Asp294His, are more
strongly associated with phenotypic traits and increased
susceptibility to skin malignancies (20).
In the current case, genetic analysis identified two
high-risk MC1R alleles (Arg151Cys and Arg160Trp), both categorized
as ‘R’ variants. Compound heterozygosity for R alleles is linked to
fair phototype, high nevus count, and increased melanoma
susceptibility. Both variants have been shown to exert
dominant-negative effects by impairing MC1R receptor trafficking
and downstream cAMP signaling (22). This genotype aligns with findings by
van der Poel et al (23),
who demonstrated a positive association between the ‘R/R’ genotype
and increased total nevus count. Consistently, the patient in the
present case had >30 nevi and a large back-located GCMN. Further
supporting this association, a more recent study involving 175
healthy carriers of MC1R variants reported that individuals with
‘R’ alleles tend to develop larger nevi, particularly on the back
and upper limbs. Moreover, their nevi often displayed dermoscopic
features such as visible vascular structures, globules, and
pigmented spots (24). These
features were also observed in the present case, reinforcing the
phenotypic impact of the compound ‘R’ genotype on nevus morphology
and distribution. The limitations of the present case report
include its basis in a single patient, the absence of functional
validation or familial segregation studies, and the analysis of
only germline DNA; somatic alterations in lesional tissue remain
unknown. Despite these limitations, the case illustrated the
integrative value of combining clinical imaging and genomic data.
In conclusion the present case emphasized that personalized
surveillance of patients with GCMN benefits from integration of
AI-assisted dermoscopy, histopathology, and germline genetics. The
compound heterozygous MC1R variants may explain the nevus burden of
the patient and justified intensified follow-up. AI technology
allowed precise, noninvasive monitoring, while histology ruled out
melanoma and guided management. An integrative, multidisciplinary
approach should be considered standard for high-risk GCMN patients,
as it enhances early detection and supports informed clinical
decisions.
Acknowledgements
Not applicable.
Funding
Funding: The present study was financed by the European
Union-NextGenerationEU, through the National Recovery and
Resilience Plan of the Republic of Bulgaria, project no.
BG-RRP-2.004-0003, and supported by Project
BG16RFPR002-1.014-0002-С001, ‘Centre of Competence in Personalized
Medicine, 3D and Telemedicine, Robotic-Assisted and Minimally
Invasive Surgery,’ funded by PRIDST 2021-2027 and co-funded by the
EU.
Availability of data and materials
The data generated in the present study may be found
in ClinVar under accession numbers SUB15686128 and SUB15686145.
Authors' contributions
PV recruited the patients. IY, PV, ZK and SP
collected clinical and biological data. ZK performed the molecular
analysis. PV and IY were responsible for diagnosis and treatment of
the patient. ZK analyzed the data. PV, ZK, SP and IY were involved
in the writing and revision of the manuscript. SP and IY reviewed
and revised the manuscript. All authors read and approved the final
manuscript. ZK and PV confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
The present study was approved (approval
no.755/19.12.2023) by the Ethics Commission of the Medical
University-Pleven, (Pleven Bulgaria). Written informed consent for
participation was obtained from the patient.
Patient consent for publication
Written informed consent for publication was
obtained from the patient.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kinsler VA, O'Hare P, Bulstrode N, Calonje
JE, Chong WK, Hargrave D, Jacques T, Lomas D, Sebire NJ and Slater
O: Melanoma in congenital melanocytic naevi. Br J Dermatol.
176:1131–1143. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Krengel S, Scope A, Dusza SW, Vonthein R
and Marghoob AA: New recommendations for the categorization of
cutaneous features of congenital melanocytic nevi. J Am Acad
Dermatol. 68:441–451. 2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Marghoob AA, Swindle LD, Moricz CZ,
Sanchez Negron FA, Slue B, Halpern AC and Kopf AW: Instruments and
new technologies for the in vivo diagnosis of melanoma. J Am Acad
Dermatol. 49:777–799. 2003.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Haenssle HA, Fink C, Schneiderbauer R,
Toberer F, Buhl T, Blum A, Kalloo A, Hassen ABH, Thomas L, Enk A,
et al: Man against machine: Diagnostic performance of a deep
learning convolutional neural network for dermoscopic melanoma
recognition in comparison to 58 dermatologists. Ann Oncol.
29:1836–1842. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Raimondi S, Sera F, Gandini S, Iodice S,
Caini S, Maisonneuve P and Fargnoli MC: MC1R variants, melanoma and
red hair color phenotype: A meta-analysis. Int J Cancer.
122:2753–2760. 2008.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Tagliabue E, Gandini S, Bellocco R,
Maisonneuve P, Newton-Bishop J, Polsky D, Lazovich D, Kanetsky PA,
Ghiorzo P, Gruis NA, et al: MC1R variants as melanoma risk factors
independent of at-risk phenotypic characteristics: a pooled
analysis from the M-SKIP project. Cancer Manag Res. 10:1143–1154.
2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Richards S, Aziz N, Bale S, Bick D, Das S,
Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al:
Standards and guidelines for the interpretation of sequence
variants: A joint consensus recommendation of the American college
of medical genetics and genomics and the association for molecular
pathology. Genet Med. 17:405–424. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Viana AC, Gontijo B and Bittencourt FV:
Giant congenital melanocytic nevus. An Bras Dermatol. 88:863–878.
2013.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Arneja JS and Gosain AK: Giant congenital
melanocytic nevi of the trunk and an algorithm for treatment. J
Craniofac Surg. 16:886–893. 2005.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Meshram GG, Kaur N and Hura KS: Giant
congenital melanocytic nevi: An update and emerging therapies. Case
Rep Dermatol. 10:24–28. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kopf AW, Bart RS and Hennessey P:
Congenital nevocytic nevi and malignant melanomas. J Am Acad
Dermatol. 1:123–130. 1979.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Krengel S, Hauschild A and Schäfer T:
Melanoma risk in congenital melanocytic naevi: a systematic review.
Br J Dermatol. 155:1–8. 2006.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Viana ACL, Goulart EMA, Gontijo B and
Bittencourt FV: A prospective study of patients with large
congenital melanocytic nevi and the risk of melanoma. An Bras
Dermatol. 92:200–205. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Barcaui C, Bakos RM, Paschoal FM,
Bittencourt FV, de Sá BCS and Miot HA: Total body mapping in the
follow-up of melanocytic lesions: Recommendations of the Brazilian
society of dermatology. An Bras Dermatol. 96:472–476.
2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Torres-Mora J, Dry S, Li X, Binder S, Amin
M and Folpe AL: Malignant melanotic schwannian tumor: a
clinicopathologic, immunohistochemical, and gene expression
profiling study of 40 cases, with a proposal for the
reclassification of "melanotic schwannoma". Am J Surg Pathol.
38:94–105. 2014.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ranson M, Lai J, Van Brenk B, McCalmont TH
and Walsh NM: Plexiform melanocytic schwannoma: Report of a second
case and overview of a rare entity. Am J Dermatopathol. 44:943–947.
2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Sturm RA: Skin colour and skin
cancer-MC1R, the genetic link. Melanoma Res. 12:405–416.
2002.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Cai M and Hruby VJ: The melanocortin
receptor system: A target for multiple degenerative diseases. Curr
Protein Pept Sci. 17:488–96. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Roberts DW, Newton RA, Beaumont KA,
Leonard JH and Sturm RA: Quantitative analysis of MC1R gene
expression in human skin cell cultures. Pigment Cell Res. 19:76–89.
2006.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Beaumont KA, Liu YY and Sturm RA: The
melanocortin-1 receptor gene polymorphism and association with
human skin cancer. Prog Mol Biol Transl Sci. 88:85–153.
2009.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Pasquali E, García-Borrón JC, Fargnoli MC,
Gandini S, Maisonneuve P, Bagnardi V, Specchia C, Liu F, Kayser M,
Nijsten T, et al: MC1R variants increased the risk of sporadic
cutaneous melanoma in darker-pigmented Caucasians: A
pooled-analysis from the M-SKIP project. Int J Cancer. 136:618–631.
2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Beaumont KA, Shekar SN, Newton RA, James
MR, Stow JL, Duffy DL and Sturm RA: Receptor function, dominant
negative activity and phenotype correlations for MC1R variant
alleles. Hum Mol Genet. 16:2249–2260. 2007.PubMed/NCBI View Article : Google Scholar
|
|
23
|
van der Poel LAJ, Bergman W, Gruis NA and
Kukutsch NA: The role of MC1R gene variants and phenotypical
features in predicting high nevus count. Melanoma Res. 30:511–514.
2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Vallone MG, Tell-Marti G, Potrony M,
Rebollo-Morell A, Badenas C, Puig-Butille JA, Gimenez-Xavier P,
Carrera C, Malvehy J and Puig S: Melanocortin 1 receptor (MC1R)
polymorphisms' influence on size and dermoscopic features of nevi.
Pigment Cell Melanoma Res. 31:39–50. 2018.PubMed/NCBI View Article : Google Scholar
|