Introduction
Botanical extracts are used for the prevention and
treatment of common conditions by 80% of the world’s population
according to estimates made by the World Health Organization
(1). Drugs are produced from
botanical sources by isolation and purification of the most active
ingredients, while other substances in the raw fraction are
separated. Synergistic interactions of mixtures of bioactive
constituents and related substances or analogs in plant extracts
have been proposed to occur with the most active ingredient. These
interactions help explain the improved effectiveness of extracts
containing multiple ingredients compared to drugs developed from
single constituents.
We previously studied the action and metabolism of
Chinese red yeast rice (Monascus purpureus Went) (CRYR), a
dietary supplement containing monacolins, one of which (Monacolin
K) is identical in structure to the statin drug lovastatin, and to
unsaturated fatty acids and phytosterols capable of lowering
low-density lipoprotein (LDL) cholesterol in humans (2–5). The
apparent bioactivity of CRYR containing 6 mg of Monacolin K was
equivalent to 20 mg of purified lovastatin. Moreover, CRYR is well
tolerated in patients who are intolerant to statin drugs (6), suggesting that the lower amounts of
active substances, enhanced action and more complete metabolism in
comparison to a drug made from a single constituent may prevent
adverse muscle side effects. Our group previously demonstrated
similar interactions for phytochemicals in cranberry, pomegranate
and green tea (7–9). In studies demonstrating synergistic
interactions of a mixture of herbs against prostate cancer by
isobolographic analysis, Rabdosia rubescens extract (RRE)
was found to be particularly active among five herbs in a PC-SPES
mixture (10). Purified oridonin
was shown to be active against a number of different types of
cancer (11), which motivated the
present study, designed to demonstrate the interaction of multiple
components in RRE in comparison to purified oridonin. In order to
understand the mechanism of the observed synergistic interactions,
we conducted gene microarray analysis of the nuclear factor-κB
(NF-κB) pathway, which is implicated in prostate carcinogenesis, in
order to identify differential gene expression as a result of
treatment with the plant extract compared to the most active
component of the plant extract.
Materials and methods
Preparation of Rabdosia rubescens
extract
An oridonin-enriched extract of Rabdosia
rubescens (Henan, China) from the aerial part of the plant was
standardized to 4% oridonin using methods established at the UCLA
Center for Human Nutrition. RRE was administered to animals at
doses based on the average amount of oridonin contained in a single
dosage of Donglingcao, a tablet currently used in China for human
consumption. The equivalent dose to that administered to a 70-kg
human was calculated to be 0.5 mg of oridonin for a mouse with a
body weight of 25 g. For RRE, which contains 4% oridonin, the dose
to be administered was calculated to deliver the same amount of
oridonin as above and was determined to be 10.4 mg per 25-g mouse.
Both oridonin and RRE were suspended in 200 μl of water with 1%
carboxymethylcellulose.
Cell culture
DU-145, CWR22Rv1, LNCaP and PC-3 prostate cancer
cells were purchased from the American Type Tissue Culture
Collection (Rockville, MD, USA) and maintained in RPMI-1640 medium
with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin
and 100 μg/ml streptomycin in a 5% CO2 atmosphere at
37°C. Confluent cells (70–80%) were treated with oridonin at 10–100
μg/ml for 48 h, dissolved in DMSO and mixed with complete cell
medium. The final concentration of DMSO used was 0.1%
(vol/vol).
Cell viability (MTT) assay
The effect of oridonin on the viability of cells was
determined based on the uptake of MTT
[3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] by
measuring the absorbance at 540 nm in a UV spectrophotometer. Cells
were plated at a density of 10,000 cells/well in 200 μl of complete
culture medium containing 10–100 μg/ml concentrations of oridonin
in 96-well microtiter plates for 48 h. After incubation for
specified times at 37°C in a humidified incubator, MTT (5 mg/ml in
PBS) was added to each well and the cells were incubated for 2 h,
after which the plate was centrifuged at 1,800 × g for 5 min at
4°C. The absorbance at 540 nm was measured with a microplate
reader. The effect of oridonin on growth inhibition was assessed as
the percentage of cell viability, where 0.1% DMSO-treated cells
were deemed 100% viable. DMSO at the concentrations used did not
affect cell viability.
In vivo tumor xenograft animal model
The UCLA Animal Research Committee approved all
animal experimental procedures. Male SCID mice (Taconic Farm,
Germantown, NY, USA) were bred in a pathogen-free colony and housed
in groups of 4 per cage under pathogen-free conditions with a 12-h
light-dark cycle. The animals were fed an autoclaved diet ad
libitum of sterilized food pellets and water. A total of 24
SCID mice (Taconic Farms) were injected subcutaneously at 5 weeks
of age with 2×105 androgen-dependent LAPC4 prostate
cancer cells (a gift from Charles Sawyers). Mice were divided into
four groups consisting of 6 animals each and were administered the
two doses of oridonin, RRE or water alone (control) by gavage 5
days/week for 4 weeks. Tumor size was measured with calipers three
times a week starting on day 7. After 4 weeks, the mice were
sacrificed, and both serum samples and tumor tissues were
harvested. Tumor volume was calculated by the formula 0.5238 × L1 ×
L2 × H, where L1 is the long diameter, L2 is the short diameter and
H is the height of the tumor.
Transfection and NF-κB luciferase
assay
Cells were transfected using Effectene transfection
reagent (Qiagen). SBE luciferase reporter gene plasmid was obtained
from Panomics Inc. (Fremont, CA, USA). SBE (SBE3X-Lux)
luciferase reporter constructs were used to monitor NF-κB
transactivation with a vector containing multiple repeat-specific
consensus binding sites. The Renilla luciferase vector pRL-CMV
(Promega) was co-transfected with the NF-κB reporter vector as a
control to assess transfection efficiency. Twelve hours after
transfection, cells were subjected to treatment with either
oridonin or RRE at various doses. Cells were then harvested, and
luminescence was measured in a Turner 20/20n single-tube
luminometer (Turner Biosystems, Sunnyvale, CA, USA).
RNA isolation and PCR arrays
Total RNA was isolated from 100 mg of tumor tissue
using TRI Reagent according to the manufacturer’s instructions. RNA
concentration and purity were measured using the NanoDrop ND-1000
Spectrophotometer (NanoDrop, Wilmington, DE, USA). RNA quality was
required to have absorbance ratios at 260 nM compared to 280 nM
ratios (nucleic acid/protein ratio) >2.0 and 260/230 nM ratios
(estimate of organic compound contamination) >2.0. RNA (5 μg)
was treated with the RT2Nano PreAmp cDNA Synthesis kit
(SA Biosciences, Frederick, MD, USA) according to the
manufacturer’s instructions in order to generate cDNA and
pre-amplify the cDNA template. Briefly, after genomic DNA
elimination, the reverse transcription reaction was performed at
42°C for 15 min and then heated at 95°C for 5 min to inactivate the
enzyme. The cDNA was then pre-amplified and mixed with
RT2 SYBR Green/ROX qPCR master mix (SA Biosciences).
Aliquots (25 μl) were loaded into each well of a Human NF-κB
Signaling PCR-array (catalog #PAHS-025; SA Biosciences) according
to the manufacturer’s instructions on a One Step Plus real-time PCR
machine (Applied Biosystems, Foster City, CA, USA). Conditions for
amplification were as follows: 1 cycle of 10 min at 95°C, followed
by 40 cycles of 15 sec at 95°C and 1 min at 60°C. A dissociation
curve from 65 to 95°C was performed on each plate immediately after
the PCR run to determine the quality of the specific products
amplified in each well. Dissociation curves with multiple peaks
were not included in the analysis.
The PCR array data were analyzed by the ΔΔCt method.
Genes with Ct values >35 cycles were considered as
non-detectable and assigned a value of 35. The average of two
housekeeping genes [glyceraldehyde-3-phosphate dehydrogenase
(GAPDH) and β-actin (ACTB)] was used to obtain the ΔCt value for
each gene of interest. The ΔΔCt value for each gene was calculated
by the difference between the ΔCt of the treated and the control
groups. The fold-change for each gene was calculated by
2ΔΔCt, and statistical analysis to determine differences
between treatments was performed using the RT2 Profiler
PCR Array Data Analysis web-based software from the SA Bioscience
website (http://www.sabiosciences.com/pcrarraydataanalysis.php).
Statistical methods
For the cell proliferation assays, data were
expressed as a percentage of untreated cells (i.e., treatment
value-blank/vehicle value-blank) with the mean ± SE of at least
three separate experiments. Statistical methods included ANOVA and
Fisher post-hoc analysis. For PCR arrays, gene expression
was calculated as fold-change relative to the average expression in
the vehicle control. Differences were evaluated by ANOVA followed
by a pairwise t-test. The level of statistical significance was set
at p<0.05.
Results
Oridonin in Rabdosia rubescens inhibits
prostate cancer cells more potently than oridonin alone
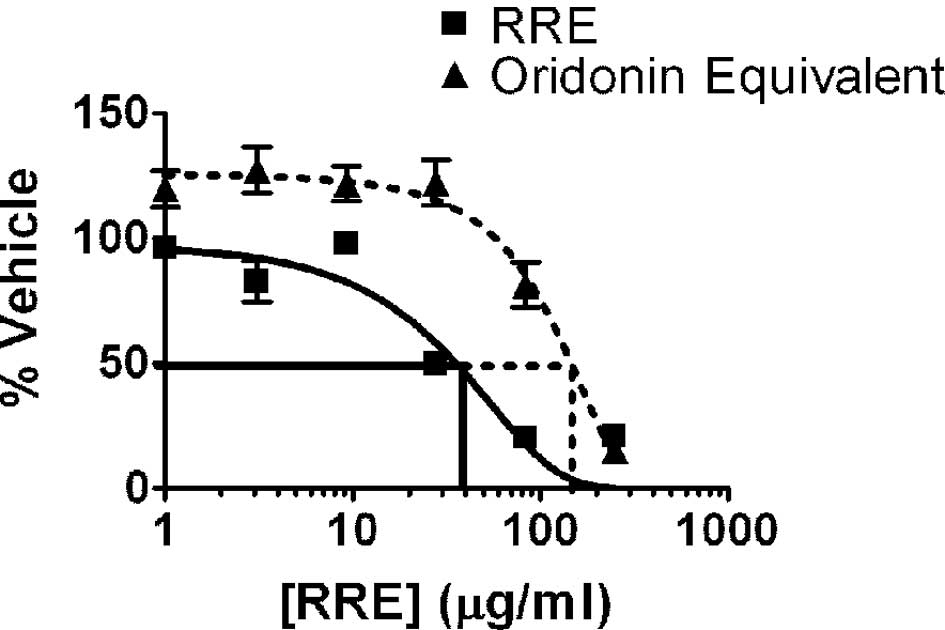
Both RRE and pure oridonin inhibited the
proliferation of DU-145, PC3, LNCaP and 22Rv1 prostate cancer cells
in vitro. As noted with the 22Rv1 cell line (Fig. 1), treatment with RRE (10–100 μg/ml
for 24 h) or oridonin (50–200 μg/ml or 20–80 μmol for 24 h)
resulted in a dose-dependent inhibition of cell growth in all four
prostate cancer cell lines. We characterized our RRE and determined
that it contained 4% oridonin. Therefore, oridonin plus other
compounds in RRE were more potent than oridonin alone in inhibiting
prostate cancer cell growth in vitro. The less aggressive
cell lines, 22Rv1 and LNCaP, were much more sensitive to the
inhibitory effects of oridonin than the more aggressive DU-145 and
PC3 cell lines (data not shown).
In the absence of RRE, five times the
oridonin concentration is required to inhibit prostate xenograft
tumor growth
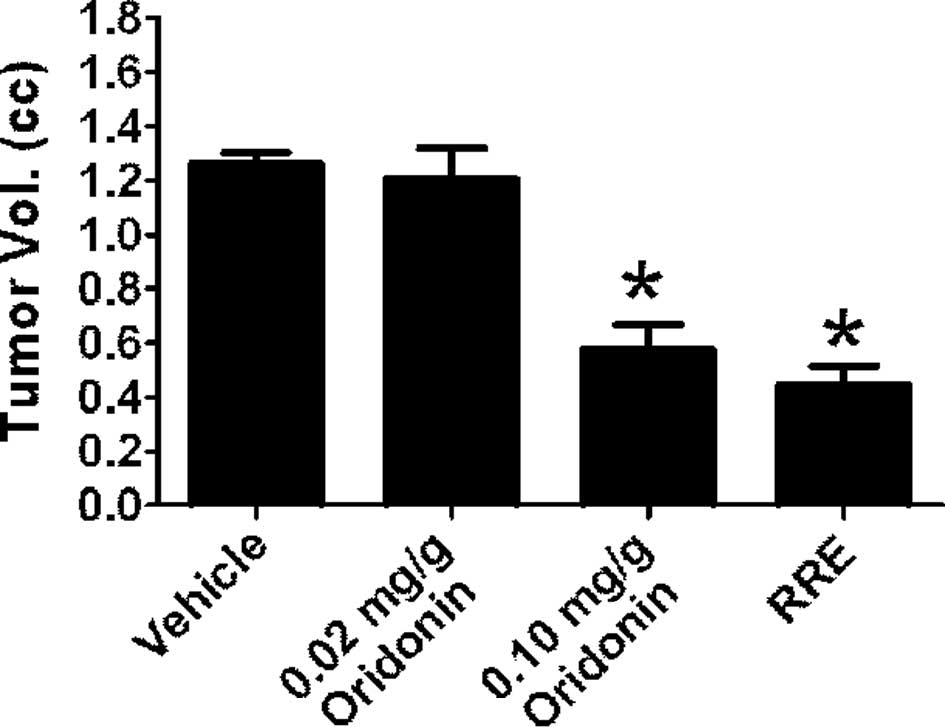
Treatment with RRE significantly inhibited tumor
growth in SCID mice implanted with LAPC-4 prostate cancer cells.
However, 0.02 mg/g oridonin failed to inhibit tumor xeno-graft
growth. When animals were administered oridonin at 0.1 mg/g, tumor
growth was inhibited to a similar degree as with administration of
RRE (0.02 mg/g oridonin equivalent). Tumor volume in mice treated
with 0.1 mg/g of oridonin was 0.600±0.295 mm3 compared
to 1.261±0.104 mm3 in control animals and 0.440±0.182
mm3 in animals treated with RRE (Fig. 2). The tumor latency period was
prolonged to 14 days in animals receiving 0.1 mg/g oridonin and to
16 days in animals receiving RRE.
Oridonin in RRE is more effective at
inhibiting the NF-κB pathway than oridonin alone
Using IκB-α protein disappearance as an indication
of activation of the NF-κB pathway, the effects of RRE or an
equivalent oridonin dose alone on the stabilization of IκB-α
protein in TNF-α-treated DU-145 cells was examined. A 15-min
incubation with 5 ng/ml of TNF-α was sufficient to degrade ∼70% of
IκB-α protein in DU-145 cells. RRE, but not pure oridonin in the
amounts found in RRE, abrogated the effect of TNF-α to cause a
decrease in IκB-α protein at 4 h (Fig.
3A).
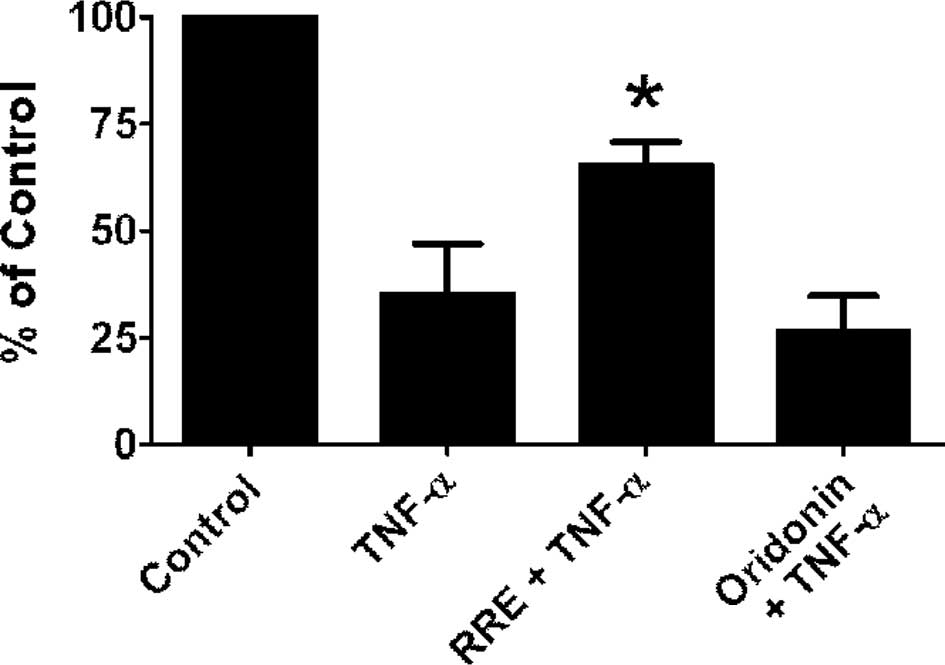
To further verify that the NF-κB pathway was
inhibited by RRE, we examined the ability of RRE or an equivalent
dose of oridonin alone to inhibit TNF-α-induced NF-κB
transactivation in DU-145 PCa cells. The cells were first
transfected with either a null vector or a luciferase reporter gene
containing NF-κB binding elements (NBE3X-Lux), and the
luciferase signal was used as an index of NF-κB transactivation.
Cells were treated with TNF-α alone, or pre-treated with RRE or an
equivalent concentration of oridonin alone for 2 h before exposure
to TNF-α, and the luciferase signal was monitored and normalized to
Renilla luciferase. Use of the null vector did not result in
spontaneous activation (data not shown). Conversely, TNF-α was
capable of inducing a very robust increase in luciferase reporter
activity at all concentrations, ranging from 0.5 to 50 ng/ml at 2
h. When cells were pre-treated with RRE before exposure to TNF-α,
NF-κB transactivation activity was significantly reduced (Fig. 3B). Equivalent concentrations of
oridonin alone had little or no effect on NF-κB activity (Fig. 3B). MTT assays were used to confirm
that the inhibitory effects were not due to toxicity of the
treatments (data not shown).
Gene microarray analysis of the NF-κB
pathway shows up-regulation of inflammatory and oxidative stress
genes unique to RRE
Gene microarrays were used to determine differences
between the regulation of NF-κB pathway genes by oridonin vs. RRE
in the LAPC-4 xenograft tumors. Oridonin treatment alone (0.02 and
0.1 mg/g) induced the transcription of 16 genes (Table I); three of these were unique to
oridonin. The three genes expressed solely after oridonin treatment
included BIRC2, interleukin 1α and LTBR. The majority of the 16
genes expressed in response to oridonin are known to play a role in
inflammation and cell death. Also unique to oridonin was the
down-regulation of interferon-γ (IFNG) gene expression. By
comparison, RRE treatment increased the expression of 23 different
genes, six of which were unique to RRE (Table I). These included ATF1, ELK1,
v-FOS, HMOX1, LTA and IKBKG, which are genes controlling
inflammation and oxidative stress.
 | Table I.Fold change in LAPC-4 xenograft tumors
between control (PBS)- vs. oridonin- or RRE-treated animals. |
Table I.
Fold change in LAPC-4 xenograft tumors
between control (PBS)- vs. oridonin- or RRE-treated animals.
| Gene symbol | Gene name | Oridonin | RRE |
|---|
| ATF1 | Activating
transcription factor 1 | | 2.9 |
| BIRC2 | Baculoviral IAP
repeat-containing 2 | 2.1 | |
| NOD1 | Nucleotide-binding
oligomerization domain containing 1 | 2.3 | 2.3 |
| CFLAR | CASP8 and FADD-like
apoptosis regulator | 1.6 | 1.8 |
| EDARADD | EDAR-associated death
domain | 1.3 | 1.6 |
| ELK1 | ELK1, member of ETS
oncogene family | | 1.7 |
| FOS | V-fos FBJ murine
osteosarcoma viral oncogene homolog | | 2.8 |
| GJA1 | Gap junction protein,
α1, 43 kDa | 1.6 | 1.4 |
| HMOX1 | Heme oxygenase
(decycling) 1 | | 1.8 |
| HTR2B | 5-Hydroxytryptamine
(serotonin) receptor 2B | 2.6 | 2.7 |
| IFNG | Interferon-γ | −2.6 | |
| IKBKG | Inhibitor of κ light
polypeptide gene enhancer in B-cells, kinase γ | | 1.6 |
| IL1A | Interleukin 1α | 2.2 | |
| IL1R1 | Interleukin 1
receptor, type I | 1.7 | 1.8 |
| IL8 | Interleukin 8 | | 2.8 |
| LTA | Lymphotoxin α (TNF
superfamily, member 1) | | 1.9 |
| MALT1 | Mucosa associated
lymphoid tissue lymphoma translocation gene 1 | 2.8 | 3.4 |
| MAP3K1 | Mitogen-activated
protein kinase kinase kinase 1 | 2.7 | 2.6 |
| PPM1A | Protein phosphatase
1A (formerly 2C), magnesium-dependent, α isoform | 1.7 | 1.6 |
| REL | V-rel
reticuloendotheliosis viral oncogene homolog (avian) | 2.7 | 2.6 |
| TRIM13 | Tripartite
motif-containing 13 | 1.3 | 1.4 |
| TICAM2 | Toll-like receptor
adaptor molecule 2 | 2.0 | 2.1 |
| TLR3 | Toll-like receptor
3 | 2.7 | 3.4 |
Discussion
Rabdosia rubescens (aka Donglingcao) is a
traditional Chinese medicine used to treat oral cancer. Oridonin,
the most biologically active molecule purified from RRE, has been
studied extensively for its effects on breast cancer (12,13),
leukemia (14), cervical cancer
(15), melanoma (16–18)
and prostate cancer (19).
Oridonin has previously been shown to inhibit the proliferation of
a wide variety of human cancer cells, including prostate (LNCaP,
DU-145 and PC3), breast (MCF-7 and MDA-MB-231) and non-small cell
lung (NCI-H520, NCI-H460 and NCI-H1299) cancers, acute
promyelocytic leukemia (NB4) and glioblastoma multiforme (U118 and
U138) (11). In the aggressive
HT1080 fibrosarcoma cell line, oridonin has been shown to induce
apoptosis through a p53-mediated mechanism that is regulated by
NF-κB (20). Another study with
the human melanoma A375-S2 cell line also demonstrated that
oridonin acts via a p53-mediated mechanism inhibited by blocking
PI3-K pathway activation (21).
Our microarray results suggest that the enhanced
inhibition of RRE to reduce prostate xenograft tumor growth may
occur via the up-regulation of genes in the inflammatory pathway
(Table I). This suggests that RRE
may have affected the immune response to the xenografts by
recruiting immune cells to the tumor site. Supporting a possible
involvement of oridonin in regulating immune cells and immune
system, a previous study showed that oridonin enhanced the
phagocytosis of apoptotic U937 cells by macrophage-like U937 cells
through the release of TNF-α and IL-1β (22).
Synergy research has, as its main aim, the
establishment of a scientific basis for the therapeutic superiority
of plant extracts from traditional medicines, fruits, vegetables
and grains, as compared to single constituents isolated and
purified as drugs to maximize potency. Synergistic effects of the
mixtures of bioactive constituents and their byproducts contained
in plant extracts may account for the apparent enhanced potency of
plant extracts compared to individual constituents (23,24).
However, the mechanisms underlying this synergy remain to be
established. We used gene expression arrays to uncover targets of
RRE that were not activated by oridonin. Future research on synergy
utilizing genomics, proteomics and systems biology may provide the
impetus for the development of new botanical dietary supplements
that fulfill the necessary quality, safety and efficacy standards
while maximizing efficacy and minimizing potential toxicity.
Acknowledgements
Support for this study was provided by
the Prostate Cancer Foundation.
References
|
1.
|
National Policy on Traditional Medicine
and Regulation of Herbal Medicines: Report of a WHO Global Survery.
WHO Press; Geneva: 2005
|
|
2.
|
Hong MY, Seeram NP, Zhang Y and Heber D:
Chinese red yeast rice versus lovastatin effects on prostate cancer
cells with and without androgen receptor overexpression. J Med
Food. 11:657–666. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Hong MY, Seeram NP, Zhang Y and Heber D:
Anticancer effects of Chinese red yeast rice versus monacolin K
alone on colon cancer cells. J Nutr Biochem. 19:448–458. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Li Z, Seeram NP, Lee R, Thames G, Minutti
C, Wang HJ and Heber D: Plasma clearance of lovastatin versus
chinese red yeast rice in healthy volunteers. J Altern Complement
Med. 11:1031–1038. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Heber D, Yip I, Ashley JM, Elashoff DA,
Elashoff RM and Go VL: Cholesterol-lowering effects of a
proprietary Chinese red-yeast-rice dietary supplement. Am J Clin
Nutr. 69:231–236. 1999.PubMed/NCBI
|
|
6.
|
Halbert SC, French B, Gordon RY, Farrar
JT, Schmitz K, Morris PB, Thompson PD, Rader DJ and Becker DJ:
Tolerability of red yeast rice (2,400 mg twice daily) versus
pravastatin (20 mg twice daily) in patients with previous statin
intolerance. Am J Cardiol. 105:198–204. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Seeram NP, Adams LS, Hardy ML and Heber D:
Total cranberry extract versus its phytochemical constituents:
antiproliferative and synergistic effects against human tumor cell
lines. J Agric Food Chem. 52:2512–2517. 2004. View Article : Google Scholar
|
|
8.
|
Seeram NP, Adams LS, Henning SM, Niu Y,
Zhang Y, Nair MG and Heber D: In vitro antiproliferative, apoptotic
and antioxidant activities of punicalagin, ellagic acid and a total
pomegranate tannin extract are enhanced in combination with other
polyphenols as found in pomegranate juice. J Nutr Biochem.
16:360–367. 2005. View Article : Google Scholar
|
|
9.
|
Henning SM, Niu Y, Liu Y, Lee NH, Hara Y,
Thames GD, Minutti RR, Carpenter CL, Wang H and Heber D:
Bioavailability and antioxidant effect of epigallocatechin gallate
administered in purified form versus as green tea extract in
healthy individuals. J Nutr Biochem. 16:610–616. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Adams LS, Seeram NP, Hardy ML, Carpenter C
and Heber D: Analysis of the interactions of botanical extract
combinations against the viability of prostate cancer cell lines.
Evid Based Complement Alternat Med. 3:117–124. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Ikezoe T, Chen SS, Tong XJ, Heber D,
Taguchi H and Koeffler HP: Oridonin induces growth inhibition and
apoptosis of a variety of human cancer cells. Int J Oncol.
23:1187–1193. 2003.PubMed/NCBI
|
|
12.
|
Hsieh TC, Wijeratne EK, Liang JY,
Gunatilaka AL and Wu JM: Differential control of growth, cell cycle
progression, and expression of NF-kappaB in human breast cancer
cells MCF-7, MCF-10A, and MDA-MB-231 by ponicidin and oridonin,
diterpenoids from the chinese herb Rabdosia rubescens.
Biochem Biophys Res Commun. 337:224–231. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Sartippour MR, Seeram NP, Heber D, Hardy
M, Norris A, Lu Q, Zhang L, Lu M, Rao JY and Brooks MN: Rabdosia
rubescens inhibits breast cancer growth and angiogenesis. Int J
Oncol. 26:121–127. 2005.
|
|
14.
|
Ikezoe T, Yang Y, Bandobashi K, Saito T,
Takemoto S, Machida H, Togitani K, Koeffler HP and Taguchi H:
Oridonin, a diterpenoid purified from Rabdosia rubescens,
inhibits the proliferation of cells from lymphoid malignancies in
association with blockade of the NF-kappa B signal pathways. Mol
Cancer Ther. 4:578–586. 2005.
|
|
15.
|
Hu HZ, Yang YB, Xu XD, Shen HW, Shu YM,
Ren Z, Li XM, Shen HM and Zeng HT: Oridonin induces apoptosis via
PI3K/Akt pathway in cervical carcinoma HeLa cell line. Acta
Pharmacol Sin. 28:1819–1826. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Ren KK, Wang HZ, Xie LP, Chen DW, Liu X,
Sun J, Nie YC and Zhang RQ: The effects of oridonin on cell growth,
cell cycle, cell migration and differentiation in melanoma cells. J
Ethnopharmacol. 103:176–180. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Wang HJ, Li D, Yang FY, Tashiro S, Onodera
S and Ikejima T: Oridonin induces human melanoma A375-S2 cell death
partially through inhibiting insulin-like growth factor 1 receptor
signaling. J Asian Nat Prod Res. 10:787–798. 2008.PubMed/NCBI
|
|
18.
|
Zhang CL, Wu LJ, Tashiro S, Onodera S and
Ikejima T: Oridonin induced A375-S2 cell apoptosis via
bax-regulated caspase pathway activation, dependent on the
cytochrome c/caspase-9 apoptosome. J Asian Nat Prod Res. 6:127–138.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Chen S, Gao J, Halicka HD, Huang X,
Traganos F and Darzynkiewicz Z: The cytostatic and cytotoxic
effects of oridonin (Rubescenin), a diterpenoid from Rabdosia
rubescens, on tumor cells of different lineage. Int J Oncol.
26:579–588. 2005.PubMed/NCBI
|
|
20.
|
Zhang Y, Wu Y, Wu D, Tashiro S, Onodera S
and Ikejima T: NF-kappa B facilitates oridonin-induced apoptosis
and autophagy in HT1080 cells through a p53-mediated pathway. Arch
Biochem Biophys. 489:25–33. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Zhang CL, Wu LJ, Zuo HJ, Tashiro S,
Onodera S and Ikejima T: Cytochrome c release from oridonin-treated
apoptotic A375-S2 cells is dependent on p53 and extracellular
signal-regulated kinase activation. J Pharmacol Sci. 96:155–163.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Liu YQ, You S, Tashiro S, Onodera S and
Ikejima T: Activation of phosphoinositide 3-kinase, protein kinase
C, and extracellular signal-regulated kinase is required for
oridonin-enhanced phagocytosis of apoptotic bodies in human
macrophage-like U937 cells. J Pharmacol Sci. 98:361–371. 2005.
View Article : Google Scholar
|
|
23.
|
Wagner H and Ulrich-Merzenich G: Synergy
research: approaching a new generation of phytopharmaceuticals.
Phytomedicine. 16:97–110. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Ulrich-Merzenich G, Panek D, Zeitler H,
Wagner H and Vetter H: New perspectives for synergy research with
‘omic’ technologies. Phytomedicine. 16:495–508. 2009.
|