Introduction
Hepatocellular carcinoma (HCC) is one of the most
common malignancies worldwide, particularly in China. HCC is the
second leading cause of cancer-related death among males (1). Although advances have been made in
the diagnosis and treatment of HCC, the long-term outcome for
patients with HCC is still extremely poor due to its high
recurrence and metastasis rate (2). The five-year recurrence rate in HCC
is as high as 50–60% (3). Thus,
the ability to predict individual risk of recurrence and subsequent
prognosis is critical to guide surgical and chemotherapeutic
treatment (4–7). Therefore, it is necessary to
investigate the molecular alterations correlated with the
recurrence or metastasis of HCC. The recurrence and metastasis of
HCC is a complex process involving various factors at each step
(8,9).
Cell migration is one characteristic of highly
aggressive metastatic cancer cells (10), and is one of the most important
steps in HCC invasion and metastasis (11–13).
Cell migration is highly regulated by spatial and temporal changes
in the actin cytoskeleton which are essential for many
physiological and pathological processes, including cancer cell
invasion. Rac1, a member of the Rho GTPase family, is a key
regulator of actin cytoskeletal dynamics and relays signals from
various stimuli such as growth factors, cytokines and adhesion
molecules to downstream effectors modulating cell migration and
invasion (14). Notably, Rac1 has
been shown to promote HCC cell migration (15–17).
Dock180 is known to be the main target of signal adaptor protein
Crk and acts as a guanine-nucleotide exchange factor for small
GTPase Rac1. Studies in Caenorhabditis elegans and
Drosophila reveal that Dock180 homologues modulate various
functions including phagocytosis, cell migration and actin
cytoskeletal organization through the activation of Rac1 (18). Dock180 facilitates nucleotide
exchange on Rac1 through its unconventional Docker GEF domain
(19), but requires binding to
engulfment and cell motility 1 (Elmo1) to achieve GDP/GTP exchange
on Rac (20). As a result, the
Dock180 family of proteins has been suggested to function as an
upstream regulator of the small GTPase Rac. The best characterized
Rac activation mediated by the Dock family is through the
Dock180-Elmo1 complex.
Although studies with experimental models have
indicated an important role of Elmo1 in cell invasion and migration
in human glioma and ovarian cancer cell lines (21), the clinical evidence of the role of
Elmo1 in human HCC is limited. Therefore, in the present study, we
evaluated the expression of Elmo1 in HCC tissues and analyzed its
correlation with clinicopathological characteristics and the
prognosis of HCC. Elmo1 expression was investigated in HepG2 cells
with low metastatic potential and HCCLM3 cells with high metastatic
potential by Western blotting. Knockdown of Elmo1 in HCCLM3 cells
caused a reduction in cell migration and invasion. In addition,
cell migration and invasion was measured by in vitro wound
healing and transwell migration assays.
Materials and methods
Patients and specimens
Prior informed consent was obtained from the
patients for the collection of liver specimens in accordance with
the guidelines of The Second Xiangya Hospital of Central South
University. The study protocols were approved by the Ethics
Committee of the Central South University, Changsha, Hunan, China.
HCC specimens were collected from 131 patients who underwent
hepatic resection for HCC at the Department of Surgery, The Second
Xiangya Hospital of Central South University from October 2005 to
November 2008. The patients did not receive transcatheter arterial
chemoembolism therapy or any local ablative therapy. The sample
included 115 males and 16 females with a median age of 49 years
(range 21–79). From these 131 HCC cases, fresh specimens of matched
adjacent non-tumorous tissues (ANTTs) were collected from 32 cases.
The samples were immediately frozen in liquid nitrogen and
subsequently stored at −80°C for reverse transcription-polymerase
chain reaction (qRT-PCR) and Western blot analysis. Five normal
liver samples were obtained from 5 patients with liver cavernous
hemoangioma as controls. The specimens were embedded in paraffin
and stained by H&E. The diagnoses of the patients were
confirmed by histopathology Histopathological examination was
conducted in a blinded manner.
Real-time reverse
transcription-polymerase chain reaction
qRT-PCR was performed as described previously
(22). The primers for Elmo1 were
as follows: forward, 5′-TGCCACA AAGTGCTGGAGATG-3′; reverse,
5′-ACGGACAGGCTCAG GTGATTC-3′. GAPDH expression was determined as a
control using primers: forward, 5′-GCACCGTCAAGGCTGAGAAC-3′;
reverse, 5′-TGGTGAAGACGCCAGTGGA-3′. The real-time PCR reaction was
carried out on an ABI 7500 thermal cycler (Applied Biosystems)
using SYBR® Green I chemistry. The PCR cycling
parameters were as follows: 50 cycles at 95°C for 5 sec and at 60°C
for 20 sec. The results were analyzed using the
2−ΔCt method according to the formula: ΔCt =
CtElmo1 - CtGAPDH.
SDS-PAGE and Western blot analysis
Tissues from HCC samples, ANTTs and the cell lines
were lysed with buffer containing 50 mM Tris-HCl, pH 7.3, 150 mM
NaCl, 2% NP-40, 0.5% deoxycholate, 2 mM EDTA, 2 mM NaF, and 1%
protease inhibitor cocktail (Pierce, Rockford, IL, USA). Total
protein (100 μg) was separated by SDS-PAGE and transferred to
polyvinylidene difluoride filters (PVDF) (Millipore, Danvers, MA,
USA). Membranes were blocked in 5 mg/ml milk, incubated with the
goat polyclonal anti-Elmo1 (1:400; Abcam UK), anti-Rac1 and
anti-fibronectin (1:500; Santa Cruz Biotechnology, Santa Cruz, CA,
USA) antibodies. HRP-conjugated mouse anti-goat IgG (1:2000; KPL,
Gaithersburg, MD, USA) was used as the secondary antibody. The
membrane was extensively washed, and the proteins were detected
with the Enhanced SuperSignal West Duro chemiluminescence system
(Pierce). β-actin (1:300; Sigma-Aldrich, St. Louis, MO, USA) was
used as a reference probe.
Immunohistochemistry and follow-up
Paraffin-embedded tissues were sectioned into serial
4-μm slices and stained with hematoxylin and eosin (H&E). In
brief, after deparaffinization and rehydration, endogenous
peroxidase was blocked with methanol containing 0.3% hydrogen
peroxide for 30 min and microwave-pretreated in EDTA buffer (1 mM,
pH 8.0) for 10 min for antigen retrieval. The concentration of goat
polyclonal anti-human Elmo1 antibody was 1:300. Immunostaining for
Elmo1 was carried out with the streptavidin-peroxidase system
(Zhongshan Golden Bridge Biotechnology, Beijing, China). Nuclei
were lightly counterstained with hematoxylin. Negative controls
were treated with phosphate-buffered saline (0.01 M, pH 7.2)
instead of the antibodies. The Elmo1 protein level in each section
was scored on a scale of 0 to 3+ to delineate low expression (0 or
1+) or high expression (2+ or 3+). Scoring was performed by two
independent investigators blinded to the clinical and follow-up
data.
Patient follow-up was carried out through written
correspondence or telephone contact. The rate of follow-up was
92.0%. A total of 131 patients were followed up post-operatively
for 30–1300 days, with a median follow-up time of 460 days. The
diagnosis of recurrence and metastasis was based on postoperative
imaging, serum AFP levels or re-operative resection. Overall
survival was calculated from the date of surgery to the date of
death or last follow-up. Disease-free survival was calculated from
the date of surgery to the date of recurrence, metastasis, death or
last follow-up. To determine factors influencing survival after
hepatic resection, the following 8 conventional clinicopathological
variables were assessed in all 131 HCC cases: gender, age, liver
cirrhosis, AFP concentration, tumor size, Edmondson-Steiner grade
and vein invasion.
Cell lines and cell culture
The HCC HepG2 and HCCLM3 cell lines, normal liver
L-02 cells and 293T retroviral packaging cells were purchased from
the Cell Institute, Shanghai, China. These cell lines were cultured
in high glucose Dulbecco's modified Eagle's media (Gibco-BRL,
Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum
(HyClone Laboratories Inc., Logan, UT, USA) and maintained in 5%
CO2 at 37°C.
Plasmids and transfection
Empty vector pSuper.retro. neo-GFP was purchased
from Invitrogen Inc. Two different Elmo1 small interfering RNA
(siRNA) DNA oligonucleotides were designed using OligoEngine RNAi
design software. siRNA for human Elmo1 targeting 19 nucleotides of
the human Elmo1 transcript (nucleotides 2921–2949,
5′-TGAGGACTGATGTGGTAGA-3′) and siRNA targeting another region of
the human Elmo1 transcript (nucleotides 1256–1298,
5′-TCCGCATTTAGTGGACAGG-3′) were designed. According to the insert
sequence of the Mission Non-Target siRNA control vector
(5′-CAACAAGAT GAAGAGCACCAA-3′; Sigma), a control non-target siRNA
was designed using two different Elmo1 siRNAs and one non-target
siRNA segment subcloned in the pSuper-GFP vector, respectively.
293T retroviral packaging cells, used for the production of
amphotropic virus, were transfected with the relevant pSuper
retroviral expression plasmid (Invitrogen, USA) at a confluence of
70% using Lipofectamine 2000 (Invitrogen) according to the
manufacturer's instructions. Clones expressing the retrovirus at
1×105 colony-forming U/ml were selected for subsequent
experiments. Multiple rounds of infection, which increase the
number of infected cells as well as the number of copies per cell,
were carried out during the transfection of the HCCLM3 cells.
Briefly, medium containing the retrovirus was added to the cells
supplied with polybrene (5 μg/ml) for 12 h; after a 12-h rest by
replacing the medium with fresh medium, the retrovirus was added
again. This double infection resulted in a 60–70% transfection
efficiency.
Migration and invasion assays
For the migration assay, HCCLM3 cells
(1×106) were seeded on 6-cm plates coated with 10 μg/ml
type I collagen. The cells were incubated for 24 h, and the
monolayer was then disrupted with a cell scraper (1.2-mm diameter).
Images were captured at 0 and 24 h under a phase contrast
microscope (Nikon ELWD 0.3). Experiments were carried out in
triplicate, and four fields for each plate were assessed. For the
invasion assay, a transwell plate with an 8-μm diameter pore
membrane (Costar) was coated with 200 μg/ml Matrigel and incubated
overnight. Twenty-thousand HCCLM3 cells were seeded into the upper
chamber of the transwell, and the lower chamber was filled with 0.8
ml DMEM supplemented with 0.5% FCS to induce chemotaxis. After 24 h
of incubation at 37°C, the cells were fixed in methanol and stained
with H&E. The number of cells that invaded through the pores to
the lower surface of the filter was counted under a microscope.
Three invasion chambers were used per condition. The values
obtained were calculated by averaging the total number of cells
from three filters.
Statistical analysis
Analysis was conducted using SPSS 11.5 software
(SPSS Inc., Chicago, IL, USA). Quantitative values were presented
as the mean ± SD or the median (range). The independent t-test was
used to compare Elmo1 mRNA and protein expression in HCC and ANTT
samples. Potential relationships between Elmo1 expression and
clinicopathological variables were assessed by analysis of variance
(ANOVA). Survival curves were plotted using the Kaplan-Meier method
and analyzed using the log-rank test. In addition, the univariate
and multivariate Cox proportional hazards regression model was used
to identify independent risk factors for survival. All tests were
two-tailed, and P<0.05 was considered to indicate statistical
significance.
Results
Expression of Elmo1 mRNA and protein in
HCC tissues and cell lines
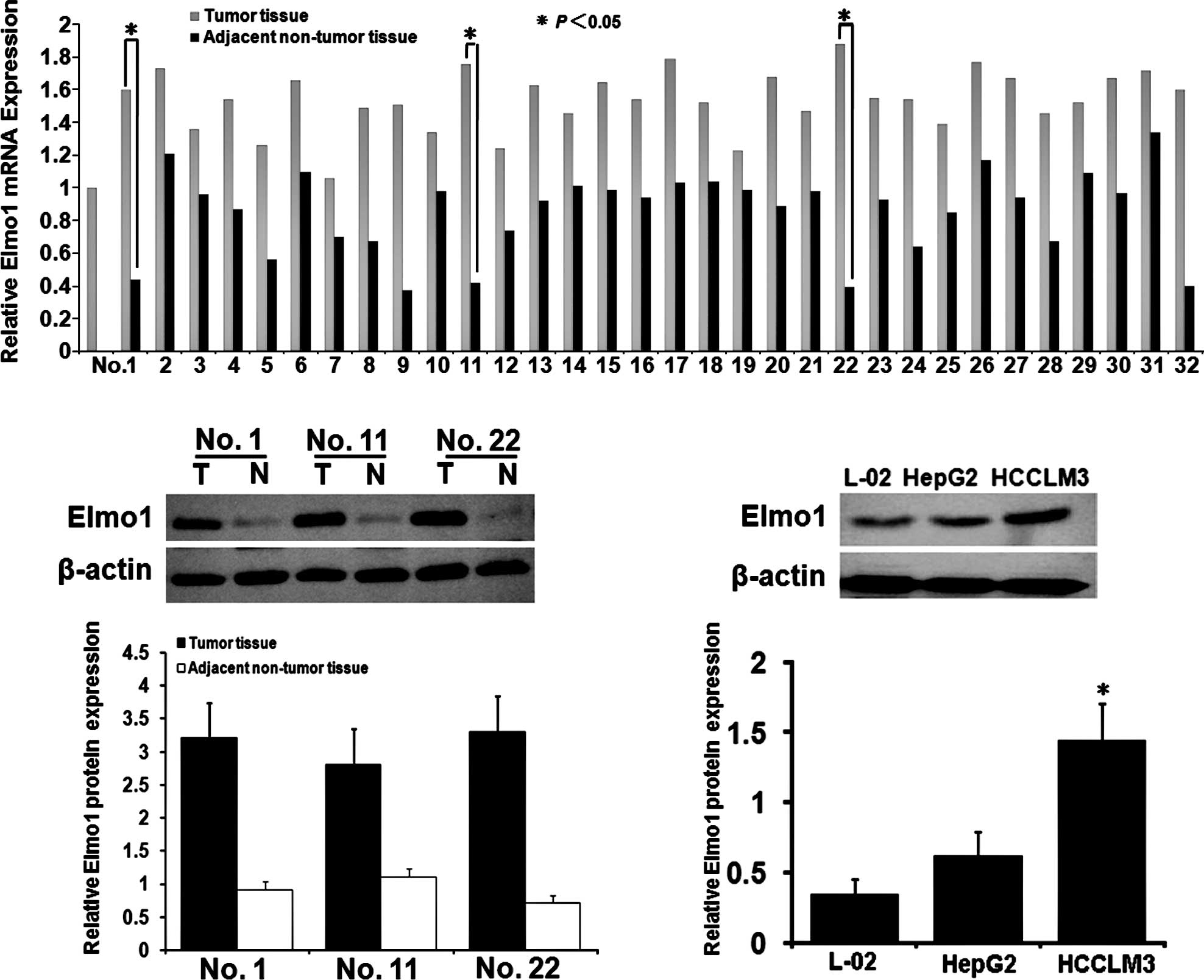
The Elmo1 mRNA levels in the HCC tissues were
significantly higher than those in the ANTTs, as determined by
qRT-PCR analysis (Fig. 1A). The
t-test showed that the expression of Elmo1 mRNA in the HCC tissues
was significantly higher than that in the ANTTs (8.3±0.8 vs.
1.7±0.7; P<0.05). The same 32 paired specimens were assessed by
Western bloting (Fig. 1B).
Consistent with the mRNA expression, the Elmo1 protein levels in
the HCC tissues were significantly higher than those in the paired
ANTTs (1.42±0.32 vs. 0.43±0.11; P<0.05). However, there were no
differences between the mRNA and protein levels the ANTTs and the
normal liver tissues (data not shown). Correlations between Elmo1
mRNA and protein expression with clinicopathological
characteristics of HCC were investigated, and Elmo1 expression was
significantly related to vein invasion of HCC (P<0.05). However,
no correlation was found between Elmo1 expression and the other
clinicopathological features of the HCC cases (P>0.05) (Table I). To determine the correlation
between Elmo1 expression and HCC metastasis, the expression of
Elmo1 was determined in the HCCLM3 and HepG2 cells and the normal
liver L-02 cell line as the control using qRT-PCR and Western
blotting. As shown in Fig. 1C, the
expression of Elmo1 protein in HCCLM3 was significantly higher than
that in the HepG2 cells (1.67±0.11 vs. 1.06±0.02; P<0.05).
Collectively, the results are in agreement with previously findings
that Elmo1 expression is correlated with metastatic potential in
HCC, and that the HCCLM3 cell line may be a suitable target for
Elmo1 suppression.
 | Table I.Elmo1 mRNA and protein expression
levels in relation to clinicopathologic parameters of 32 cases of
HCC. |
Table I.
Elmo1 mRNA and protein expression
levels in relation to clinicopathologic parameters of 32 cases of
HCC.
| Clinicopathologic
variables | No. | mRNA | P-value | Protein | P-value |
|---|
| Gender | | | | | |
| Male | 30 | 1.35±0.52 | | 0.92±0.24 | |
| Female | 2 | 1.04±0.13 | 0.608 | 0.89±0.12 | 0.415 |
| Age | | | | | |
| ≤60 years | 26 | 1.28±0.48 | | 0.90±0.21 | |
| >60 years | 6 | 1.51±0.61 | 0.528 | 0.92±0.32 | 0.758 |
| Liver cirrhosis | | | | | |
| Absence | 6 | 1.44±0.55 | | 1.03±0.22 | |
| Presence | 26 | 1.29±0.49 | 0.393 | 0.87±0.22 | 0.423 |
| Tumor size | | | | | |
| ≤5 cm | 6 | 1.11±0.34 | | 0.75±0.14 | |
| >5 cm | 26 | 1.36±0.51 | 0.332 | 0.93±0.23 | 0.236 |
| Tumor nodules | | | | | |
| Multiple (≥2) | 20 | 1.47±0.52 | | 1.00±0.20 | |
| Solitary | 12 | 1.06±0.31 | 0.164 | 0.73±0.15 | 0.153 |
| Cell
differentiation | | | | | |
| I–II | 15 | 1.12±0.35 | | 0.79±0.17 | |
| III–IV | 17 | 1.53±0.52 | 0.059 | 1.01±0.21 | 0.061 |
| Vein invasion | | | | | |
| Absence | 17 | 1.08±0.37 | | 0.74±0.22 | |
| Presence | 15 | 1.55±0.53 | 0.004a | 1.09±0.26 | 0.006a |
Correlation of Elmo1 expression with the
prognosis of HCC
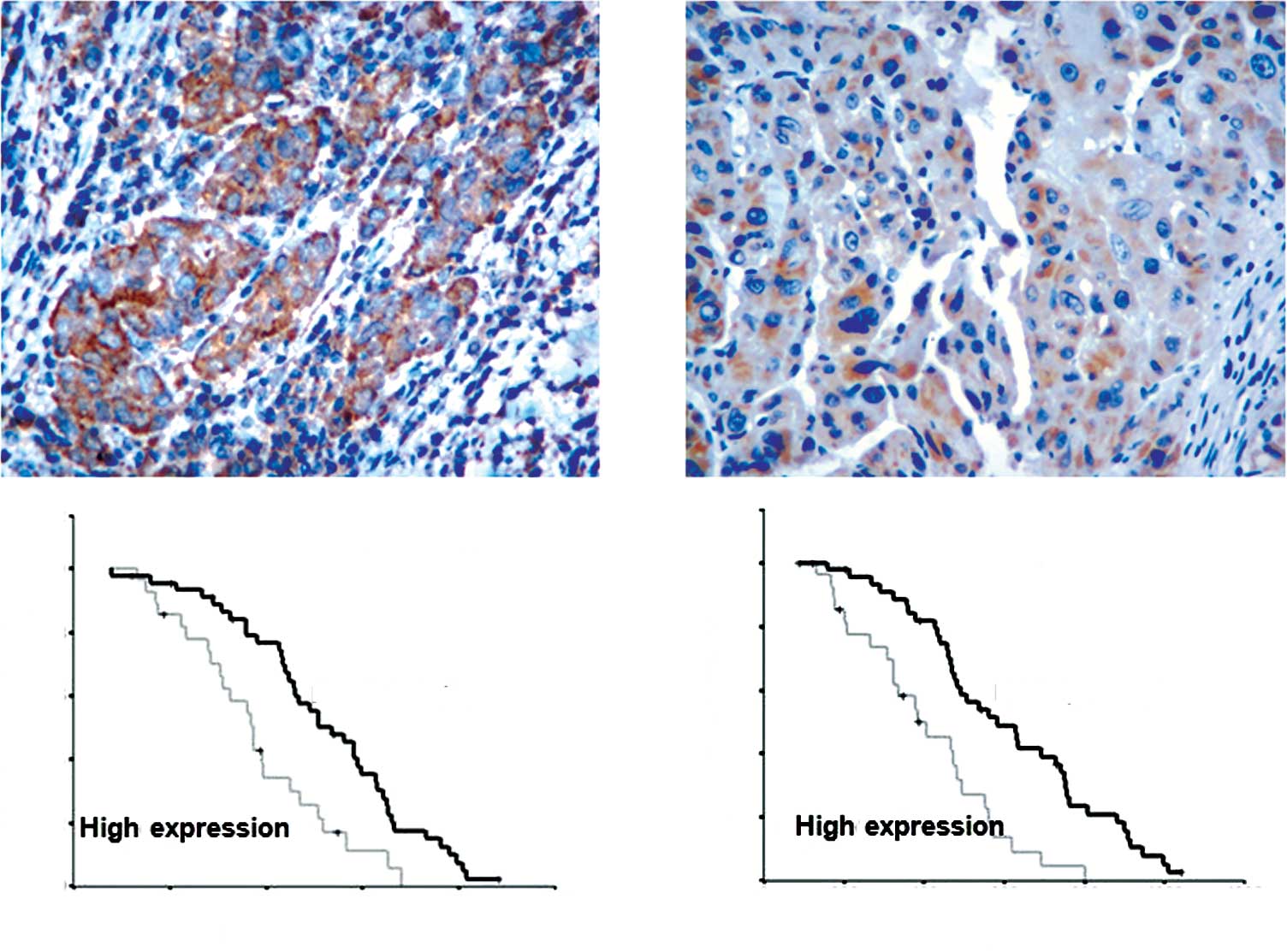
Immunohistochemical staining was carried out in all
131 paired HCC sample. Among the cases, 85.7% displayed positive
staining primarily in the cytoplasm of the cells within the tumor
tissues (Fig. 2A), while 22.9%
exhibited positive staining in the non-tumor tissues (P<0.05)
(Fig. 2B). We further examined
whether positive Elmo1 expression was correlated with the prognosis
of HCC. Low expression of Elmo1 was correlated with a longer median
survival time compared with high expression. The overall survival
time between the patients with low or high Elmo1 expression levels
was significantly different (1010 vs. 597 days; P=0.013) (Fig. 2D), as was the median overall
disease-free survival time between the patients with low or high
Elmo1 levels (690 vs. 921 days; P=0.002, Fig. 2C). To assess whether the expression
level of Elmo1 is an independent prognostic factor for the survival
of HCC patients, univariate and multivariate Cox regression
analyses were used to identify factors that may predict survival
after hepatic resection. Univariate Cox regression analysis
indicated that the Elmo1 expression level, tumor size,
Edmondson-Steiner grade, Child-Pugh classification, serum AFP
level, liver cirrhosis and vein invasion were all significantly
associated with survival. Using multivariate Cox regression
analysis, the Elmo1 expression level (HR 3.32; P=0.001) and
microscopic vein invasion (HR 2.95; P=0.02) were found to be
independent prognostic factors of survival (Table II).
 | Table II.Multivariate Cox analysis for the
prediction of survival factors of HCC. |
Table II.
Multivariate Cox analysis for the
prediction of survival factors of HCC.
| Variables | No. | Multivariable
analysis
|
|---|
| Relative risk (95%
CI) | P-value |
|---|
| Age (years) | | | |
| <60 | 106 | 1 | |
| ≥60 | 25 | 1.042
(1.008–1.077) | 0.280 |
| Cirrhosis | | | |
| Absence | 56 | 1 | |
| Presence | 75 | 1.25
(0.83–1.52) | 0.330 |
| Tumor size | | | |
| <5 cm | 56 | 1 | |
| ≥5 cm | 75 | 1.22
(0.76–1.82) | 0.380 |
| Serum AFP | | | |
| ≤20 ng/ml | 47 | 1 | |
| >20 ng/ml | 84 | 0.86
(0.44–1.16) | 0.490 |
| Cell
differentiationa | | | |
| I–II | 50 | 1 | |
| III–IV | 81 | 1.05
(0.94–1.13) | 0.190 |
| Microscopic venous
invasion | | | |
| Absence | 87 | 1 | |
| Presence | 44 | 2.95
(2.03–3.78) | 0.020b |
| Elmo1
expression | | | |
| Low | 52 | 1 | |
| High | 79 | 3.32
(2.09–4.36) | 0.001c |
Elmo1 affects HCCLM3 invasion and
migration
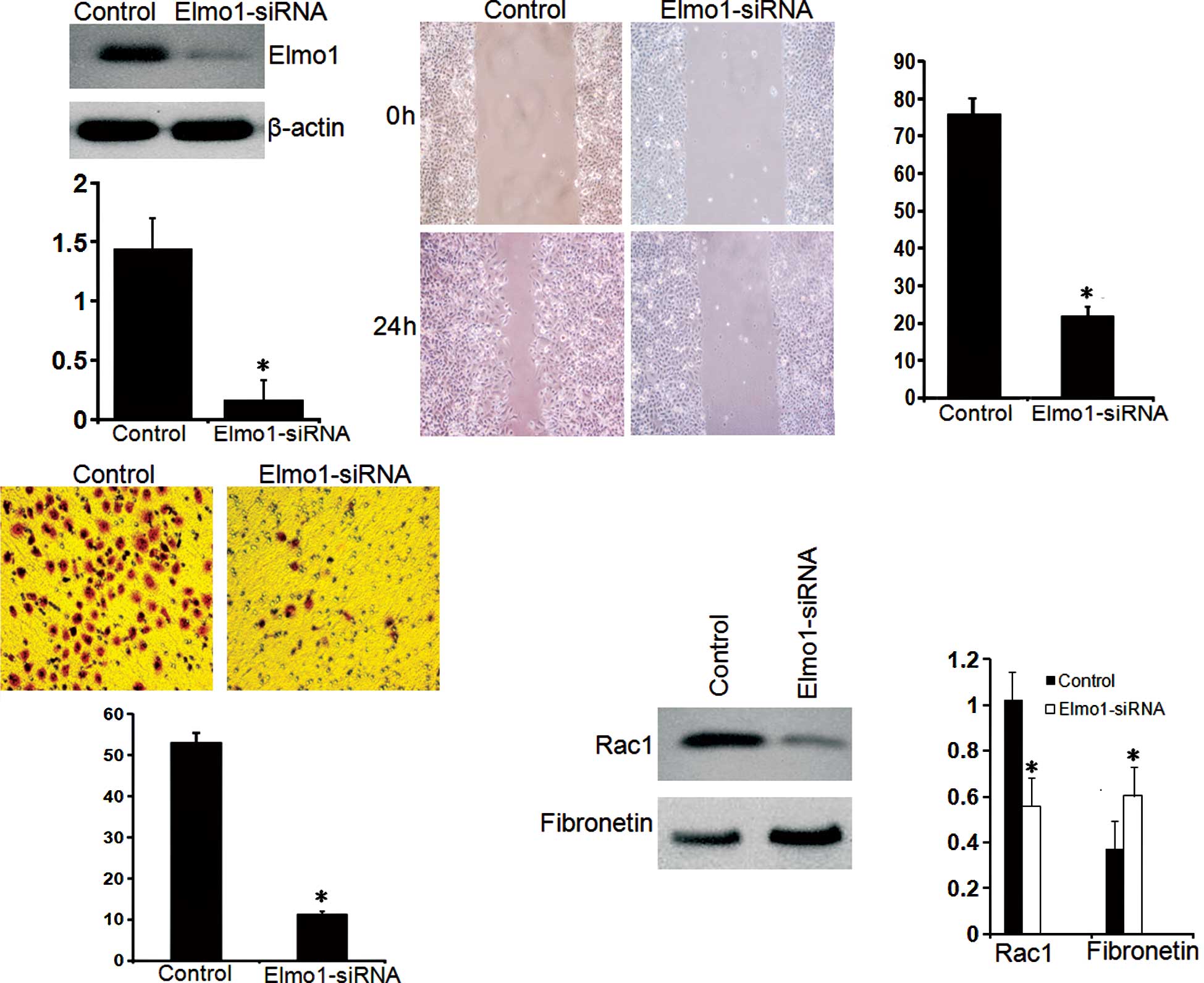
siRNA treatment was employed to inhibit the
expression of Elmo1 in HCCLM3 cells with high metastatic potential.
As shown in Fig. 3A, the candidate
sequence inhibited the protein expression of Elmo1 by >60%.
After a 3-week selection in medium supplemented with G418, the
HCCLM3 cell line Elmo1-siRNA was successfully contructed, which
stably expressed decreased Elmo1. We first investigated the effect
of Elmo1 on the invasion and migration of HCCLM3 cells. As shown in
Fig. 3B, Elmo1-siRNA cells
exhibited an 85% decrease in wound healing ability compared to the
control cells (P<0.05), suggesting a role for Elmo1 in the
migration of HCCLM3 cells. To substantiate this observation, a
Matrigel invasion assay in transwell culture chambers was carried
out to determine the effect of Elmo1 on the in vitro
invasion of HCCLM3 cells. As shown in Fig. 3C, the proportion of Elmo1-siRNA
cells that passed through the Matrigel was only 13% compared with
that of the control cells (P<0.05). Together, these results
support a critical role for Elmo1 in the invasion of HCCLM3 cells.
Meanwhile, the reduced expression of Rac1 and increased expression
of fibronectin were also noted in the Elmo1-siRNA HCCLM3 cells,
indicating that Elmo1 expression is involved in cell adhesion to
the extracellular matrix (Fig.
3D). Up-regulation of Elmo1 expression may be involved in the
invasive phenotype of HCC cells by promoting cell migration and
adhesion (23).
Discussion
Management of metastasis contributes to an
improvement in the prognosis of HCC patients. The development of
cancer such as HCC is characterized by multi-step processes
associated with alterations in the expression of genes involved in
cell proliferation, invasion and metastasis (24). Previous findings indicate that
invasion and metastasis are, to a large extent, attributable to the
ability of cells to migrate (25).
In a previous study, the association between Dock180
and Elmo1 was confirmed by immunoprecipitation experiments. The
results showed that Dock180 interacted predominantly with Crk1, and
that inhibition of endogenous Dock180 expression attenuated the
invasive behavior of SKOV3 cell lines concomitant with a reduction
in activated Rac1, which is a key regulator of actin cytoskeletal
dynamics modulating cell migration and invasion (26). However, the role of Elmo1 in human
HCC remains unknown. Thus, the present study aimed to investigate
the role of Elmo1 in HCC. Elmo1 mRNA and protein expression was
assessed in HCC tissues, and the expression levels of Elmo1 mRNA
and protein were found to be significantly higher in HCC tissues
than in the corresponding ANTTs. These results were consistent with
a previous study in gliomas and ovarian cancer cell lines, which
also suggested that the Elmo1 gene may play an important role in
invasion and metastasis (21).
In the present study, levels of Elmo1 mRNA and
protein in HCC tissues as determined by qRT-PCR and Western blot
analysis were investigated for a correlation with
clinicopathological features of HCC. The expression of Elmo1 mRNA
and protein was significantly correlated with vein invasion and the
Edmondson-Steiner grade of the HCC tissues. Substantial evidence
indicates that vein invasion and angiogenesis are highly correlated
with invasion and metastasis, as well as with the prognosis of HCC.
Consistent with these results, a highly significantly association
was noted between the Elmo1 expression level and
prognosis/metastasis of HCC. Multivariate Cox regression analyses
indicated that Elmo1 expression was an independent risk factor for
the prognosis of HCC patients, in addition to Edmondson-Steiner
grade and venous invasion. These results suggest that Elmo1 may
play a key role in HCC development.
These findings were confirmed in three HCC cell
lines with different spontaneous metastatic potential. HepG2 cells
exhibit a moderate metastatic potential, whereas HCCLM3 cells show
highly invasive potential, as indicated by extensive metastases via
orthotopic inoculation (27).
Based on this difference in metastatic potential, this cell model
system may serve as a useful platform for the study of HCC
metastasis. The protein expression of Elmo1 was previously found to
be significantly higher in the HCCLM3 cell line compared to the
HepG2 cell line (28) as verified
in our study. To evaluate the role of Elmo1 expression in
regulating cell migration in HCC, the HCCLM3 cell line was treated
with Elmo1 siRNA, which successfully blocked Elmo1 expression. The
results indicate that Elmo1 regulates cell migration in HCC cells,
and that increased expression of Elmo1 is associated with the
metastatic potential of HCC.
In the present study, we demonstrated that the
knockdown of Elmo1 was associated with the inhibition of invasion
and the migration of HCCLM3 cells, consistent with the critical
role of Elmo1 in the control of cellular motility. Thus, Elmo1 may
serve as a possible powerful therapeutic target for metastatic
HCC.
References
|
1.
|
He J, Gu D, Wu X, Reynolds K, Duan X, Yao
C, Wang J, Chen CS, Chen J, Wildman RP, Klag MJ and Whelton PK:
Major causes of death among men and women in China. N Engl J Med.
353:1124–1134. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Fan ST, Lo CM, Liu CL, Lam CM, Yuen WK,
Yeung C and Wong J: Hepatectomy for hepatocellular carcinoma:
toward zero hospital death. Ann Surg. 229:322–330. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Hubert C, Sempoux C, Rahier J, Horsmans Y,
Geubel A, van Beers BE, Annet L, Zech F, Leonard D and Gigot JF:
Prognostic risk factors of survival after resection of
hepatocellular carcinoma. Hepatogastroenterology. 54:1791–1797.
2007.PubMed/NCBI
|
|
4.
|
Ibrahim S, Roychowdhury A and Hean TK:
Risk factors for intrahepatic recurrence after hepatectomy for
hepatocellular carcinoma. Am J Surg. 194:17–22. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Mann CD, Neal CP, Garcea G, Manson MM,
Dennison AR and Berry DP: Prognostic molecular markers in
hepatocellular carcinoma: a systematic review. Eur J Cancer.
43:979–992. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Murray CJ and Lopez AD: Mortality by cause
for eight regions of the world: global burden of disease study.
Lancet. 349:1269–1276. 1997. View Article : Google Scholar
|
|
7.
|
Giannelli G and Antonaci S: Novel concepts
in hepatocellular carcinoma: from molecular research to clinical
practice. J Clin Gastroenterol. 40:842–846. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Yang LY, Tao YM, Ou DP, Wang W, Chang ZG
and Wu F: Increased expression of Wiskott-Aldrich syndrome protein
family verprolin-homologous protein 2 correlated with poor
prognosis of hepatocellular carcinoma. Clin Cancer Res.
12:5673–5679. 2006. View Article : Google Scholar
|
|
9.
|
Farinati F, Rinaldi M, Gianni S and
Naccarato R: How should patients with hepatocellular carcinoma be
staged? Validation of a new prognostic system. Cancer.
89:2266–2273. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Wong CC, Wong CM, Au SL and Ng IO:
RhoGTPases and Rho-effectors in hepatocellular carcinoma
metastasis: ROCK N'Rho move it. Liver Int. 30:642–656.
2010.PubMed/NCBI
|
|
11.
|
Chae S, Jun HO, Lee EG, Yang SJ, Lee DC,
Jung JK, Park KC, Yeom YI and Kim KW: Osteopontin splice variants
differentially modulate the migratory activity of hepatocellular
carcinoma cell lines. Int J Oncol. 35:1409–1416. 2009.PubMed/NCBI
|
|
12.
|
Feng H, Zhang J, Li X and Chen WN:
HBX-mediated migration of HBV-replicating HepG2 cells: insights on
development of hepatocellular carcinoma. J Biomed Biotechnol. Sep.
16–2009.(Epub ahead of print).
|
|
13.
|
Grise F, Bidaud A and Moreau V: Rho
GTPases in hepatocellular carcinoma. Biochim Biophys Acta.
1795:137–151. 2009.PubMed/NCBI
|
|
14.
|
Fukui K, Tamura S, Wada A, Kamada Y, Sawai
Y, Imanaka K, Kudara T, Shimomura I and Hayashi N: Expression and
prognostic role of RhoA GTPases in hepatocellular carcinoma. J
Cancer Res Clin Oncol. 132:627–633. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Carmona-Cuenca I, Roncero C, Sancho P,
Caja L, Fausto N and Fernández M: Upregulation of the NADPH oxidase
NOX4 by TGF-beta in hepatocytes is required for its pro-apoptotic
activity. J Hepatol. 49:965–976. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Lee TK, Poon RT, Yuen AP, Man K, Yang ZF,
Guan XY and Fan ST: Significance of the Rac signaling pathway in
HCC cell motility: implications for a new therapeutic target.
Carcinogenesis. 26:681–687. 2005.PubMed/NCBI
|
|
17.
|
Hiramoto K, Negishi M and Katoh H: Dock4
is regulated by RhoG and promotes Rac-dependent cell migration. Exp
Cell Res. 312:4205–4216. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Epting D, Wendik B, Bennewitz K, Dietz CT,
Driever W and Kroll J: The Rac1 regulator ELMO1 controls vascular
morphogenesis in zebrafish. Circ Res. 107:45–55. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Hope H, Schmauch C, Arkowitz RA and
Bassilana M: The Candida albicans ELMO homologue functions
together with Rac1 and Dck1, upstream of the MAP Kinase Cek1, in
invasive filamentous growth. Mol Microbiol. 76:1572–1590. 2007.
|
|
20.
|
Komander D, Patel M, Laurin M, Fradet N,
Pelletier A and Barford D: An alpha-helical extension of the ELMO1
pleckstrin homology domain mediates direct interaction to DOCK180
and is critical in Rac signaling. Mol Biol Cell. 19:483748–483751.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Bouschet T, Martin S, Kanamarlapudi V,
Mundell S and Henley JM: The calcium-sensing receptor changes cell
shape via a beta-arrestin-1 ARNO ARF 6 ELMO protein network. J Cell
Sci. 120:2489–2497. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Ip WK, Lai PB, Wong NL, Sy SM, Beheshti B,
Squire JA and Wong N: Identification of PEG10 as a progression
related biomarker for hepatocellular carcinoma. Cancer Lett.
250:284–291. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Shimazaki A, Tanaka Y, Shinosaki T, Ikeda
M, Watada H, Hirose T, Kawamori R and Maeda S: ELMO1 increases
expression of extracellular matrix proteins and inhibits cell
adhesion to ECMs. Kidney Int. 70:1769–1776. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Jarzynka MJ, Hu B, Hui KM, Bar-Joseph I,
Gu W, Hirose T, Haney LB, Ravichandran KS, Nishikawa R and Cheng
SY: ELMO1 and Dock180, a bipartite Rac1 guanine nucleotide exchange
factor, promote human glioma cell invasion. Cancer Res.
67:7203–7211. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Hsu SC, Kuo CL, Lin JP, Lee JH, Lin CC, Su
CC, Yang MD and Chung JG: Crude extracts of Euchresta
formosana radix inhibit invasion and migration of human
hepatocellular carcinoma cells. Anticancer Res. 27:2377–2384.
2007.
|
|
26.
|
Wong CC, Wong CM, Ko FC, Chan LK, Ching YP
and Yam JW: Deleted in liver cancer 1 (DLC1) negatively regulates
Rho/ROCK/MLC pathway in hepatocellular carcinoma. PLoS One.
23:E27792008. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Tian J, Tang ZY, Ye SL, Liu YK, Lin ZY,
Chen J and Xue Q: New human hepatocellular carcinoma (HCC) cell
line with highly metastatic potential (MHCC97) and its expressions
of the factors associated with metastasis. Br J Cancer. 81:814–821.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Zhang SZ, Pan FY, Xu JF, Yuan J, Guo SY,
Dai G, Xue B, Shen WG, Wen CJ, Zhao DH and Li CJ: Knockdown of
c-Met by adenovirus-delivered small interfering RNA inhibits
hepatocellular carcinoma growth in vitro and in vivo.
Mol Cancer Ther. 4:1577–1584. 2005. View Article : Google Scholar : PubMed/NCBI
|