Introduction
Hepatocellular carcinoma (HCC) is one of the most
common malignant tumors worldwide, especially in East Asia,
including China. However, the pathogenesis of hepatocarcinogenesis
is far from clear. microRNAs (miRNAs) are a type of highly
conserved non-coding small RNAs which regulate gene expression at
the post-transcriptional level. It is now clear that miRNAs can
potentially regulate every aspect of cellular activity, including
differentiation and development, metabolism, proliferation, and
apoptotic and viral infection. Recently, miRNAs have been found to
play pivotal roles in many malignancies including HCC development
(1–9). The presence of a molecular prognostic
miRNA signature in primary HCC clinical specimens has also been
confirmed by several recent studies (4,6,7,10–12),
and many miRNAs have been found to play important regulatory roles
in hepatocarcinogenesis.
Our previous study found that miRNA-602 has an
important regulatory activity in HBV-mediated hepatocarcinogenesis
by inhibiting the tumor-suppressive gene RASSF1A from very early
stages of chronic HBV hepatitis to HBV-positive cirrhosis to HCC
(13). In this study, the
expression of miRNA-450a and its potential target, DNA
methyltransferase 3a (DNMT3a), was investigated in HCC.
Materials and methods
Patients and cell lines
Histologically normal liver samples were obtained by
biopsy during surgery from eight patients with gallbladder stones.
Thirty-four HCC and corresponding non-malignant para-tumorous
specimens were collected by radical hepatectomy. All tissues were
obtained with informed consent from the patients, and were verified
by biochemistry and pathological examination. The study was
approved by the Institutional Review Board of Tongji Medical
College, Huazhong University of Science and Technology (China). The
cell lines, HepG2 and L02, were cultured in RMPI-1640 with 10%
fetal bovine serum.
microRNA arrays
miR-450a from 8 normal livers, 34 HCC, and
corresponding non-tumorous tissues was analyzed. microRNA arrays
were performed as described previously (13). Briefly, 100 ng RNA of each specimen
was extracted using TRIzol (Invitrogen) and an RNeasy Mini kit
(Qiagen) according to the manufacturer's instructions. The samples
were hybridized on a hybridization station. Scanning was performed
with an Axon GenePix 4000B microarray scanner.
Quantitative real-time PCR
Total-RNA was extracted from the tissues and cell
lines by TRIzol reagent (Invitrogen), according to the
manufacturer's instructions. For miRNA qPCR, reverse transcription
was performed using the QuantiMir RT kit (System Biosciences).
Primers for miR-450a were forward, 5′-TTTTGCGATGTGTTCC-3′ and
reverse, 5′-GTGCAG GGTCCGAGGT-3′; and for control U6 forward,
5′-CTCGCT TCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATT TGCGT-3′.
Primers for DNMT3a were forward, 5′-CAATGA CCTCTCCATCGTCAAC-3′ and
reverse, 5′-CATGCAGGA GGCGGTAGAA-3′; and for β-actin forward,
5′-GAACGG TGAAGGTGACAG-3′ and reverse, 5′-TAGAGAGAAGTG
GGGTGG-3′.
The amplification of miR-450a was performed as
follows: denaturation at 95°C for 10 min, followed by 40 cycles of
95°C for 10 sec, 60°C for 20 sec, and 72°C for 10 sec.
Amplification of DNMT3a was performed as follows: denaturation at
95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C
for 1 min. U6 RNA was used as an miRNA internal control, and
β-actin was used to normalize the amount of total-mRNA in each
sample. All values were calculated as ratios normalized to U6 or
β-actin.
Transfection of miR-450a mimics into
HepG2 cells
Synthesized miR-450a mimics were purchased from
Dharmacon (Lafayette, CA). HepG2 cells were cultured in RPMI-1640
plus 10% fetal bovine serum. After reaching 30 to 50% confluency,
the cells were transfected with 60 nM of the miR-450a mimics or an
miRNA mimic control. Cell proteins were harvested and measured 72 h
after transfection.
Western blot analyses
Western blotting was performed as described
previously (13). Briefly, cell
lysates were electrophoresed on 10 to 20% polyacrylamide gels
(Bio-Rad) and transferred to Immobilon-PSQ membranes (Millipore,
MA). The membranes were blocked with TBS containing 5% skim milk
and 0.1% Tween-20, and then incubated with a primary antibody.
Anti-DNMT3a and anti-GAPDH antibodies (CST for DNMT3a and ProMab
for GAPDH) were used according to the manufacturer's instructions.
GAPDH was used as an internal control, and all values were
calculated as ratios normalized to GAPDH.
Luciferase activity assay
The 3′UTR of DNMT3a containing an intact miR-450a
recognition sequence was amplified by PCR and inserted into a pGL3
vector (Promega) immediately downstream of the luciferase gene. A
pGL3 construct containing DNMT3a 3′UTR with point mutations in seed
sequence was synthesized using a site-directed mutagenesis kit
(Stratagene, USA) according to the manufacturer's instructions. The
primer for DNMT3a was forward, 5′-GCTCTAGACGAAAAGGGTTGGACATCAT-3′
and reverse, 5′-GCTCTAGAGCCGAGGGAGTCTCCTTTTA-3′.
Cells (2×105) were co-transfected with
500 ng of the pGL3-DNMT3a-WT or pGL3-DNMT3a-MUT constructs with
miR-450a mimics or a negative control. Each sample was
co-transfected with 50 ng of pRL-TK plasmid expressing Renilla
luciferase to monitor the transfection efficiency (Promega). A
luciferase activity assay was performed 48 h after transfection
with the dual luciferase reporter assay system (Promega). The
relative luciferase activity was normalized with Renilla luciferase
activity.
Cell apoptosis and proliferation
assays
Apoptosis was detected by Annexin V-FITC/PI (KeyGen,
China) double-staining. Briefly, 72 h after transfection, cells
(2×106/ml) were harvested and stained with anti-Annexin
V conjugated with FITC and propidium iodide (PI) for 15 min, and
then detection was carried out using FACScan. Data were analyzed
using CellQuest software.
Cell proliferation was measured using the Cell
Counting Kit-8 (CCK-8) (Beyotime, China). Cells at 0, 10, 20, 30,
40, 50, 60 and 70 h after transfection were treated with 10 μl
CCK-8 per well according to the manufacturer's instructions. The
absorbance in each well was measured at 450 nm by a microplate
reader.
Statistics
All experiments were repeated 3 times, and data were
recorded as mean ± SD and analyzed using the Student's t-test and
one-way ANOVA by SPSS 13.0 software. P<0.05 was considered to be
significantly significant.
Results
Down-regulation of miR-450a is validated
in HCC tissues and cell lines
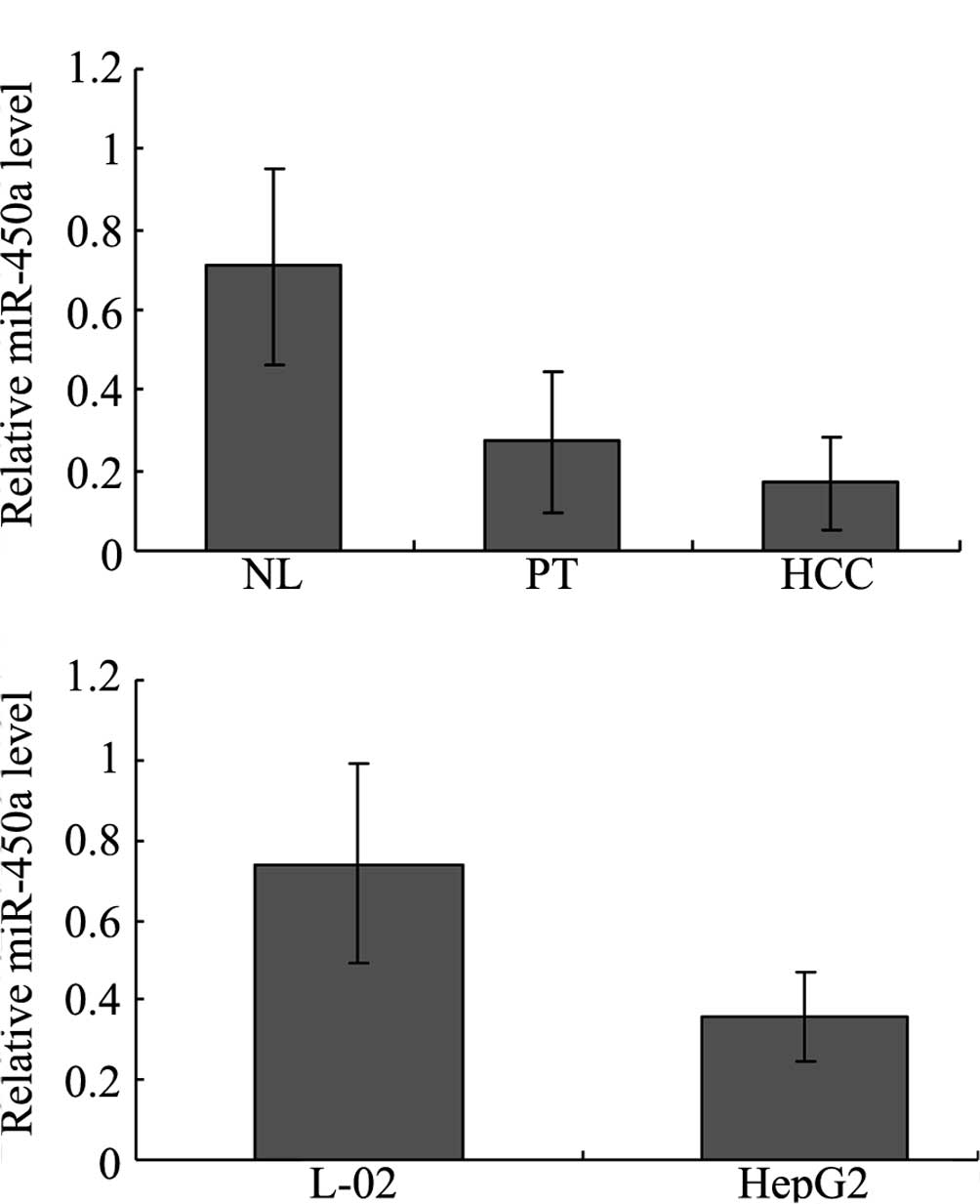
To validate miR-450a expression in the HCC tissues,
miR-450a levels were quantified using real-time PCR. Our results
showed that the expression of miR-450a was significantly
down-regulated in the HCC tissues compared with that in the normal
liver (NL) and para-tumorous (PT) tissues. In NL, PT and HCC
tissues, the levels were 0.710±0.245, 0.271±0.175, 0.168±0.114,
respectively (P<0.05 for NL vs. PT and NL vs. HCC) (Fig. 1A). miR-450a levels were not
significantly different between the PT and HCC groups (P>0.05,
Fig. 1A). The levels of miR-450a
in the L02 and HepG2 cells were also evaluated using real-time PCR.
The miR-450a level was significantly lower in the HepG2 than that
in the L02 cells. In the L02 and HepG2 cells, the levels were
0.740±0.251 and 0.358±0.112, respectively (P<0.01, Fig. 1B), which indicated that miR-450a
levels may be associated with hepatocellular carcinogenesis.
In silico prediction of an miR-450a
target
Bioinformatics analysis suggested that the key
enzyme in DNA methylation, DNMT3a, was one of the potential targets
of miR-450a. (http://www.ebi.ac.uk/enright-srv/microcosm/cgi-bin/targets/v5/hit_list.pl?genome_id=&mirna_id=hsa-mir-450a&external_name=DNMT3a&gene_id=&go_class=function&go_term=&logic=phrase&terms=).
The predicted binding of miR-450a with DNMT3a 3′UTR is illustrated
below:
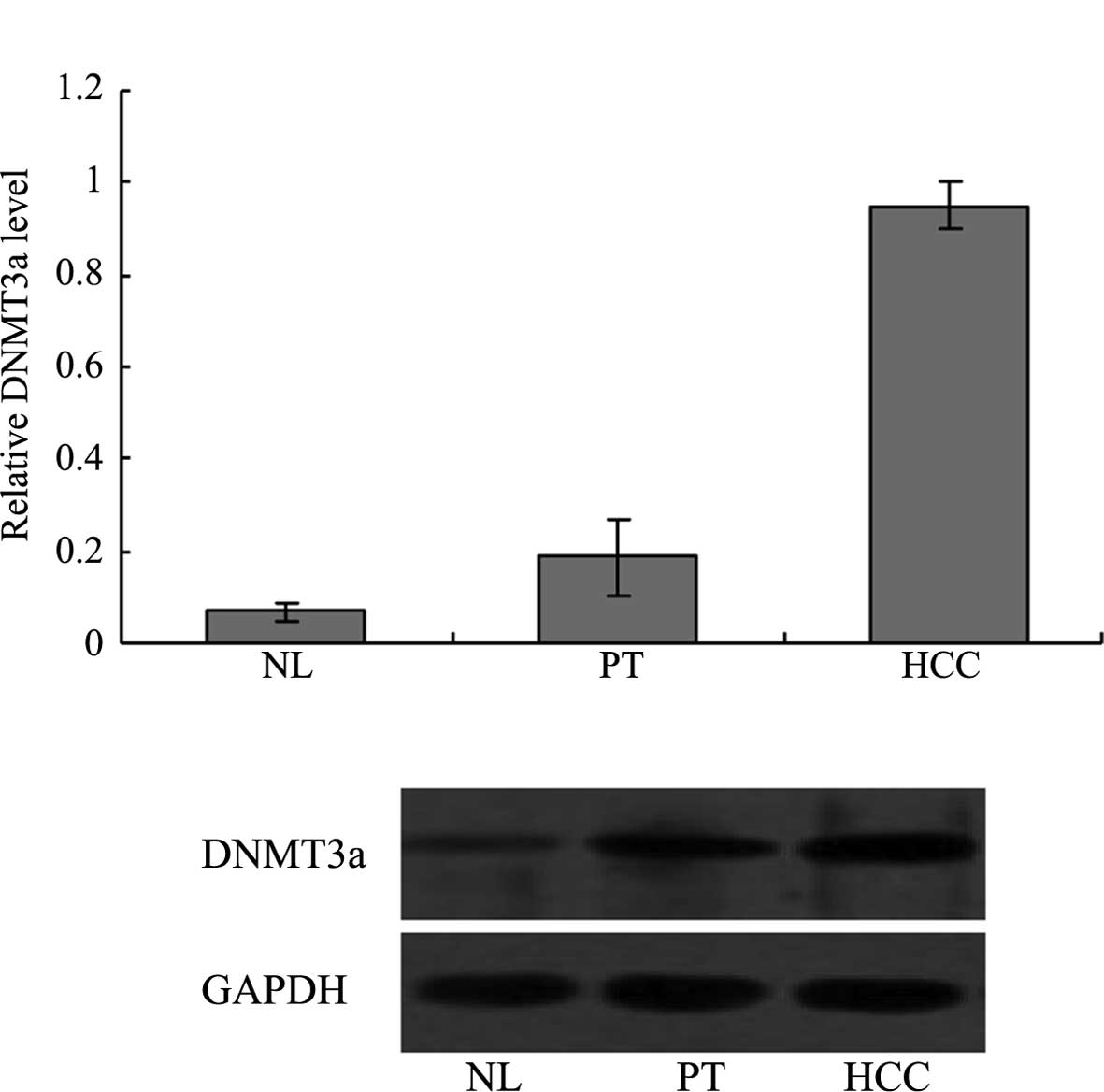
Expression of the potential miR-450a
target gene DNMT3a is higher in HCC compared with normal liver
Expression of the miR-450a potential target gene
DNMT3a in various liver tissues was investigated. The DNMT3a mRNA
level in HCC (0.950±0.053) was significantly higher than levels in
the NL (0.068±0.017) or PT (0.186±0.082) tissues (P<0.05 for HCC
vs. NL and HCC vs. PT) (Fig. 2A).
The DNMT3a protein level in HCC (0.522±0.014) was significantly
higher than levels in the NL (0.214±0.097) and PT (0.400±0.018)
tissues (P<0.05 for HCC vs. NL and HCC vs. PT) (Fig. 2B).
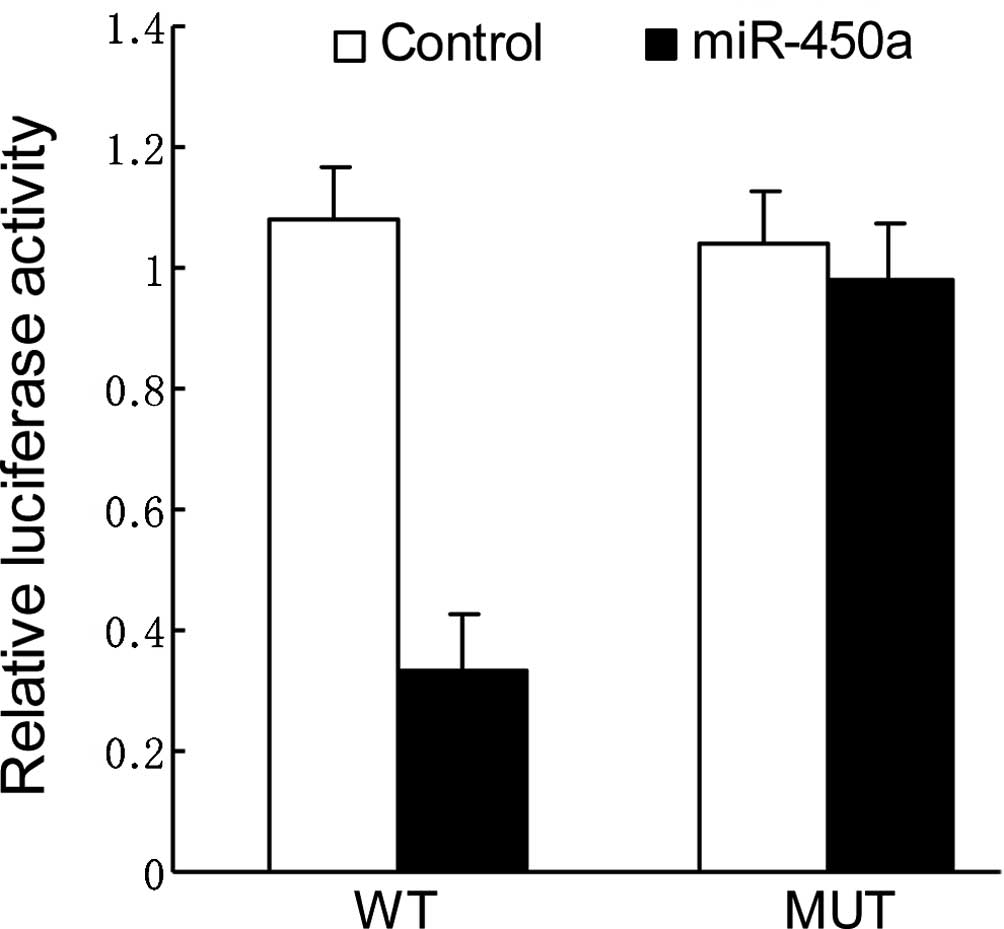
DNMT3a is the direct target of
miR-450a
To validate the miRNA-target interactions, the
DNMT3a complementary sites, with or without mutations, were cloned
into the 3′UTR of the firefly luciferase gene and co-transfected
with miR-450a mimics or a negative control in HepG2 cells. The
relative luciferase activity of the wild-type (WT) construct of
DNMT3a 3′UTR in HepG2 cells was significantly reduced in the
presence of miR-450a whereas such a suppressive effect was not
observed in the cells with the mutant (MUT) construct of DNMT3a
3′UTR (Fig. 3).
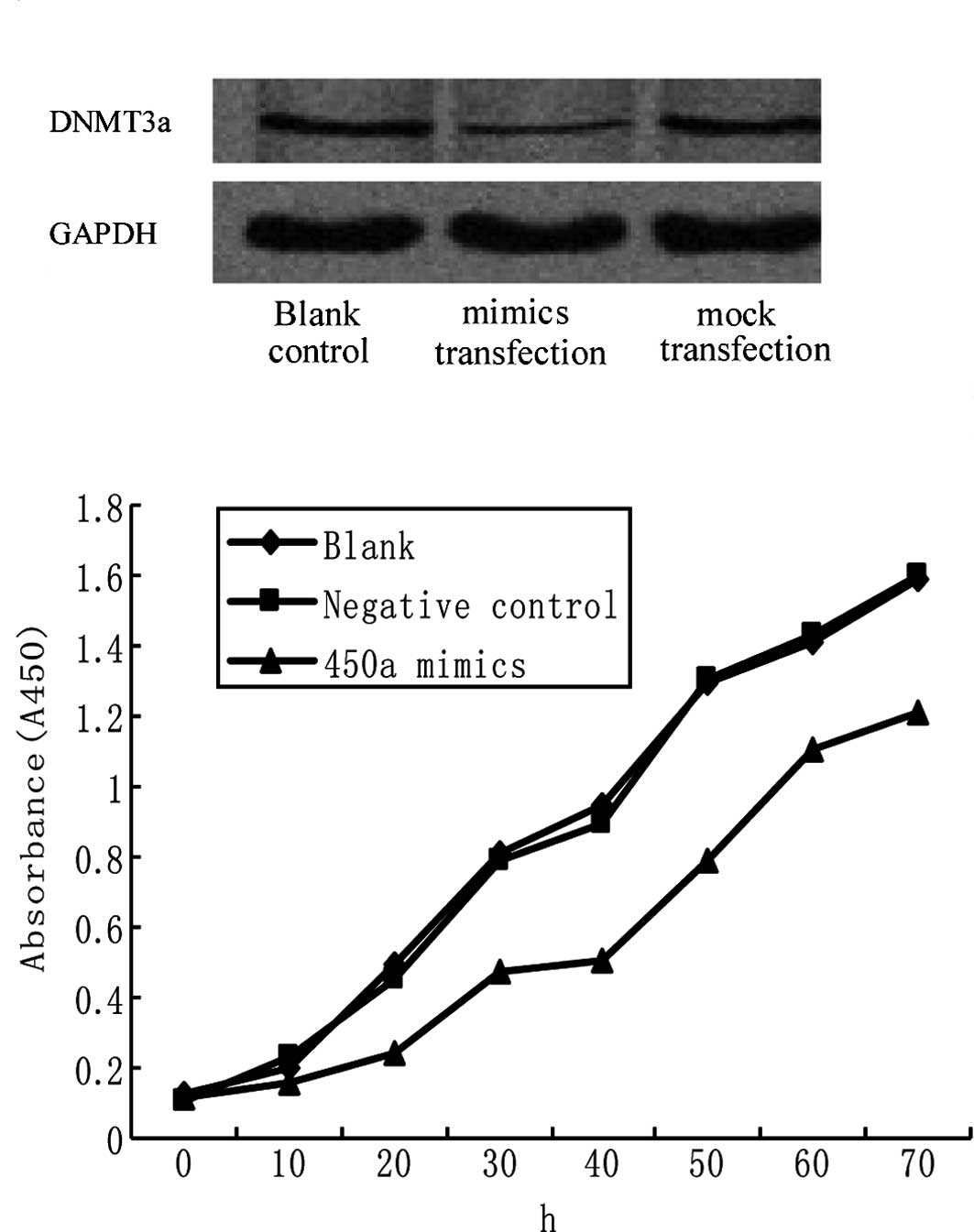
Ectopic miR-450a expression
down-regulates DNMT3a and inhibits HepG2 cell proliferation
miRNA-450a mimics were transfected into HepG2 cells,
and protein levels of the target gene, DNMT3a, were measured.
DNMT3a protein level was much lower in the mimic group than those
in the blank control or mock group. Levels in the blank control,
mimic and mock groups were 0.347±0.053, 0.114±0.011 and
0.335±0.033, respectively (P<0.05 for mimic vs. blank control
and mock) (Fig. 4A).
In order to clarify whether miR-450a deregulation
plays a role in hepatocarcinogenesis, the proliferation and
apoptosis rates of HepG2 cells were measured after miR-450a
exposure. An MTT assay indicated that at 20, 30 and 40 h after
miR-450a mimic exposure, the proliferation rate of HepG2 was
inhibited to 52, 42 and 46.3% of the control, respectively
(Fig. 4B). The differences between
the intervention and control groups at 20, 30, and 40 h were
significant (P<0.05 for all points) (Fig. 4B), while the difference between the
blank and control groups was not significant (P>0.05; Fig. 4B).
The apoptosis rate of HepG2 72 h after transfection
with the miR-450a mimics was not significantly different from that
of the HepG2 cells without miR-450a exposure (data not shown).
Discussion
Maintenance of genomic stability is regulated by
both genetic and epigenetic mechanisms. It is well known that
promoter hypermethylation mediated by DNA methyltransferases
(DNMTs) is the main reason for epigenetic inactivation of
tumor-suppressor genes (TSGs). Increasing evidence has revealed
that viral genes are important in regulating DNA methylation
(14). The epigenetic mechanisms
involved in virus-associated cancers are poorly understood.
Hypermethylation is responsible for the silencing of TSGs involved
in hepatocarcinogenesis. Several recent studies have suggested that
HBx is involved in epigenetic regulation during
hepatocarcinogenesis (15,16). Previous studies support a role for
miRNAs as both targets and effectors in aberrant mechanisms of DNA
hypermethylation (17,18). miRNAs are non-coding RNAs, 19–25
nucleotides in length, that regulate gene expression by inducing
translational inhibition or cleavage of their target mRNAs through
base pairing at partially or fully complementary sites (19). Research groups have shown that
miRNAs are altered in human malignancies and can function as
tumor-suppressor genes or oncogenes through regulation of
expression of their target genes (19). Several studies have shown that
specific miRNAs are aberrantly expressed in malignant HCC cells or
tissues compared to non-malignant hepatocytes or tissue (4,11,20–23).
In hepatocarcinogenesis proceeding from normal liver
to chronic hepatitis/cirrhosis to HCC, increased expression of DNMT
mRNA is correlated with a progressive increase in the number of
methylated genes (24). Presently
three catalytically active DNMTs, namely DNMT1, DNMT3a and DNMT3b,
have been identified. Emerging evidence has revealed that levels of
DNMT1, DNMT3a and DNMT3b mRNA are reportedly increased in various
malignancies, including colorectal, liver, and gastric cancers
(25,26). DNMTs and demethyltransferase were
found to cooperate with each other, leading to genetic instability
that eventually promotes cancer progression (27). Recently, one study demonstrated
that a high level of DNMT3a protein was significantly associated
with a lower overall survival in lung cancer (18). It has been demonstrated that DNMT3a
mediates tumor promotion through its interaction with p53 and the
resultant suppression of p53-mediated transcription of
tumor-suppressor genes (28).
In the present study, we characterized the role of
miR-450a in the regulation of DNA methylation in HCC for the first
time. The results revealed that miR-450a expression was
down-regulated to a greater extent in HCC tissues than in
corresponding non-cancerous liver tissues, and miR-450a expression
in HepG2 cells was significantly lower than that in L02 cells.
Additionally our findings indicate that there is a vital link
between miR-450a and DNMT3a. First, bioinformatics analysis
suggests that the key enzyme in DNA methylation, DNMT3a, is one of
the potential targets of miR-450a. Second, our findings indicate
that miR-450a expression is inversely correlated with DNMT3a
expression in HCC. Down-regulation of miR-450a resulted in an
up-regulation of DNMT3a. More importantly, we also provide evidence
from the luciferase activity assay that DNMT3a is a direct target
of miR-450a. Taken together, our findings confirm that miR-450a
regulates DNMT3a expression and may have a tumor-suppressive role
in HCC development.
We also provide insights concerning the biological
function of miR-450a in HepG2 cells. DNMT3a protein expression was
decreased significantly more by restoring miR-450a expression in
HepG2 cells transfected with the miR-450a mimics than in the
control group. The CCK-8 assay and flow cytometric analysis were
also performed in the HepG2 cells. Ectopic miR-450a expression in
HepG2 cells caused an inhibition of cell proliferation. These
results indicate that the enhanced expression of miR-450a by gene
transfer may reverse the malignant phenotypes of HCC cell lines by
inhibiting DNMT3a expression.
As the number of samples used for investigating
miR-450a down-regulation and the correlation between DNMT3a and
miR-450a expression was small, further validation in large cohorts
and in independent studies are necessary. Additional studies are
needed to investigate the regulatory mechanism of miR-450a
expression in order to better understand the mechanism by which
miR-450a is down-regulated in HCC. In conclusion, the results from
the current study suggest that miR-450a plays an important
regulatory role in hepatocarcinogenesis through inhibition of
DNMT3a expression, and miR-450a may be a potential target for the
treatment of HCC.
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
DNMT3a
|
DNA methyltransferase 3a
|
|
miRNA
|
microRNA
|
|
UTR
|
untranslated region
|
Acknowledgements
The study was supported by the
National Natural Science Foundation grants of China (NSFC nos.
30672067, 30700190, 30900663) and a grant from the New-Century
Excellent Talent Program of the Education Ministry of China (no.
NCET-07-318). We also thank Medjaden Bioscience Limited for
assisting in the preparation of this manuscript.
References
|
1.
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Gregory RI and Shiekhattar R: MicroRNA
biogenesis and cancer. Cancer Res. 65:3509–3512. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Gramantieri L, Ferracin M, Fornari F, et
al: Cyclin G1 is a target of miR-122a, a microRNA frequently
downregulated in human hepatocellular carcinoma. Cancer Res.
67:6092–6099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Murakami Y, Yasuda T, Saigo K, et al:
Comprehensive analysis of microRNA expression patterns in
hepatocellular carcinoma and non-tumorous tissues. Oncogene.
25:2537–2745. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Budhu A, Jia HL, Forgues M, et al:
Identification of metastasis-related microRNAs in hepatocellular
carcinoma. Hepatology. 47:897–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Shah YM, Morimura K, Yang Q, Tanabe T,
Takagi M and Gonzalez FJ: Peroxisome proliferator-activated
receptor alpha regulates a microRNA-mediated signaling cascade
responsible for hepatocellular proliferation. Mol Cell Biol.
27:4238–4247. 2007. View Article : Google Scholar
|
|
9.
|
Huang YS, Dai Y, Yu XF, et al: Microarray
analysis of microRNA expression in hepatocellular carcinoma and
non-tumorous tissues without viral hepatitis. J Gastroenterol
Hepatol. 23:87–94. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Jiang J, Gusev Y, Aderca I, et al:
Association of microRNA expression in hepatocellular carcinomas
with hepatitis, cirrhosis, and patient survival. Clin Cancer Res.
14:419–427. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Varnholt H, Drebber U, Schulze F, et al:
MicroRNA gene expression profile of hepatitis C virus-associated
hepatocellular carcinoma. Hepatology. 47:1223–1232. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Ladeiro Y, Couchy G, Balabaud C, et al:
MicroRNA profiling in hepatocellular tumors is associated with
clinical features and oncogene/tumor-suppressor gene mutations.
Hepatology. 47:1955–1963. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Yang L, Ma Z, Wang D, Zhao W, Chen L and
Wang G: MicroRNA-602 regulating tumor suppressive gene RASSF1A is
overexpressed in hepatitis B virus-infected liver and
hepatocellular carcinoma. Cancer Biol Ther. 9:1–6. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Li HP, Leu YW and Chang YS: Epigenetic
changes in virus-associated human cancers. Cell Res. 15:262–271.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Park IY, Sohn BH, Yu E, et al: Aberrant
epigenetic modifications in hepatocarcinogenesis induced by
hepatitis B virus X protein. Gastroenterology. 132:1476–1494. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Tong A, Gou L, Lau QC, et al: Proteomic
profiling identifies aberrant epigenetic modifications induced by
hepatitis B virus X protein. J Proteome Res. 8:1037–1046. 2009.
View Article : Google Scholar
|
|
17.
|
Saito Y, Liang G, Egger G, et al: Specific
activation of microRNA-127 with downregulation of the
proto-oncogene BCL6 by chromatin-modifying drugs in human cancer
cells. Cancer Cell. 9:435–443. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Fabbri M, Garzon R, Cimmino A, et al:
MicroRNA-29 family reverts aberrant methylation in lung cancer by
targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA.
104:15805–15810. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Garzon R, Fabbri M, Cimmino A, Calin GA
and Croce C: MicroRNA expression and function in cancer. Trends Mol
Med. 12:580–587. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Kutay H, Bai S, Datta J, et al:
Downregulation of miR-122 in the rodent and human hepatocellular
carcinomas. J Cell Biochem. 99:671–678. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Jacob JR, Sterczer A, Toshkov IA, et al:
Integration of woodchuck hepatitis and N-myc rearrangement
determine size and histologic grade of hepatic tumors. Hepatology.
39:1008–1016. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Harada H, Nagai H, Ezura Y, et al:
Down-regulation of a novel gene, DRLM, in human liver malignancy
from 4q22 that encodes a NAP-like protein. Gene. 296:171–177. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Wong QW, Lung RW, Law PT, et al:
MicroRNA-223 is commonly repressed in hepatocellular carcinoma and
potentiates expression of Stathmin1. Gastroenterology. 135:257–269.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Oh BK, Kim H, Park HJ, et al: DNA
methyltransferase expression and DNA methylation in human
hepatocellular carcinoma and their clinicopathological correlation.
Int J Mol Med. 20:65–73. 2007.PubMed/NCBI
|
|
25.
|
Eads CA, Danenberg KD, Kawakami K, Saltz
LB, Danenberg PV and Laird PW: CpG island hypermethylation in human
colorectal tumors is not associated with DNA methyltransferase
over-expression. Cancer Res. 59:2302–2306. 1999.PubMed/NCBI
|
|
26.
|
Ding WJ, Fang JY, Chen XY and Peng YS: The
expression and clinical significance of DNA methyltransferase
proteins in human gastric cancer. Dig Dis Sci. 53:2083–2089. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Fang JY, Cheng ZH, Chen YX, et al:
Expression of DNMT1, demethylase, MeCP2 and methylation of
tumor-related genes in human gastric cancer. World J Gastroenterol.
10:3394–3398. 2004.PubMed/NCBI
|
|
28.
|
Wang YA, Kamarova Y, Shen KC, et al: DNA
methyltransferase-3a interacts with p53 and represses p53-mediated
gene expression. Cancer Biol Ther. 4:1138–1143. 2005. View Article : Google Scholar : PubMed/NCBI
|