Introduction
Small-cell lung cancer (SCLC) patients are commonly
classified into limited stage and extensive stage groups by the
Veterans Administration Lung Cancer Study Group (VALSG) and SCLC
makes up approximately 15% of all lung cancer cases and is markedly
associated with cigarette smoking (1–3). The
prognosis of SCLC, which has been linked to the extent of disease
as well as other factors, is poor. The life expectancy of those
with untreated SCLC is approximately 3.5 months for the limited
group and 6 weeks for the extensive group (4–6).
Certain genetic factors have been identified as prognostic factors
for SCLC; however, the underlying mechanism of this cancer remains
unknown (7,8).
microRNAs (miRNAs) are RNA molecules that are
approximately 22 nucleotides in length that are implicated in a
number of biological processes, such as embryonic development,
cellular differentiation, proliferation, apoptosis, cancer
development and insulin secretion (9,10).
More than 700 miRNAs have been identified in humans, and these
miRNAs are responsible for regulating at least 30% of
protein-coding gene expression (11). Specifically, miRNAs target
nucleotides 2–8 at the 5′ end, which is known as the ‘seed region’
of the 3′ UTR of the target messenger RNA (mRNA). Perfect
complementarity between the miRNA and its target mRNA sequence
reduces protein levels due to RNA silencing (12,13).
There is increasing evidence indicating that single nucleotide
polymorphisms (SNPs) in the 3′ UTR region, that is targeted by
miRNAs, alter the expression of target genes and thereby affect an
individual's cancer risk, and miRNA-binding SNPs have been
extensively examined in recent genotyping studies (14–19).
PR-Set7/Set8/KMT5a (SET8), which is regulated by
miR-502 via the binding site in the SET8 3′ UTR, encodes a histone
H4 lysine 20 monomethyltransferase that is implicated in normal
cell cycle progression (20–22).
Previous studies have suggested that the SNP rs16917496, which is
located within the miR-502 binding site in the SET8 3′ UTR,
modulates SET8 expression and contributes to risk and age-at-onset
of cancer (16,23). We also found that this SNP modifies
hepatocellular carcinoma (HCC) outcome by altering SET8 expression,
which depends, at least in part, on its binding affinity with
miR-502 (24). In this study, we
genotyped this SNP in SCLC patients to assess its association with
cancer risk and disease outcome.
Materials and methods
DNA extraction
Blood samples were collected at the Fourth Hospital
of Hebei University from 44 SCLC patients who received treatment at
the Department of Respiratory Medicine between 2005 and 2009. Blood
samples were also collected from 44 healthy female controls. The
genomic DNA was immediately extracted using the Wizard Genomic DNA
extraction kit (Promega, Madison, WI, USA). All of the patients
received and signed consent forms, and all procedures were
supervised and approved by the hospital's Human Tissue Research
Committee.
Genotyping of rs16917496
The SNP rs16917496 was genotyped using the ligation
detection reaction (LDR) method with the forward and reverse
primers, 5′-CCTGGTCAGTGGTCA GCAAAT-3′ and
5′-CTGGGAAACACGCTCAAAATC-3′, respectively, to amplify the DNA
fragments flanking rs16917496 in the SET8 3′ UTR using the
sequence in the NCBI database (http://www.ncbi.nlm.nih.gov/snp/). PCR was performed
using a PCR Master Mix kit according to the manufacturer's
instructions (Promega). The ligation was performed using the probes
S1 (5′-TTGTGGTTTAGCTTTG TATTTAAAC-3′), S2 (5′-TTTTTGTGGTTTAGCTTTGTA
TTTAAAT-3′) and S3 (5′-AAGGAAATAAACTTGAAAAT TATTT-3′), and the
ligated products were separated using the ABI PRISM Genetic
Analyzer 3730XL (Applied Biosystems, Foster City, CA, USA).
Polymorphisms were confirmed based on the 3-bp difference in length
for different alleles of rs16917496.
Statistical analysis
The χ2 test was used to analyze
dichotomous values, such as the presence or absence of an
individual SNP in the SCLC patients and healthy controls. Survival
curves were calculated using the Kaplan-Meier method, and
comparisons between the curves were carried out using the log-rank
test. Multivariate survival analysis was performed using a Cox
proportional hazards model. All of the the statistical analyses
were performed using the SPSS 18.0 software package (SPSS Company,
Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
Clinical characteristics of the SCLC
patients
A total of 44 SCLC patients, including 13 limited
stage and 31 extensive stage cases, were enrolled in this study. A
review of the patients was performed every 3 months for 2 years. No
patients were lost during follow-up. The correlation between the
data collected during the 2-year follow-up and patient clinical
characteristics was analysed using the Kaplan-Meier method. As
shown in Table I, treatment was
associated with overall survival of the SCLC patients, while
gender, age, smoking and VALSG classification were not
statistically significant predictors of post-operative survival
time (Table I). The treatment
included platinum-based chemotherapy and chemotherapy combined with
radiotherapy, but no survival difference existed between these two
treatments, therefore we treated the two groups together for
comparison with no-treatment patients.
 | Table I.Univariate analysis of clinical
characteristics associated with the overall survival of the SCLC
patients. |
Table I.
Univariate analysis of clinical
characteristics associated with the overall survival of the SCLC
patients.
| Characteristics | No. of cases | 2-year survival rate
(%) | P-value |
|---|
| Treatment | | | <0.001 |
| Yes | 41 | 26.8 | |
| No | 3 | 0.0 | |
| Gender | | | 0.135 |
| Male | 28 | 17.9 | |
| Female | 16 | 37.5 | |
| Age (years) | | | 0.157 |
| ≤55 | 22 | 31.8 | |
| >55 | 22 | 18.2 | |
| VALSG
classification | | | 0.936 |
| Limited
disease | 13 | 23.1 | |
| Extensive
disease | 31 | 25.8 | |
| Smoking | | | 0.243 |
| Yes | 23 | 17.4 | |
| No | 21 | 33.3 | |
| SET8 genotype | | | 0.088 |
| CC+CT | 22 | 36.4 | |
| TT | 22 | 13.6 | |
Association of SET8 polymorphisms with
SCLC outcome
A total of 44 SCLC patients and healthy controls
were genotyped for the SNP analysis of the rs16917496 polymorphism.
The SET8 CC, CT and TT genotype frequencies in the control
samples were 6, 12 and 24, which were comparable to the genotype
frequencies in the SCLC patients (8, 12 and 22 for CC, CT and TT).
No statistically significant association with cancer risk was
detected for the distribution of the rs16917496 polymorphism
between the 44 SCLC cases and healthy controls (data not shown). We
subsequently assessed the correlation between rs16917496 and
overall survival of these SCLC patients.
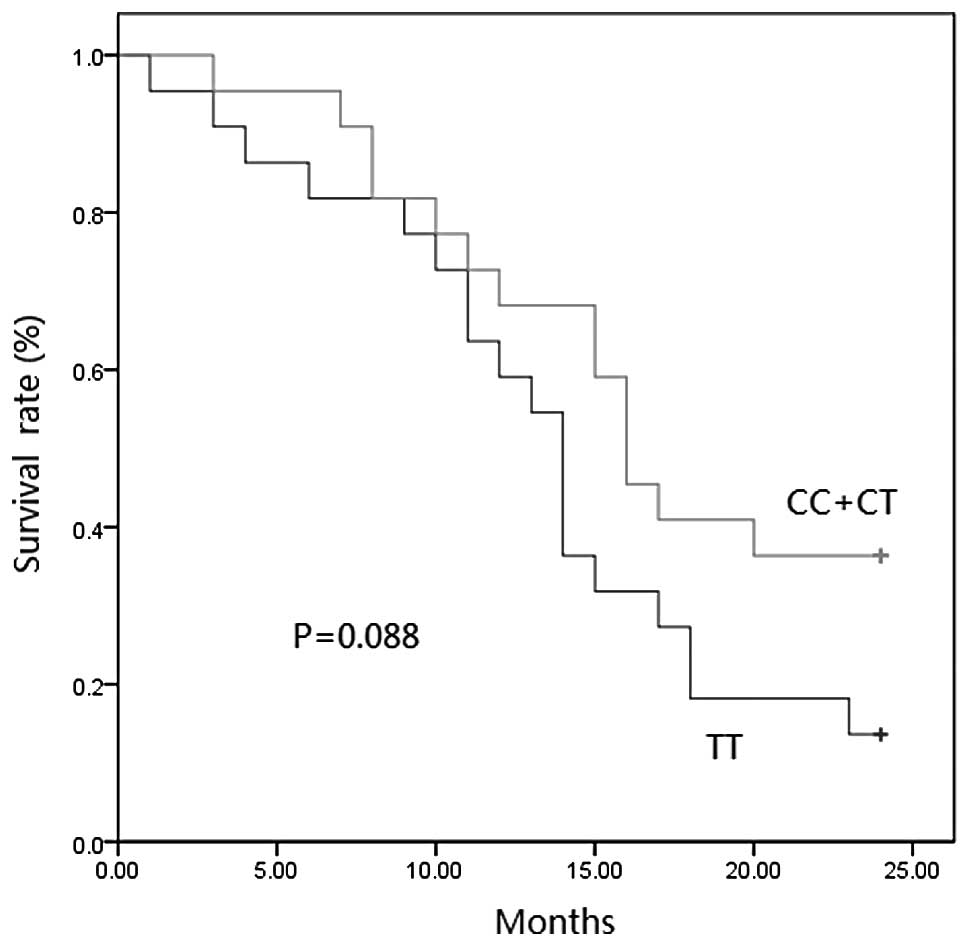
Since only 8 SCLC patients carried C/C alleles, we
combined the C/C and C/T carriers together for further analysis.
The survival curves of the SCLC patients were plotted using the
Kaplan-Meier method and analysed by log-rank test. The 2-year
survival rate of the C/C+C/T and T/T patients were 36.4 and 13.6%,
respectively. A borderline difference with a p-value of 0.088 for
the survival rate of the two genotypes was found, with the T allele
linking with shorter survival time (Fig. 1).
Multivariate analysis with the Cox proportional
hazards model was performed for these survival predictive factors.
As shown in Table II, a
statistical difference in survival rate appeared for the rs16917496
SNP, and following adjustment for the predictive factor of
treatment, this SNP was identified as an independent predictor of
SCLC outcome (relative risk, 0.453; 95% CI 0.217–0.944;
p=0.035).
 | Table II.Multivariate analysis of prognostic
factors associated with overall survival in SCLC patients with Cox
proportional hazards model. |
Table II.
Multivariate analysis of prognostic
factors associated with overall survival in SCLC patients with Cox
proportional hazards model.
| Factors | Relative risk | 95% CI | P-value |
|---|
| Treatment | 0.050 | 0.011–0.231 | <0.001 |
| SET8 genotype | 0.453 | 0.217–0.944 | 0.035 |
Discussion
Yu et al found that 12 miRNA binding site
SNPs displayed an aberrant allelic frequency in human cancers using
case-control association studies (16). The presently studied SNP has also
been proven to be associated with the early onset of breast cancer
(23). In this study, we
elucidated its association with SCLC cancer risk and outcome, and
revealed that the T/T genotype of rs16917496 is associated with
shorter survival time. These data are consistent with our previous
study, which revealed that the T/T allele was associated with
shorter survival time in post-operative HCC patients (24). No statistical differences were
detected for SCLC cancer risk and age-at-onset among the genotypes
of this SNP (data not shown).
The limited stage SCLC patients displayed a longer
survival rate than that of extensive stage patients in previous
reports (4,5), but the lack of statistical difference
for overall survival between these two groups may be due to the
small sample size that we used.
As a methyltransferase, SET8 modulates p53
expression by specifically methylating lysine 382 of histones that
are associated with the p53 genomic sequence (25). The correlation between the
methylation status of p53 in response to SET8 and their correlation
with SCLC carcinogenesis requires further study.
Although SNP studies in miRNA binding sites are at
an early stage, our results indicate that the alleles of SNPs in
miRNA binding sites have an effect on cancer outcome. However, the
results from this study require validation in other populations and
laboratory-based functional studies. miRNAs are key factors
associated with the susceptibility of a patient to therapeutic
responses in a number of complex diseases, including cancer
(26). The analysis of genetic
polymorphisms in miRNA binding sites may help to identify patient
subgroups with a high risk of poor outcome.
Acknowledgements
This work was supported by the
National Natural Science Foundation of China, no. 30801384.
References
|
1.
|
Adjei AA, Marks RS and Bonner JA: Current
guidelines for the management of small cell lung cancer. Mayo Clin
Proc. 74:809–816. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Simon GR and Wagner H: For the American
College of Chest Physicians. Small cell lung cancer. Chest.
123(Suppl): 259S–271S. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
National Comprehensive Cancer Network:
National Comprehensive Cancer Network practice guidelines in
oncology v. 1.2007: small cell lung cancer. Available at:
http://www.nccn.org/professionals/physician_gls/PDF/sclc.pdfuri.
Accessed January 9, 2007.
|
|
4.
|
Hyde L, Yee J, Wilson R and Panto ME: Cell
type and the natural history of lung cancer. JAMA. 193:52–54. 1965.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Lassen U, Osterlind K, Hansen M,
Dombernowsky P, Bergman B and Hansen HH: Long term survival in
small-cell lung cancer: posttreatment characteristics in patients
surviving 5 to 18+ years – an analysis of 1,714 consecutive
patients. J Clin Oncol. 13:1215–1220. 1995.PubMed/NCBI
|
|
6.
|
Rawson NS and Peto J: An overview of
prognostic factors in small cell lung cancer. A report from the
Subcommittee for the Management of Lung Cancer of the United
Kingdom Coordinating Committee on Cancer Research. Br J Cancer.
61:597–604. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Knez L, Sodja E, Kern I, Košnik M and
Cufer T: Predictive value of multidrug resistance protein,
topoisomerases II and ERCC1 in small cell lung cancer: A systematic
review. Lung Cancer. 72:271–279. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Kin YH, Ishii G, Goto K, Ota S, Kubota K,
Murata Y, Mishima M, Saijo N, Nishiwaki Y and Ochiai A: Expression
of the breast cancer resistance protein is associated with a poor
clinical outcome in patients with small-cell lung cancer. Lung
Cancer. 65:105–111. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicated that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Zeng Y, Yi R and Cullen BR: MicroRNAs and
small interfering RNAs can inhibit mRNA expression by similar
mechanisms. Proc Natl Acad Sci USA. 100:9779–9784. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Zeng Y, Wagner EJ and Cullen BR: Both
natural and designed microRNAs can inhibit the expression of
cognate mRNAs when expressed in human cells. Mol Cell. 9:1327–1333.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Chin LJ, Ratner E, Leng S, et al: A SNP in
a let-7 microRNA complementary site in the KRAS 3′ untranslated
region increases non-small cell lung cancer risk. Cancer Res.
68:8535–8540. 2008.PubMed/NCBI
|
|
15.
|
Brendle A, Lei H, Brandt A, Johansson R,
Enquist K, Henriksson R, Hemminki K, Lenner P and Försti A:
Polymorphisms in predicted microRNA-binding sites in integrin genes
and breast cancer: ITGB4 as prognostic marker. Carcinogenesis.
29:1394–1399. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Yu Z, Li Z, Jolicoeur N, Zhang L, Fortin
Y, Wang E, Wu M and Shen SH: Aberrant allele frequencies of the
SNPs located in microRNA target sites are potentially associated
with human cancers. Nucleic Acids Res. 35:4535–4541. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Landi D, Gemignani F, Naccarati A, Pardini
B, Vodicka P, Vodickova L, Novotny J, Forsti A, Hemminki K, Canzian
F and Landi S: Polymorphisms within microRNA-binding sites and risk
of sporadic colorectal cancer. Carcinogenesis. 29:579–584. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Saunders MA, Liang H and Li W-H: Human
polymorphism at microRNA and microRNA target sites. Proc Natl Acad
Sci USA. 9:3300–3305. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Gao Y, He Y, Ding J, et al: An
insertion/deletion polymorphism at miRNA-122-binding site in the
interleukin-1alpha 3′ untranslated region confers risk for
hepatocellular carcinoma. Carcinogenesis. 30:2064–2069.
2009.PubMed/NCBI
|
|
20.
|
Fang J, Feng Q, Ketel CS, Wang H, Cao R,
Xia L, Erdjument-Bromage H, Tempst P, Simon JA and Zhang Y:
Purification and functional characterization of SET8, a nucleosomal
histone H4-lysine 20-specific methyltransferase. Curr Biol.
12:1086–1099. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Nishioka K, Rice JC, Sarma K,
Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P,
Tempst P, Steward R, Lis JT, Allis CD and Reinberg D: PR-Set7 is a
nucleosome-specific methyltransferase that modifies lysine 20 of
histone H4 and is associated with silent chromatin. Mol Cell.
9:1201–1213. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Wu S, Wang W, Kong X, Congdon LM, Yokomori
K, Kirschner MW and Rice JC: Dynamic regulation of the PR-Set7
histone methyltransferase is required for normal cell cycle
progression. Genes Dev. 24:2531–2542. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Song F, Zheng H, Liu B, Wei S, Dai H,
Zhang L, Calin GA, Hao X, Wei Q, Zhang W and Chen K: An
miR-502-binding site single-nucleotide polymorphism in the
3′-untranslated region of the SET8 gene is associated with early
age of breast cancer onset. Clin Cancer Res. 19:6292–6300.
2009.
|
|
24.
|
Guo Z, Wu C, Wang X, Wang C, Zhang R and
Shan B: A polymorphism at the miR-502 binding site in the
3’-untranslated region of the histone methyltransferase SET8 is
associated with hepatocellular carcinoma outcome. Int J Cancer. Nov
18–2011. View Article : Google Scholar : (E-pub ahead of
print).
|
|
25.
|
Shi X, Kachirskaia I, Yamaguchi H, West
LE, Wen H, Wang EW, Dutta S, Appella E and Gozani O: Modulation of
p53 function by SET8-mediated methylation at lysine 382. Mol Cell.
27:636–46. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Merritt WM, Lin YG, Han LY, et al: Dicer,
Drosha, and outcomes in patients with ovarian cancer. N Engl J Med.
359:2641–2650. 2008. View Article : Google Scholar : PubMed/NCBI
|