Introduction
Lung cancer is the most frequent cancer and one of
the leading causes of cancer-related deaths worldwide, accounting
for 30% of all cancer-related deaths. An epidemiological study
estimated that the number of deaths due to lung cancer in 2010 was
1.5 million, rendering lung cancer a major public health challenge
(1,2). The annual mortality rate of lung
cancer in China is high with approximately 400 thousand deaths
(3). Non-small cell lung cancer
(NSCLC) accounts for 89% of all lung cancers and approximately one
third of NSCLC patients are diagnosed at a locally advanced stage
(4,5). Despite aggressive treatment, the
prognosis of NSCLC patients is still poor with a 5-year survival
rate of approximately 10% and a median survival time of 16–18
months (6,7). Many clinical factors, such as tumor
stage, metastasis, gender and weight loss, are predictors of
prognosis of NSCLC patients (8),
but there are few studies concerning the relationship between
oxidative markers and NSCLC prognosis (9).
Lung cancer carcinogenesis is associated with
increased oxidative stress which results in DNA damage (10,11).
The human mitochondrial genome is a 16-kb closed-circular duplex
molecule that contains 37 genes, including two ribosomal RNAs and a
complete set of 22 tRNAs (12).
Mitochondrial DNA (mtDNA) is believed to be more susceptible to DNA
damage and acquires mutations at a higher rate than nuclear DNA
because of high levels of reactive oxygen species (ROS), lack of
protective histones and the limited capacity for DNA repair in
mitochondria (13–15). Somatic mtDNA mutations and
polymorphisms are associated with a wide variety of degenerative
diseases and cancers (16,17), and can be homoplasmic by clonal
expansion (18,19), or heteroplasmic in tumor tissues
(20,21). In many cancers, somatic mutations
and polymorphisms are located in an mtDNA non-coding region called
the displacement loop (D-loop) (22,23),
which contains 1122 bp (nucleotides 16024–16569 and 1–576;
www.mitomap.org). This region is important for the
regulation of both replication and expression of the mitochondrial
genome as it contains the leading-strand origin of replication and
the main promoter for transcription (24).
Sequence changes have been examined extensively in
the D-loop in cancers, but few single-nucleotide polymorphisms
(SNPs) have been selected for predicting cancer risk and outcome;
their predictive values are still unclear (25–29).
In this study, we assessed the prediction power of these SNPs on
the outcome of NSCLC patients.
Materials and methods
Tissue specimens and DNA extraction
Blood samples were collected at The Fourth Hospital
of Hebei University from NSCLC patients who received treatment at
the Department of Respiratory Medicine between 2001 and 2009. The
genomic DNA was immediately extracted using the Wizard Genomic DNA
extraction kit (Promega, Madison, WI, USA) and stored at −20°C. All
procedures were supervised and approved by the Human Tissue
Research Committee of our hospital, and an informed consent was
obtained from all participants.
PCR amplification and sequence
analysis
The forward primer, 5′-CCCCATGCTTACAAGCAAGT-3′
(nucleotide 16190–16209) and reverse, 5′-GCTTTGAGGAGGTAAGC TAC-3′
(nucleotide 602–583) were used for amplification of a 982-bp
product from the mtDNA D-loop region. PCR was performed according
to the protocol included in the PCR Master Mix kit (Promega) and
purified prior to sequencing. Cycle sequencing was carried out with
the Dye Terminator Cycle Sequencing Ready Reaction kit (Applied
Biosystems, Foster City, CA, USA) and the products were then
separated on the ABI PRISM Genetic Analyzer 3100 (Applied
Biosystems). Polymorphisms were confirmed by repeated analyses from
both strands.
Statistical analysis
Survival curves were calculated using the
Kaplan-Meier method, and compared with the log-rank test.
Multivariate survival analysis was performed using a Cox
proportional hazards model. All of the statistical analysis was
carried out with the SPSS 13.0 software package (SPSS Co., Chicago,
IL, USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
A total of 80 patients were enrolled in this study
and a follow-up review was conducted every 3 months for 2 years.
One patient was lost to follow-up at the first year and 3 patients
were lost at the second year. The remaining 76 patients shared the
same performance status (ECOG score, 0). Of these, 20 patients were
at stage III, 56 at stage IV and 56 died during follow-up. The data
collected during the 2-year follow-up were analyzed for clinical
characteristics using the Kaplan-Meier method and were compared by
the log-rank test. Gender, age, TNM classification, smoking and
histology were not statistically significant predictors of the
length of overall survival, however treatment was correlated with
survival in these patients (Table
I).
 | Table I.Univariate analysis of clinical
characteristics associated with overall survival in the NSCLC
patients. |
Table I.
Univariate analysis of clinical
characteristics associated with overall survival in the NSCLC
patients.
| Characteristics | No. of cases | 2-year survival rate
(%) | p-value |
|---|
| Gender | | | 0.382 |
| Male | 48 | 77.1 | |
| Female | 28 | 67.9 | |
| Age (years) | | | 0.715 |
| ≤45 | 7 | 28.6 | |
| >45 | 69 | 26.1 | |
| TNM
classification | | | 0.520 |
| III | 20 | 25.0 | |
| IV | 56 | 26.8 | |
| Smoking | | | 0.253 |
| Yes | 36 | 19.4 | |
| No | 40 | 32.5 | |
| Treatmenta | | | 0.000 |
| Yes | 67 | 29.9 | |
| No | 9 | 0 | |
| Histology | | | 0.669 |
| SQ | 25 | 28.0 | |
| AC | 51 | 25.5 | |
We sequenced the D-loop region in all of the 76
NSCLC patients and 121 SNPs were identified. The relationship
between survival and the 121 SNPs was examined. The NSCLC patients
were divided into two groups on the basis of their genotype at each
SNP site, and overall survival curve was plotted using the
Kaplan-Meier method for all NSCLC patients at these sites. A
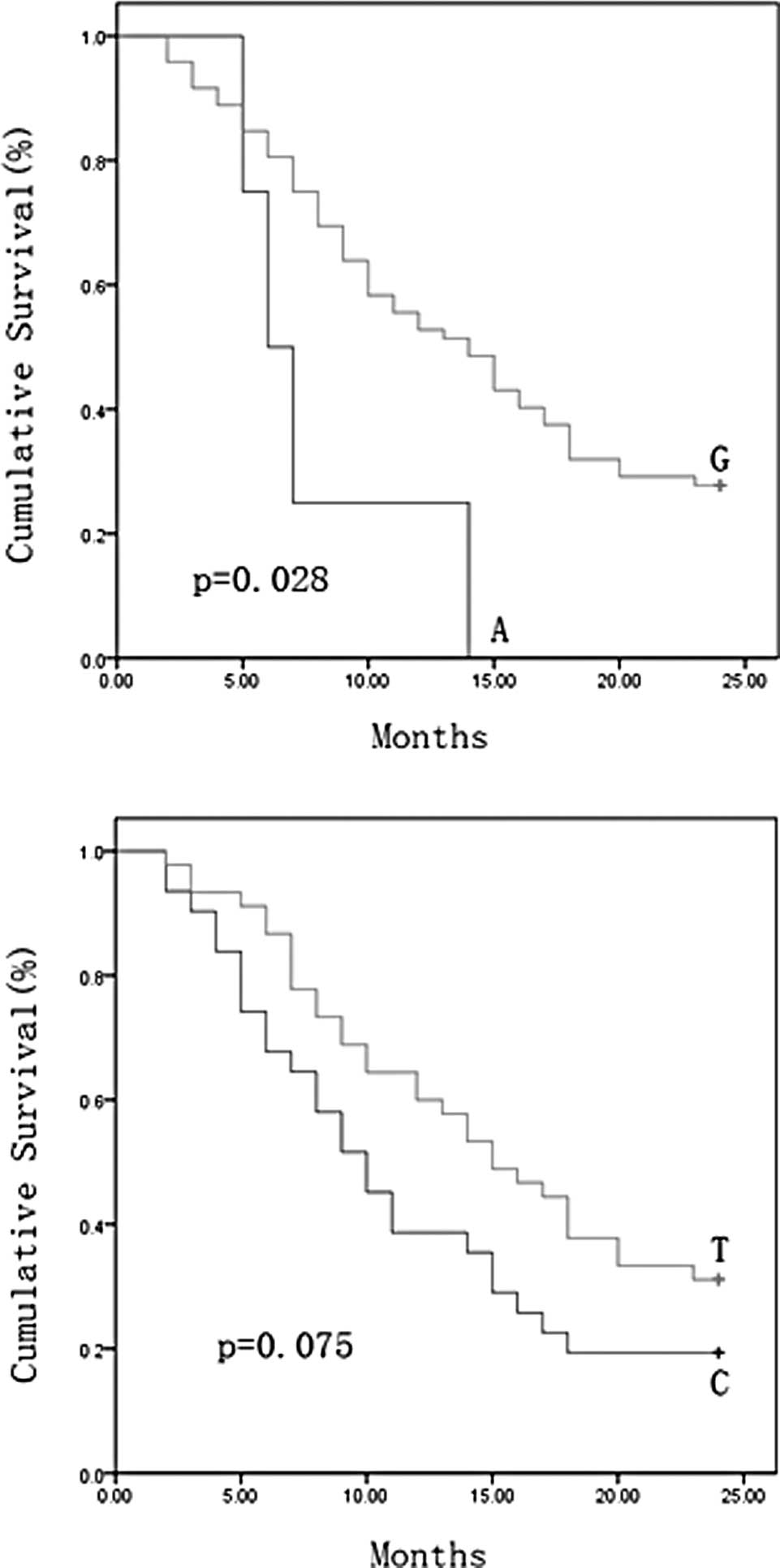
dramatic difference in survival rate was found for nucleotide 16390
(p=0.028) and another nucleotide of 16519 was also identified with
borderline level of difference (p=0.075). The minor allele 16390A
and 16519C were associated with a shorter length of survival
(Fig. 1A and B). We performed
multivariate analysis for these predictors including these two SNPs
and treatment with the Cox proportional hazards model. As shown in
Table II, the 16390 alleles and
treatment were identified as independent predictors for NSCLC
outcome. The length of survival for patients with the minor allele
16390A genotype was significantly less than that for patients with
the common allele 16390G (relative risk, 0.323; 95% CI,
0.109–0.951; p=0.040) at the 16390 site. These data demonstrated
the strong prediction power of nucleotide 16390 on outcome for
NSCLC patients.
 | Table II.Multivariate analysis of prognostic
factors associated with overall survival in NSCLC patients with Cox
proportional hazards model. |
Table II.
Multivariate analysis of prognostic
factors associated with overall survival in NSCLC patients with Cox
proportional hazards model.
| Factors | Relative risk | 95% CI | p-value |
|---|
| Treatment | 0.143 | 0.065–0.315 | 0.000 |
| Nucleotide | | | |
| 16390 | 0.323 | 0.109–0.951 | 0.040 |
| 16519 | 0.628 | 0.361–1.093 | 0.100 |
Discussion
We previously identified cancer risk and
outcome-associated SNPs of the D-loop in several types of cancer
(25,26,30).
In this study, selected SNPs in the mtDNA D-loop were examined for
their ability to predict cancer outcome in NSCLC patients. Two
SNPs, 16390 (G/A) and 16519 (T/C), were identified by the log-rank
test for their association with overall survival. Multivariate
survival analysis identified 16390 (G/A) as independent prediction
markers for NSCLC outcome.
The mtDNA D-loop is important for regulation of
mitochondrial genome replication and expression. SNPs in this
region may affect mtDNA replication so as to alter the electron
transport chain, which is responsible for the high ROS release and
nuclear genome damage as well as cancer initiation and promotion
(31–33). Overexpression of protein binding to
the D-loop region has been proven to increase ROS and affect tumor
progression (34).
In conclusion, SNPs in the mtDNA D-loop were found
to be independent prognostic markers for NSCLC outcome. The
analysis of genetic polymorphisms in the D-loop may help to
identify patient subgroups at a high risk for poor disease outcome,
thereby helping to refine therapeutic decisions in NSCLC.
Acknowledgements
This study was supported by the
National Natural Science Foundation of China (no. 30801384).
References
|
1.
|
Parkin DM, Bray FI and Devesa SS: Cancer
burden in the year 2000. The global picture. Eur J Cancer. 37(Suppl
8): S4–S66. 2001.PubMed/NCBI
|
|
2.
|
Cao C, Zhang YM, Wang R, Sun SF, Chen ZB,
Ma HY, Yu YM, Ding QL, Shu LH and Deng ZC: Excision repair cross
complementation group 1 polymorphisms and lung cancer risk: a
meta-analysis. Chin Med J (Engl). 124:2203–2208. 2011.PubMed/NCBI
|
|
3.
|
Yang L, Yang G, Zhou M, Smith M, Ge H,
Boreham J, Hu Y, Peto R, Wang J and Chen Z: Body mass index and
mortality from lung cancer in smokers and nonsmokers: a nationally
representative prospective study of 220,000 men in China. Int J
Cancer. 125:2136–2143. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Gandara D, Narayan S, Lara PN Jr, Goldberg
Z, Davies A, Lau DH, Mack P, Gumerlock P and Vijayakumar S:
Integration of novel therapeutics into combined modality therapy of
locally advanced non-small cell lung cancer. Clin Cancer Res.
11:5057s–5062s. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Yin M, Liao Z, Huang YJ, Liu Z, Yuan X,
Gomez D, Wang LE and Wei Q: Polymorphisms of homologous
recombination genes and clinical outcomes of non-small cell lung
cancer patients treated with definitive radiotherapy. PLoS One.
6:e200552011. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Yang P, Allen MS, Aubry MC, Wampfler JA,
Marks RS, Edell ES, Thibodeau S, Adjei AA, Jett J and Deschamps C:
Clinical features of 5,628 primary lung cancer patients: experience
at Mayo Clinic from 1997 to 2003. Chest. 128:452–462. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Cullen MH, Billingham LJ, Woodroffe CM,
Chetiyawardana AD, Gower NH, Joshi R, Ferry DR, Rudd RM, Spiro SG,
Cook JE, et al: Mitomycin, ifosfamide, and cisplatin in
unresectable non-small-cell lung cancer: effects on survival and
quality of life. J Clin Oncol. 17:3188–3194. 1999.PubMed/NCBI
|
|
8.
|
Bi N, Yang M, Zhang L, Chen X, Ji W, Ou G,
Lin D and Wang L: Cyclooxygenase-2 genetic variants are associated
with survival in unresectable locally advanced non-small cell lung
cancer. Clin Cancer Res. 16:2383–2390. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Gupta A, Srivastava S, Prasad R, Natu SM,
Mittal B, Negi MP and Srivastava AN: Oxidative stress in non-small
cell lung cancer patients after chemotherapy: association with
treatment response. Respirology. 15:349–356. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Loft S, Svoboda P, Kawai K, Kasai H,
Sørensen M, Tjønneland A, Vogel U, Møller P, Overvad K and
Raaschou-Nielsen O: Association between 8-oxo-7,8-dihydroguanine
excretion and risk of lung cancer in a prospective study. Free
Radic Biol Med. Oct 20–2011.(E-pub ahead of print).
|
|
11.
|
Lawless MW, O’Byrne KJ and Gray SG:
Oxidative stress induced lung cancer and COPD: opportunities for
epigenetic therapy. J Cell Mol Med. 13:2800–2821. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Shadel GS and Clayton DA: Mitochondrial
DNA maintenance in vertebrates. Annu Rev Biochem. 66:409–435. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
DiMauro S and Schon EA: Mitochondrial DNA
mutations in human disease. Am J Med Genet. 106:18–26. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Beal MF: Mitochondia, free radicals, and
neurodegeneration. Curr Opin Neurobiol. 6:661–666. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Lightowlers RN, Chinnery PF, Turnbull DM
and Howell N: Mammalian mitochondrial genetics: heredity,
heteroplasmy and disease. Trends Genet. 13:450–455. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Wallace DC: Mouse models for mitochondrial
disease. Am J Med Genet. 106:71–93. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Fliss MS, Usadel H, Caballero OL, Wu L,
Buta MR, Eleff SM, Jen J and Sidransky D: Facile detection of
mitochondrial DNA mutations in tumors and bodily fluids. Science.
287:2017–2019. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Nomoto S, Yamashita K, Koshikawa K, Nakao
A and Sidransky D: Mitochondrial D-loop mutation as clonal markers
in multicentric hepatocellular carcimona and plasma. Clin Cancer
Res. 8:481–487. 2002.PubMed/NCBI
|
|
19.
|
Mambo E, Gao X, Cohen Y, Guo Z, Talalay P
and Sidransky D: Electrophile and oxidant damage of mitochondrial
DNA leading to rapid evolution of homoplasmic mutations. Proc Natl
Acad Sci USA. 100:1838–1843. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Yoneyama H, Hara T, Kato Y, Yamori T,
Matsuura ET and Koike K: Nucleotide sequence variation is frequent
in the mitochondrial DNA displacement loop region of individual
human tumor cells. Mol Cancer Res. 3:14–20. 2005.PubMed/NCBI
|
|
21.
|
Jakupciak JP, Maragh S, Markowitz ME,
Greenberg AK, Hoque MO, Maitra A, Barker PE, Wagner PD, Rom WN,
Srivastava S, Sidransky D and O’Connell CD: Performance of
mitochondrial DNA mutations detecting early stage cancer. BMC
Cancer. 8:2852008. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Nashikawa M, Nishiguchi S, Shiomi S,
Tamori A, Koh N, Takeda T, Kubo S, Hirohashi K, Kinoshita H, Sato E
and Inoue M: Somatic mutation of mitochondrial DNA in cancerous and
noncancerous liver tissue in individuals with hepatocellular
carcinoma. Cancer Res. 61:1843–1845. 2001.PubMed/NCBI
|
|
23.
|
Sanchez-Cespedes M, Parrella P, Nomoto S,
Cohen D, Xiao Y, Esteller M, Jeronimo C, Jordan RC, Nicol T and
Koch WM: Identification of a mononucleotide repeat as a major
target for mitochondrial DNA alterations in human tumors. Cancer
Res. 61:7015–7019. 2001.PubMed/NCBI
|
|
24.
|
Taanman JW: The mitochondrial genome:
structure, transcription, translation and replication. Biochim
Biophys Acta. 1410:103–123. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Zhang R, Zhang F, Wang C, Wang S, Shiao YH
and Guo Z: Identification of sequence polymorphism in the D-loop
region of mitochondrial DNA as a risk factor for hepatocellular
carcinoma with distinct etiology. J Exp Clin Cancer Res.
29:1302010. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Wang C, Zhang F, Fan H, Peng L, Zhang R,
Liu S and Guo Z: Sequence polymorphisms of mitochondrial D-loop and
hepatocellular carcinoma outcome. Biochem Biophys Res Commun.
406:493–496. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Navaglia F, Basso D, Fogar P, Sperti C,
Greco E, Zambon CF, Stranges A, Falda A, Pizzi S, Parenti A,
Pedrazzoli S and Plebani M: Mitochondrial DNA D-loop in pancreatic
cancer: somatic mutations are epiphenomena while the germline 16519
T variant worsens metabolism and outcome. Am J Clin Pathol.
126:593–601. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Wang L, Bamlet WR, de Andrade M, Boardman
LA, Cunningham JM, Thibodeau SN and Petersen GM: Mitochondrial
genetic polymorphisms and pancreatic cancer risk. Cancer Epidemiol
Biomarkers Prev. 16:1455–1459. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Wang L, McDonnell SK, Hebbring SJ,
Cunningham JM, St Sauver J, Cerhan JR, Isaya G, Schaid DJ and
Thibodeau SN: Polymorphisms in mitochondrial genes and prostate
cancer risk. Cancer Epidemiol Biomarkers Prev. 17:3558–3566. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Zhang R, Wang R, Zhang F, Wu C, Fan H, Li
Y, Wang C and Guo Z: Single-nucleotide polymorphisms in the
mitochondrial displacement loop and outcome of esophageal squamous
cell carcinoma. J Exp Clin Cancer Res. 29:1552010. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Bandy B and Davison AJ: Mitochondrial
mutations may increase oxidative stress: implications for
carcinogenesis and aging? Free Radic Biol Med. 8:523–539. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Gille JJ and Joenje H: Cell culture models
for oxidative stress: superoxide and hydrogen peroxide versus
normobaric hyperoxia. Mutat Res. 275:405–414. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Shigenaga MK, Hagen TM and Ames BN:
Oxidative damage and mitochondrial decay in aging. Proc Natl Acad
Sci USA. 91:10771–10778. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Dement GA, Maloney SC and Reeves R:
Nuclear HMGA1 nonhistone chromatin proteins directly influence
mitochondrial transcription, maintenance, and function. Exp Cell
Res. 313:77–87. 2007. View Article : Google Scholar
|