Introduction
Malignant melanoma is the most aggressive neoplasm
with severe metastatic potential. In recent decades, the incidence
of malignant melanoma has steadily increased. A particularly
worrying feature of the tumor is its increasing incidence and its
capacity for rapid metastatic spread. microRNAs represent a class
of endogenously expressed, small non-coding RNAs that regulate gene
expression (1,2). As a consequence of translation, these
mRNAs are inhibited or destabilized, resulting in downregulation of
the encoded protein. A few microRNAs have been classified as
oncogenes or tumor-suppressor genes as their expression is altered
in tumors, which in some cases has been shown to contribute to the
phenotypes of cancer cells. Recently, the microRNA-34 (miR-34)
family was identified as the mediator of tumor suppression by p53
(1). Many reports suggest that the
miR-34s contribute to inhibition of invasion or metastasis in
various tumors. These facts suggest that miR-34s play an important
role as inhibitors of tumor growth. However, the biological
characteristics of miR-34s in human malignant melanoma are not well
understood (3). In this study, we
evaluated the expression of miR-34 family members in four human
melanoma cell lines (A375, G361, C32TG and SK-MEL-24) which have
the wild-type p53 gene using real-time reverse transcription PCR.
We also examined their generative and invasive characteristics
using the cell proliferation assay and the invasion/migration
assay.
Materials and methods
Cell culture
Human melanoma cell lines A375 and G361 were
obtained from Dainippon Pharmaceutical Co., Ltd. (Osaka, Japan) and
the Health Science Research Resources Bank (Osaka, Japan),
respectively. SK-MEL-24 and C32TG were obtained from the American
Type Culture Collection (Manassas, VA, USA). These four cell lines
were confirmed to have the wild-type p53 gene status (4–6).
Cell lines were maintained in Dulbecco’s modified minimal essential
medium (DMEM) supplemented with 10% heat-inactivated fetal bovine
serum (FBS), 100 U/ml penicillin and 100 μg/ml
streptomycin.
Quantitative evaluation of miR-34s
Total RNA containing microRNA was extracted using a
mirVana™ miRNA Isolation Kit (Applied Biosystems, Foster City, CA,
USA) and cDNA was synthesized using a TaqMan® MicroRNA
Reverse Transcription Kit (Applied Biosystems). Quantitative
reverse transcription PCR (RT-PCR) for miR-34a/b/c and U6
snRNA(RNU6B) was performed according to the manufacturer’s
recommendations. The primers for miR-34a (MI0000268), miR-34b
(MI0000742), miR-34c (MI0000743) and RNU6B (NR002752) were
purchased from TaqMan® MicroRNA Assays (Applied
Biosystems). We used TaqMan® Universal PCR Master Mix,
No AmpErase® UNG (Applied Biosystems) for the real-time
PCR. Real-time RT-PCR assays were run on Thermal Cycler
Dice® TP800 (Takara Bio, Inc., Japan) with the
comparative ΔCt method (7,8). All samples were assayed in
quadruplicate and values were normalized by the respective amounts
of RNU6B expression as an endogenous control. The positive control
standard was T5, a thrombospondin-2-overexpressing human melanoma
cell line established by our laboratory (9).
Growth analyses of the cell lines
The cells were seeded at 1.0×103
cells/well in 2-cm dishes. The cell number was determined with a
Coulter Counter (Beckman Coulter, Fullerton, CA). Quadruplicate
cultures of each cell line were prepared at all time points
(10).
In vitro invasion/migration assays
Cell invasion was assayed in BD BioCoat™ Matrigel™
Invasion Chambers (24-well, 8-μm pore, Becton-Dickinson
Labware, Bedford, MA, USA). Control insert chambers were used for
migration assays. DMEM supplemented with 5% FBS was used as a
chemoattractant. Cells (2.5×103) were suspended in
serum-free DMEM and seeded onto the invasion chambers and control
chambers. After 24 h of incubation, cells were fixed with methanol
and stained with crystal violet for 15 min. Cells remaining on the
upper face of the membranes were scraped off and those on the lower
face were counted using an inverted microscope. All assays were
performed in triplicate. The results were calculated by using the
following formula: %Invasion = (mean count of invading cells)/(mean
count of migrating cells) ×100.
Statistical analysis
Statistical comparisons of data sets were analyzed
by one-way factorial ANOVA and post-hoc test (Dunnett). Data are
shown as means ± standard error of the mean (SEM). These analyses
were performed using JMP version 8 software (SAS Institute Inc.,
Cary, NC, USA). P-values <0.05 were considered to denote
statistical significance.
Results
Expression of miR-34s in the melanoma
cell lines
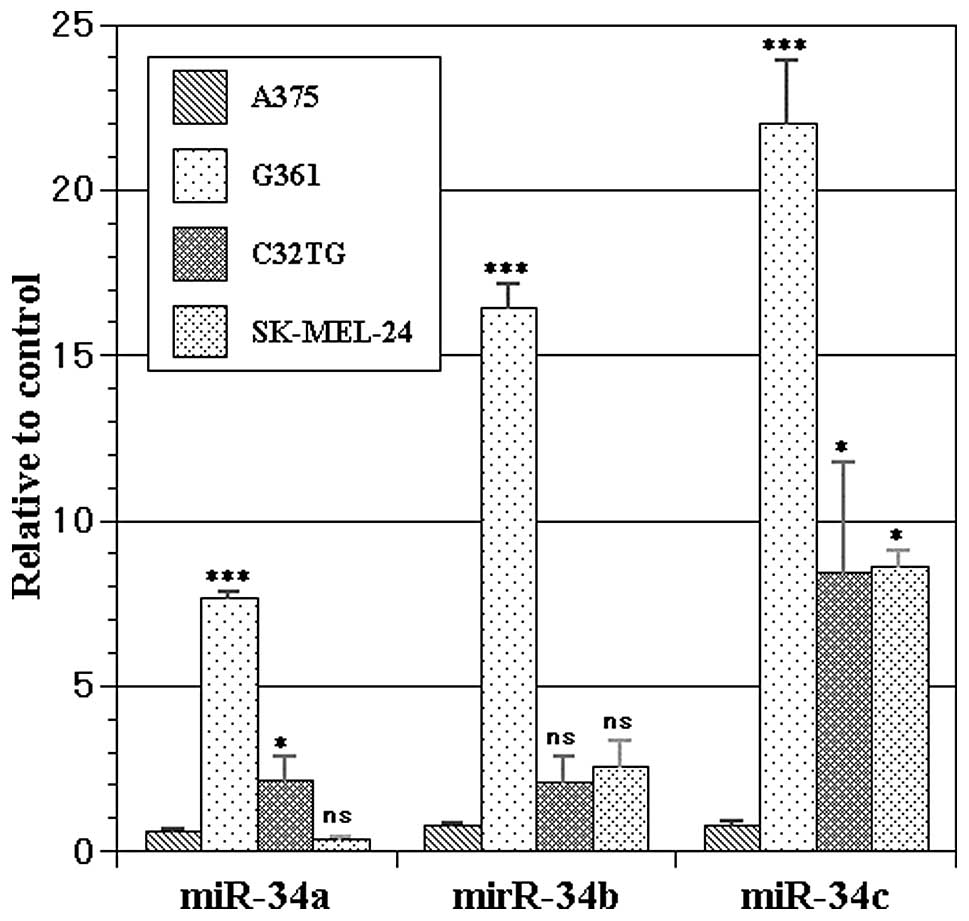
All four melanoma cell lines showed significant
expression of miR-34s as follows – A375: miR-34a 0.6176±0.0295,
miR-34b 0.7625±0.0630, miR-34c 0.7877±0.1126; G361: 7.6424±0.2011,
16.4127±0.7376, 22.0332±1.8522; C32TG: 2.1630±0.7064,
2.1091±0.7209, 8.4425±3.3104; SK-MEL-24: 0.3621±0.0559,
2.5659±0.7612, 8.5907±0.5193 (Fig.
1). Significant differences were noted in the expression of
each microRNA (ANOVA, p<0.0001 in miR-34a/b/c). Dunnett’s
post-hoc test against A375 revealed significant overexpression of
miR-34a in G361 (p<0.001) and C32TG (p=0.0302), that of miR-34b
in G361 (p<0.001), and that of miR-34c in G361 (p<0.001),
C32TG (p=0.0387) and SK-MEL-24 cells (p=0.0351).
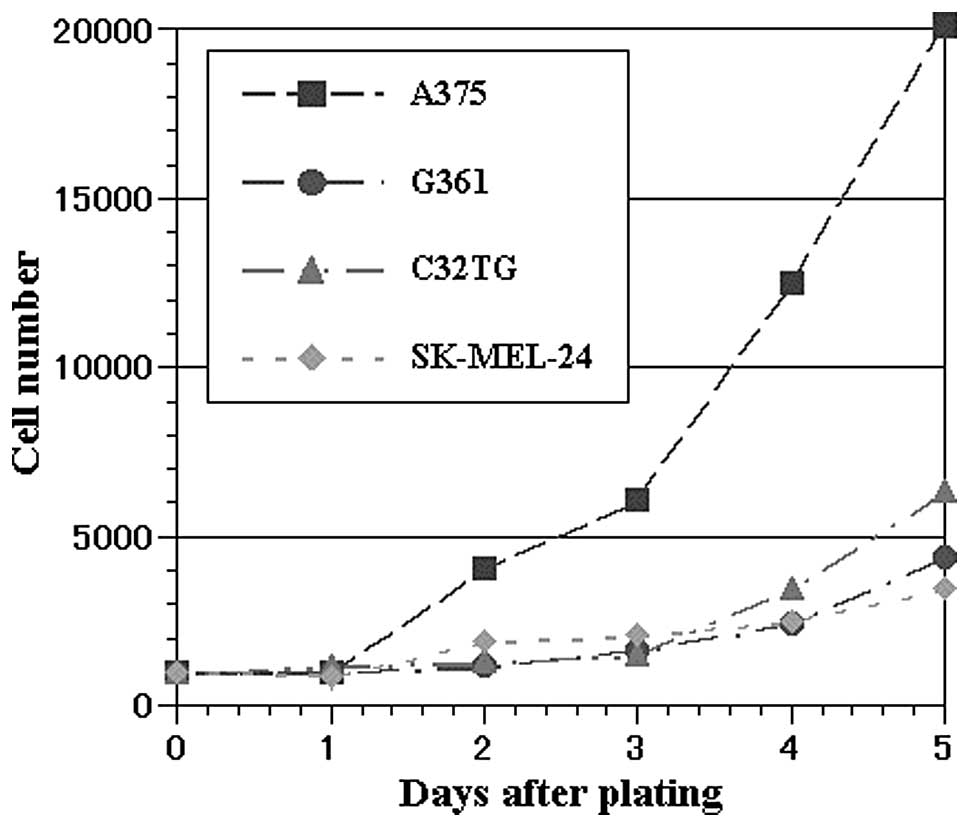
Growth characteristics of malignant
melanoma cell lines
We examined the growth characteristics of the four
cell lines in cell culture conditions (Fig. 2). Their cell doubling times were as
follows: A375 23:23, G361 68:24, C32TG 47:22 and SK-MEL-24 67:03
(values shown are h:min).
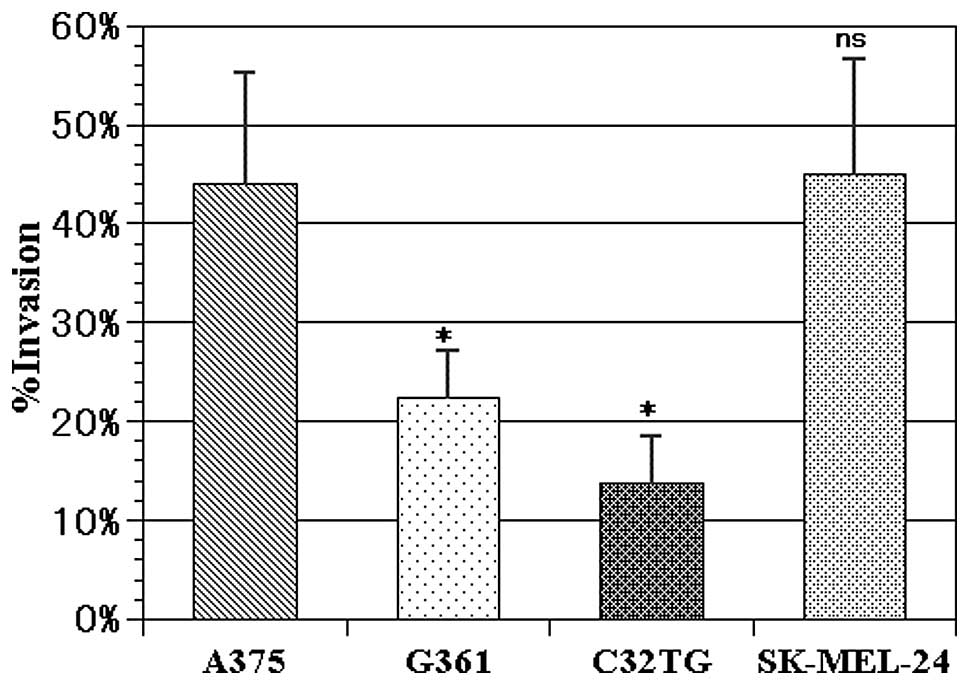
In vitro invasion/migration assays
To investigate the invasive ability of the four cell
lines, Matrigel™ invasion assays were performed (Fig. 3). The percent invasion of each cell
line was as follows: A375, 44.00±6.52%; G361, 22.37±2.71%; C32TG,
13.76±2.75%; SK-MEL-24, 45.05±6.71%. These results revealed
significant differences (ANOVA, p=0.005). Dunnett’s post-hoc test
against A375 showed significant differences in G361 (p=0.0405) and
C32TG (p=0.0074).
Discussion
In the present study, we examined the biological
role of the miR-34 family in four human malignant melanoma cell
lines (A375, G361, C32TG, SK-MEL-24). Real-time PCR revealed that
all four melanoma cell lines showed significant expression of the
miR-34s, although expression levels of all miR-34s in the A375 cell
line were extremely low. Compared with A375, the expression levels
of miR-34a in G361 and C32TG cells, miR-34b in the G361 cells, and
miR-34c in the G361, C32TG and SK-MEL-24 cell lines were
significantly high. The proliferative ability of A375 was higher
than that of the other three cell lines. The in vitro
invasiveness of A375 and SK-MEL-24 was greater than that of the
G361 and C32TG cell lines. These results suggest that
overexpression of miR-34a and c suppresses the invasive and
generative potential, respectively, of human malignant
melanoma.
p53 is activated by the deregulated expression of
oncogenes, which induce replication stress and thereby DNA damage
(11). Apart from the direct
repressive effects of p53 on core promoters, the induction of
microRNAs represents an attractive mechanism for the downregulation
of proteins observed after p53 activation (12). MicroRNAs form a class of
endogenously expressed, small non-coding RNAs with a recently
established key role in the post-transcriptional regulation of gene
expression (13–15). microRNA family miR-34s, known for
their role in the p53 tumor-suppressor network, are controlled in a
tissue-specific manner by p53 directly, inducing apoptosis, cell
cycle arrest and senescence (1,2,16–24).
Several target genes of miR-34s have been identified (18,25,26).
The miR-34 family comprises three processed microRNAs that are
encoded by two different genes: miR-34a is encoded by its own
transcript, whereas miR-34b and miR-34c share a common primary
transcript (1).
In malignant melanoma, the interplay between miR-137
and miR-182 was reported to play a key role in the MITF
(microphthalmia-associated transcription factor) regulating
network, resulting in degradation of the extracellular matrix and
controlling migration/invasion ability (3,27,28).
However, Bemis et al suggested that there may be more
microRNAs regulating MITF (27).
Lodygin et al showed that expression of miR-34a is silenced
in various tumors including malignant melanoma due to aberrant CpG
methylation of the corresponding promoter region (29). Migliore et al demonstrated
that reduced expression of miR-34b or miR-34c represents an
additional pathway for regulating the expression of the MET
oncogene in melanocytic cells (30). It was also reported that miR-34a
regulates uveal melanoma cell migration through its target gene,
c-Met (31).
Ectopic expression of miR-34a was found to cause
cell cycle arrest in the G1 phase (16,19,21).
In human colon cancer cells, tumor cells showed signs of senescence
after introduction of ectopic miR-34a (22). It is also suggested that miR-34a
inhibits cell growth and enhances chemosensitivity, as well as cell
cycle and apoptosis regulators, in prostate cancer cell lines
(32). miR-34b/c, which is also
induced by p53, was able to regulate CpG methylation in oral
squamous cell carcinoma and colorectal carcinoma (33,34).
It was also reported that miR-34b and c represent novel effectors
mediating suppression of such critical components of neoplastic
growth as cell proliferation and adhesion-independent colony
formation of neoplastic epithelial ovarian cells (18). In this study, miR-34a was inversely
correlated with invasiveness, and miR-34c reduced the proliferative
potential. Many reports have shown that miR-34b and c have similar
biological characteristics. Our results showed that expression
levels of miR-34b/c were similar to those in other studies,
although statistical differences were larger in miR-34c than b.
Consequently, there have been many hypotheses regarding the
function of miR-34a/b/c in various tumor types. The biological
characteristics of miR-34a/b/c closely resemble each other, but are
by no means identical; they are suggested to interact closely and
overlap with each other.
We showed herein that the miR-34 family reduced the
tumorigenesis of malignant melanoma, although the detailed
mechanisms are still unclear. However, there is potential to
develop new therapeutic approaches based on microRNA biology. These
methods are expected to improve the notably poor prognosis and
complete lack of effective standard therapies for malignant
melanoma.
References
|
1.
|
Hermeking H: p53 enters the microRNA
world. Cancer Cell. 12:414–418. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Fabbri M, Croce CM and Calin GA:
MicroRNAs. Cancer J. 14:1–6. 2008. View Article : Google Scholar
|
|
3.
|
Mueller DW and Bosserhoff AK: Role of
miRNAs in the progression of malignant melanoma. Br J Cancer.
101:551–556. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Yamashita T, Tokino T, Tonoki H, et al:
Induction of apoptosis in melanoma cell lines by p53 and its
related proteins. J Invest Dermatol. 117:914–919. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Min FL, Zhang H, Li WJ, Gao QX and Zhou
GM: Effect of exogenous wild-type p53 on melanoma cell death
pathways induced by irradiation at different linear energy
transfer. In Vitro Cell Dev Biol Anim. 41:284–288. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Miyato Y and Ando K: Apoptosis of human
melanoma cells by a combination of lonidamine and radiation. J
Radiat Res (Tokyo). 45:189–194. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Monney L, Sabatos CA, Gaglia JL, et al:
Th1-specific cell surface protein Tim-3 regulates macrophage
activation and severity of an autoimmune disease. Nature.
415:536–541. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Chijiwa T, Abe Y, Ikoma N, et al:
Thrombospondin 2 inhibits metastasis of human malignant melanoma
through microenvironment-modification in NOD/SCID/γCnull (NOG)
mice. Int J Oncol. 34:5–13. 2009.PubMed/NCBI
|
|
10.
|
Ikoma N, Yamazaki H, Abe Y, et al: S100A4
expression with reduced E-cadherin expression predicts distant
metastasis of human malignant melanoma cell lines in the
NOD/SCID/γCnull (NOG) mouse model. Oncol Rep. 14:633–637.
2005.PubMed/NCBI
|
|
11.
|
Dominguez-Sola D, Ying CY, Grandori C, et
al: Non-transcriptional control of DNA replication by c-Myc.
Nature. 448:445–451. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Ho J and Benchimol S: Transcriptional
repression mediated by the p53 tumour suppressor. Cell Death
Differ. 10:404–408. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Chen K and Rajewsky N: The evolution of
gene regulation by transcription factors and microRNAs. Nat Rev
Genet. 8:93–103. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Genes Dev. 20:515–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Bommer GT, Gerin I, Feng Y, et al:
p53-mediated activation of miRNA34 candidate tumor-suppressor
genes. Curr Biol. 17:1298–1307. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Chang TC, Wentzel EA, Kent OA, et al:
Transactivation of miR-34a by p53 broadly influences gene
expression and promotes apoptosis. Mol Cell. 26:745–752. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Corney DC, Flesken-Nikitin A, Godwin AK,
Wang W and Nikitin AY: MicroRNA-34b and microRNA-34c are targets of
p53 and cooperate in control of cell proliferation and
adhesion-independent growth. Cancer Res. 67:8433–8438. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
He L, He X, Lim LP, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Raver-Shapira N, Marciano E, Meiri E, et
al: Transcriptional activation of miR-34a contributes to
p53-mediated apoptosis. Mol Cell. 26:731–743. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Tarasov V, Jung P, Verdoodt B, et al:
Differential regulation of microRNAs by p53 revealed by massively
parallel sequencing: miR-34a is a p53 target that induces apoptosis
and G1-arrest. Cell Cycle. 6:1586–1593. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Tazawa H, Tsuchiya N, Izumiya M and
Nakagama H: Tumor-suppressive miR-34a induces senescence-like
growth arrest through modulation of the E2F pathway in human colon
cancer cells. Proc Natl Acad Sci USA. 104:15472–15477. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Bagchi A and Mills AA: The quest for the
1p36 tumor suppressor. Cancer Res. 68:2551–2556. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Zhou Z, Flesken-Nikitin A, Corney DC, et
al: Synergy of p53 and Rb deficiency in a conditional mouse model
for metastatic prostate cancer. Cancer Res. 66:7889–7898. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Sun F, Fu H, Liu Q, et al: Downregulation
of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett.
582:1564–1568. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Pigazzi M, Manara E, Baron E and Basso G:
miR-34b targets cyclic AMP-responsive element binding protein in
acute myeloid leukemia. Cancer Res. 69:2471–2478. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Bemis LT, Chen R, Amato CM, et al:
MicroRNA-137 targets microphthalmia-associated transcription factor
in melanoma cell lines. Cancer Res. 68:1362–1368. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Segura MF, Hanniford D, Menendez S, et al:
Aberrant miR-182 expression promotes melanoma metastasis by
repressing FOXO3 and microphthalmia-associated transcription
factor. Proc Natl Acad Sci USA. 106:1814–1819. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Lodygin D, Tarasov V, Epanchintsev A, et
al: Inactivation of miR-34a by aberrant CpG methylation in multiple
types of cancer. Cell Cycle. 7:2591–2600. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Migliore C, Petrelli A, Ghiso E, et al:
MicroRNAs impair MET-mediated invasive growth. Cancer Res.
68:10128–10136. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Yan D, Zhou X, Chen X, et al: MicroRNA-34a
inhibits uveal melanoma cell proliferation and migration through
downregulation of c-Met. Invest Ophthalmol Vis Sci. 50:1559–1565.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Fujita Y, Kojima K, Hamada N, et al:
Effects of miR-34a on cell growth and chemoresistance in prostate
cancer PC3 cells. Biochem Biophys Res Commun. 377:114–119. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Kozaki K, Imoto I, Mogi S, Omura K and
Inazawa J: Exploration of tumor-suppressive microRNAs silenced by
DNA hypermethylation in oral cancer. Cancer Res. 68:2094–2105.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Toyota M, Suzuki H, Sasaki Y, et al:
Epigenetic silencing of microRNA-34b/c and B-cell translocation
gene 4 is associated with CpG island methylation in colorectal
cancer. Cancer Res. 68:4123–4132. 2008. View Article : Google Scholar : PubMed/NCBI
|