Introduction
Carcinoma of the lung is the most common malignancy
worldwide; in 2007, it was found to have the highest incidence
among malignant diseases in the Chinese population (1–3).
Moreover, lung cancer causes over 1 million deaths worldwide each
year (4). Despite the advent of
new diagnostic techniques, most lung cancers are detected at a late
stage, and the 5-year survival rate of lung cancer is less than 15%
in the US (5). Once tumor cells
have spread, the long-term prognosis is poor since no curative
treatments are available. Thus, the development of biomarkers for
effective early diagnosis of lung cancer is clearly necessary. The
molecular biomarker is a new diagnostic technique for tumors
(6). Aberrant CpG island
methylation in the promoter region of tumor-suppressor genes is
suspected of participating in the pathogenesis and progression of
lung cancer, and its use as a biomarker offers a new approach to
ensure the early diagnosis of lung cancer (7–10).
The Ras association domain family 1 A (RASSF1A)
gene, located on chromosome 3 at band p21.3 (3p21.3), is a frequent
target for aberrant methylation in lung cancer, and
hypermethylation of the RASSF1A promoter was reported in up to 60%
of non-small cell lung cancer and 100% of small-cell lung cancer
cases (11–13). These findings suggest that RASSF1A
is a putative tumor-suppressor gene and is likely to be involved in
the genesis of lung cancer, and plays an important role in the
progression of tumorigenesis. Epigenetic modification in the short
arm of chromosome 3p loci genes, including RASSF1A and RARβ,
together have been implicated in the progression of lung
tumorigenesis in one study in a Chinese population (14).
In the present study, we used the
methylation-specific polymerase chain reaction (MSP) method to
examine the methylation status of RASSF1A and RARβ in a
South-Central Chinese Han population. We analyzed the relationship
of gene methylation patterns with clinical features and lung cancer
risk.
Materials and methods
Patients
A total of 56 patients diagnosed with lung cancer
and 52 controls without cancer were included in the present study.
All patients were recruited from the Hunan Provincial Tumor
Hospital in Changsha, China. Histological classification was
conducted according to ‘Histological Typing of Lung and Pleural
Tumors, 3rd edition’ of the World Health Organization (WHO), 1999,
and the tumor stage was determined according to the TNM staging
guideline suggested by the American Joint Committee on Cancer
(AJCC) and the Union Internationale Contre le Cancer (UICC) in
2003. Fifteen of the tumors were stage I, 7 were stage II, 20 were
stage III and 14 were stage IV histologically; 29 of the 56 tumors
were squamous cell carcinomas, 17 were adenocarcinomas and 10
included other carcinoma types. The mean age of the controls was
52.6±16.2 years.
All study subjects were of South-Central Chinese
population Han ethnicity and provided written consents for
participation; the research protocol was approved by the
Institutional Review Board of the Hunan Provincial Tumor Hospital,
Changsha, China.
DNA extraction and bisulfite
treatment
Genomic DNAs were extracted from peripheral blood
lymphocytes using a standard kit-based method (Gentra Systems,
Minneapolis, MN, USA). Genomic DNA was treated with sodium
bisulfite using the EZ DNA methylation-Gold kit (Zymo Research,
USA) to modify unmethylated cytosines to uracil. The
bisulfite-modified DNA was used immediately for PCR or stored at
−70°C.
Positive control for methylation
Lung cancer patient DNAs were treated in
vitro with excess SssI methyltransferase (New England Biolabs,
Beverly, MA, USA) to generate completely methylated DNA at all CpGs
and was used as positive control for methylated alleles of each
gene. DNA from a healthy control sample was used as the control for
unmethylated alleles. Genomic DNA was treated with sodium
bisulfite.
Methylation-specific PCR
Two sets of primers were used to amplify methylated
and unmethylated alleles, as shown in Table I. The PCR condition for MSP assays
were derived from several reports (15,16).
Lymphocyte DNA, original or methylation treated in vitro
with excess SssI methyltransferase (New England Biolabs), was used
as the unmethylation- and methylation-positive controls,
respectively. Water blank was used as a negative control.
 | Table I.Sequences of primers used in MSP. |
Table I.
Sequences of primers used in MSP.
| Gene | Primer sequence
(5′-3′) | Annealing temperature
(°C) | Product size
(bp) |
|---|
| RASSF1A | | | |
| M F |
GGGTTTTGCGAGAGCGCG | 64 | 169 |
| M R |
GCTAACAAACGCGAACCG | | |
| U F |
GGTTTTGTGAGAGTGTGTTTAG | 59 | 169 |
| U R |
CACTAACAAACACAAACCAAAC | | |
| RARβ | | | |
| M F |
TCGAGAACGCGAGCGATTCG | 62 | 146 |
| M R |
GACCAATCCAACCGAAACGA | | |
| U F |
TTGAGAATGTGAGTGATTTGA | 62 | 146 |
| U R |
AACCAATCCAACCAAAACAA | | |
Statistical analysis
Statistical analyses were performed using the
SPSS13.0 statistical software. The association between the
methylation status of the two genes and clinicopathological
parameters was analyzed using the Fisher’s or Chi-square exact
test. The association between the methylation of the two genes and
lung cancer was determined using the logistic regression method to
assess odds ratio (ORs) and 95% confidence intervals (95% CI).
p<0.05 was considered to indicate statistical significance.
Results
RASSF1A and RARβ gene promoter
hypermethylation profile
We analyzed the methylation patterns of RASSF1A and
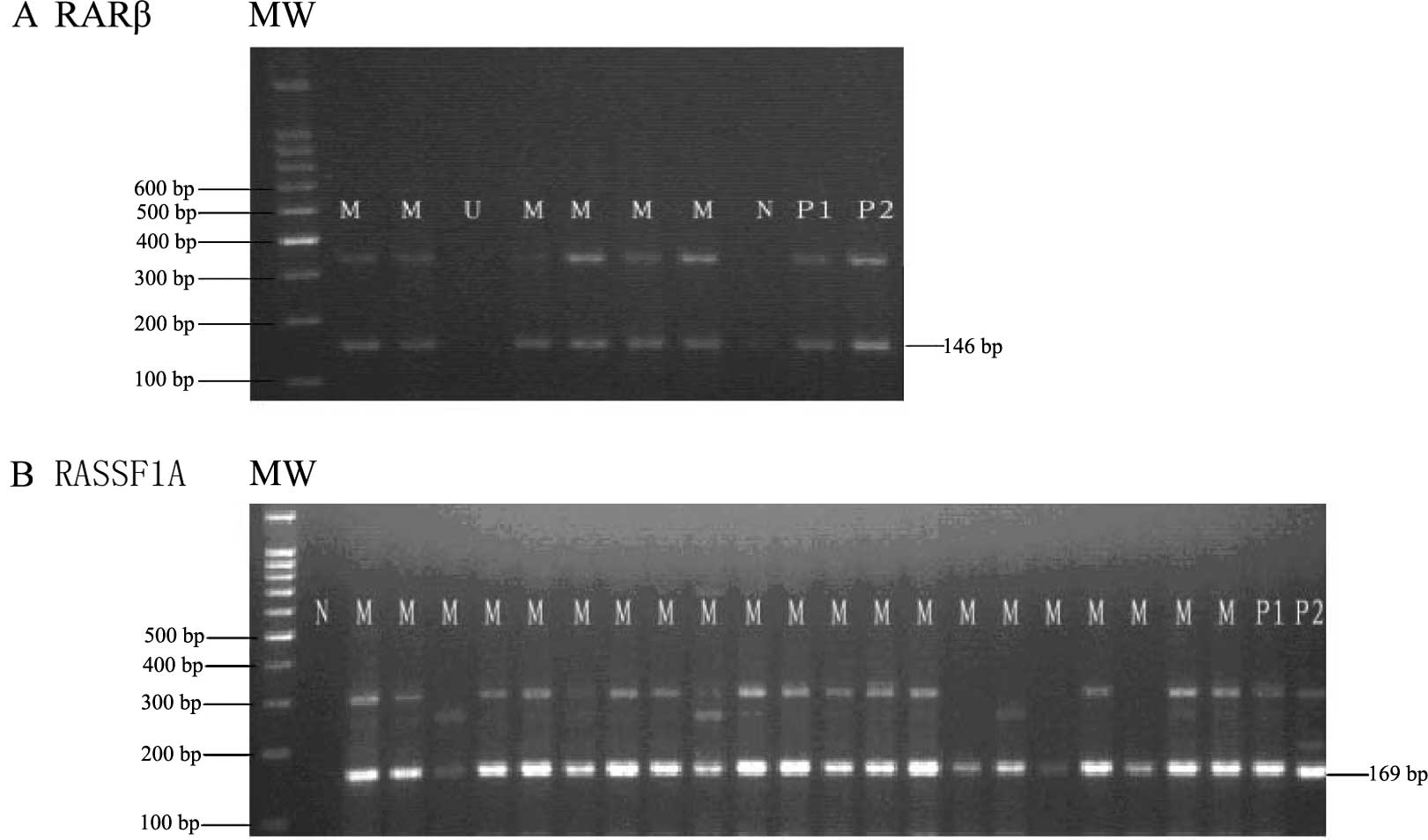
RARβ promoter regions in lung cancer cases and controls. Fig. 1 shows a typical example of the MSP
products analyzed on agarose gel for the RASSF1A and RARβ genes. We
found that the aberrant promoter methylation of the RASSF1A gene
was detected in 85.71% (48/56) of cases and the RARβ gene was
detected in 80.36% (45/56) of cases. The promoter methylation of
both genes was found in 75% (42/56) of lung cancers (Tables II and III). By contrast, none of the 52 controls
was detected to exhibit methylation in either of the two genes
(Table III). There was a
significant statistical association of the promoter methylation of
the RASSF1A gene with lung cancer risk (adjusted OR=7.50; 95% CI,
3.935–14.296; p<0.001). Similar results were obtained for
methylation of the RARβ gene (adjusted OR=5.727; 95% CI,
3.348–9.797; p<0.001). Moreover, methylation of both genes was
significantly associated with cancer risk in the cases when
compared with the controls (adjusted OR=8.429; 95% CI,
4.205–16.896; p<0.001)
 | Table II.Methylation status of RASSF1A and RARβ
genes according to clinical staging and histological grading in the
lung cancer cases. |
Table II.
Methylation status of RASSF1A and RARβ
genes according to clinical staging and histological grading in the
lung cancer cases.
| No. of cases | RASSF1A methylation
(%) | RARβ methylation
(%) | Both RASSF1A and RARβ
methylation (%) |
|---|
| Clinical stage | | | | |
| I | 15 | 13 (86.70) | 10 (66.70) | 10 (66.67) |
| II | 7 | 7 (100.0) | 6 (85.70) | 6 (85.70) |
| III | 20 | 17 (85.00) | 16 (80.00) | 15 (75.00) |
| IV | 14 | 11 (78.60) | 13 (90.90) | 11 (78.60) |
| Total | 56 | 48 (85.71) | 45 (80.36) | 42 (75.00) |
| Histological
grade | | | | |
| Squamous cell
carcinoma | 29 | 26 (89.70) | 23 (79.30) | 21 (72.40) |
| Adenocarcinoma | 17 | 14 (82.40) | 15 (88.20) | 14 (82.40) |
| Other carcinoma
typesa | 10 | 8 (80.00) | 7 (70.00) | 7 (70.00) |
| Total | 56 | 48 (85.71) | 45 (80.36) | 42 (75.00) |
 | Table III.Methylation status of the RASSF1A and
RARβ genes in the lung cancer cases and controls. |
Table III.
Methylation status of the RASSF1A and
RARβ genes in the lung cancer cases and controls.
| Gene | Methylation
status | Cases (%) | Controls (%) | p-value | OR (95% CI) |
|---|
| RASSF1A | Methylated | 48 (85.71) | 0 | <0.01a | 7.500
(3.935–14.296)a |
| Unmethylated | 8 (14.29) | 52 (100) | <0.01b | 1.929
(1.608–2.313)b |
| RARβ | Methylated | 45 (80.36) | 0 | <0.01a | 5.727
(3.348–9.797)a |
| Unmethylated | 11 (19.64) | 52 (100) | <0.01b | 1.929
(1.608–2.313)b |
| RASSF1A + RARβ (both
genes) | Methylated | 42 (75.00) | 0 | <0.01a | 8.429
(4.205–16.896)a |
| Unmethylated | 7 (12.25) | 52 (100) | <0.01b | 2.061
(1.686–2.520)b |
Clinicopathological correlation
The relationship between methylation of these two
genes and clinicopathlogical characteristics of the lung cancer
cases was analyzed and the results are documented in Table II. There was no relationship
between RASSF1A methylation status and clinicopathological
features. Similar results were obtained for methylation of the RARβ
gene. Moreover, there was no relationship between the status of
both genes being methylated and clinicopathological features.
Discussion
It is known that methylation is a major epigenetic
modification in mammals, and changes in methylation patterns play a
key role in tumorigenesis in humans. In particular, promoter CpG
island hypermethylation is closely related to inactivation and
silencing, resulting in tumor suppressor loss of gene expression
and X-chromosome inactivation, and affects the development of
carcinogenesis (17,18). Aberrant promoter region methylation
of tumor-suppressor genes is association with the mechanism for
carcinogenesis. Aberrant methylation of RASSF1A within the promoter
region has been reported in various tumor types, including lung
cancers, similar to the RARβ gene (9,11–13).
There are few studies reporting hyper-methylation of both the
RASSF1A and RARβ genes together in cancers, particularly in lung
cancer (19,20,21).
Thus, in the present study we aimed to determine whether aberrant
promoter methylation of the RASSF1A and RARβ genes is of potential
use as a molecular biomarker for lung cancer in a South-Central
Chinese Han population using MSP.
Several studies have shown separately that
methylation of CpG islands of the RASSF1A and RARβ genes has a
significant role in the development of lung cancer (20–22),
but no report in a South-Central Chinese Han population has
examined the two genes simultaneously. In the present study, we
found that the RASSF1A gene was hypermethylated in 48 out of 56
lung cancer samples. The frequency was consistent with previous
studies (9,11–13),
but higher than that found in the research of Wang et al in
primary lung cancer in a Chinese population (14). The frequency of methylation was
over 70% in all clinical stages. In agreement with this study,
several reports have shown that there is no relationship with
clinical stage and histological grade (11,14,19).
The data suggest that aberrant promoter methylation of the RASSF1A
gene is highly significantly associated with lung cancer when
compared with normal samples (p<0.01). More importantly, the
results showed that RASSF1A gene-positive carriers had a 7.5-fold
increased risk of lung cancer (adjusted OR=7.50; 95% CI,
3.935–14.296; p<0.001) in a South-Central Chinese Han
population. Thus, our data strongly support the theory that
methylation of the RASSF1A gene in lung cancer cases in a
South-Central Chinese Han population is a late event which may be
associated with carcinogenesis. In addition, promoter methylation
of the RASSF1A gene presented a significant relationship (adjusted
OR=2.00; 95% CI, 1.662–2.407; p<0.001) with lung cancer when
compared to the unmethylated and vs. the total of both (methylated
+ unmethylated) genes, thereby strengthening the relationship
between lung cancer and methylation.
Several studies have suggested that epigenetic event
of the RARβ gene may play a role in the development of lung cancer
(23–25). Our study revealed that methylation
of the RARβ gene was found in 80.36% of cases, higher than that
found in the research of Virmani et al (26), but similar to that in small-cell
lung cancers in a study by Zöchbauer-Müller et al (27). Likewise, there was no relationship
between aberrant promoter region methylation of the RARβ gene and
clinical stage/histological grade as well as the RASSF1A gene. No
methylation was detected in the controls, and the aberrant promoter
methylation of the RARβ gene had a highly significant correlation
between the cases and controls (p<0.01). Moreover, the results
showed that RARβ gene-positive carriers had a 5.7-fold increased
risk of lung cancer (adjusted OR=5.727; 95% CI, 3.348–9.797;
p<0.001) in our South-Central Chinese Han population. Thus, our
data demonstrated that RARβ methylation was also important in the
pathogenesis of lung cancer in our population. Similar to the
RASSF1A gene, promoter methylation of the RARβ gene had a
significant (adjusted OR=2.00; 95% CI, 1.662–2.407; p<0.001)
association with lung cancer when ompared to the unmethylated and
vs. the total of both (methylated + unmethylated) genes.
Aberrant methylation of the promoter region of RARβ
and RASSF1A genes is a common epigenetic event in chromosome 3 in
lung cancer. In the present study, the aberrant promoter
methylation of both genes was found in 75% (42/56) of lung cancer
cases. By researching the methylation profile of the RARβ and
RASSF1A genes for different clinical stages/ histological grades of
lung cancer, no significant difference in methylation frequency was
found. The association remained significant between cases and
controls (p<0.01). The results demonstrated that positive
carriers of both genes had an 8.4-fold increased risk of lung
cancer (adjusted OR=8.429; 95% CI, 4.205–16.896; p<0.001) in our
South-Central Chinese Han population. The risk of lung cancer for
positive carriers with both genes was higher than for positive
carriers of a single gene in the South-Central Chinese Han
population. In addition, promoter methylation of both genes had a
significant (adjusted OR=2.061; 95% CI, 1.686–2.520; p<0.001)
relationship with lung cancer compared to the unmethylated and vs.
the total of both (methylated + unmethylated) genes.
In conclusion, to our knowledge this is the first
study to indicate an association between the results of the MSP
analysis of RARβ and RASSF1A genes with lung cancer in a
South-Central Chinese Han population. The high percentage of
promoter methylation in the RARβ and RASSF1A genes indicates their
important role in the development of lung cancer in the studied
population. The risk of lung cancer for positive carriers of both
genes was higher than for positive carriers of a single gene. This
offers a potential marker for the prognosis as well as a target for
the treatment of lung cancer.
Acknowledgements
This study was supported by the
National Natural Science Foundation of China (60871007 and
61171061), the Natural Science Foundation of Hunan Province of
China (10JJ2049 and 10JJ3083), and the Science and Technology
Planning Project of Hunan Province (2011NK2006 and 2011SK3132).
References
|
1.
|
Jemal A, Siegel R, Ward E, Murray T, Xu J,
Smigal C and Thun MJ: Cancer Statistics. CA Cancer J Clin.
56:106–130. 2006.
|
|
2.
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar
|
|
3.
|
Jin YT, Xu YC, Yang RD, Huang CF, Xu CW
and He XZ: Familial aggregation of lung cancer in a high incidence
area in China. Br J Cancer. 92:1321–1325. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Estimating the world cancer burden: Globocan 2000. Int J Cancer.
94:153–156. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Jemal A, Tiwael RC, Murray T, et al:
Cancer statistics. CA Cancer J Clin. 54:92–119. 2004.
|
|
6.
|
Hirsch FR, Merrick DT and Franklin WA:
Role of biomarkers for early detection of lung cancer and
chemoprevention. Eur Respir J. 19:1151–1158. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Anglim PP, Alonzo TA and Laird-Offringa
IA: DNA methylation-based biomarkers for early detection of
non-small cell lung cancer: an update. Mol Cancer. 23:812008.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Nephew KP: What will it take to obtain DNA
methylation markers for early cervical cancer detection. Gynecol
Oncol. 112:291–292. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Zhang Y, Wang R, Song H, Huang G, Yi J,
Zheng Y, Wang J and Chen L: Methylation of multiple genes as a
candidate biomarker in non-small cell lung cancer. Cancer Lett.
303:21–28. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Choi JE, Kim DS, Kim EJ, Chae MH, Cha SI,
Kim CH, Jheon S, Jung TH and Park JY: Aberrant methylation of
ADAMTS1 in non-small cell lung cancer. Cancer Genet Cytogenet.
187:80–84. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Burbee DG, Forgacs E, Zochbauer-Muller S,
Shivakumar L, Fong K and Gao B: Epigenetic inactivation of RASSF1A
in lung and breast cancers and malignant phenotype suppression. J
Natl Cancer Inst. 93:691–699. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Honorio S, Agathanggelou A, Schuermann M,
Pankow W, Viacava P and Maher ER: Detection of RASSF1A aberrant
promoter hypermethylation in sputum from chronic smokers and ductal
carcinoma in situ from breast cancer patients. Oncogene.
22:147–150. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Grote HJ, Schmiemann V, Geddert H, Bocking
A, Kappes R and Gabbert HE: Methylation of RAS association domain
family protein 1A as a biomarker of lung cancer. Cancer.
108:129–134. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Wang Y, Yu Z, Wang T, Zhang J, Hong L and
Chen L: Identification of epigenetic aberrant promoter methylation
of RASSF1A in serum DNA and its clinicopathological significance in
lung cancer. Lung Cancer. 56:289–294. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Maruyama R, Toyooka S, Toyooka KO, et al:
Aberrant promoter methylation profile of prostate cancers and its
relationship to clinicopathological features. Clin Cancer Res.
8:514–519. 2002.PubMed/NCBI
|
|
16.
|
Neyaz MK, Kumar RS, Hussain S, et al:
Effect of aberrant promoter methylation of FHIT and RASSF1A genes
on susceptibility to cervical cancer in a North Indian population.
Biomarkers. 13:597–606. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Baylin SB: DNA methylation and gene
silencing in cancer. Nat Clin Pract Oncol. 2(Suppl 1): 4–11. 2005.
View Article : Google Scholar
|
|
18.
|
Hesson LB, Cooper WN and Latif F: The role
of RASSF1A methylation in cancer. Dis Markers. 23:73–87. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Fischer JR, Ohnmacht U, Rieger N, Zemaitis
M, Stoffregen C, Kostrzewa M, Buchholz E, Manegold C and Lahm H:
Promoter methylation of RASSF1A, RARbeta and DAPK predict poor
prognosis of patients with malignant mesothelioma. Lung Cancer.
54:109–116. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Toyooka S, Suzuki M, Maruyama R, Toyooka
KO, Tsukuda K, Fukuyama Y, Iizasa T, Aoe M, Date H, Fujisawa T,
Shimizu N and Gazdar AF: The relationship between aberrant
methylation and survival in non-small-cell lung cancers. Br J
Cancer. 91:771–774. 2004.PubMed/NCBI
|
|
21.
|
Toyooka S, Suzuki M, Tsuda T, Toyooka KO,
Maruyama R, Tsukuda K, Fukuyama Y, Iizasa T, Fujisawa T, Shimizu N,
Minna JD and Gazdar AF: Dose effect of smoking on aberrant
methylation in non-small cell lung cancers. Int J Cancer.
110:462–464. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Toyooka S, Maruyama R, Toyooka KO,
McLerran D, Feng Z, Fukuyama Y, Virmani AK, Zochbauer-Muller S,
Tsukuda K, Sugio K, et al: Smoke exposure, histologic type and
geography-related differences in the methylation profiles of
non-small cell lung cancer. Int J Cancer. 103:153–160. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Shaw RJ, Liloglou T, Rogers SN, Brown JS,
Vaughan ED, Lowe D, Field JK and Risk JM: Promoter methylation of
P16, RARbeta, E-cadherin, cyclin A1 and cytoglobin in oral cancer:
quantitative evaluation using pyrosequencing. Br J Cancer.
94:561–568. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Chan EC, Lam SY, Tsang KW, Lam B, Ho JC,
Fu KH, Lam WK and Kwong YL: Aberrant promoter methylation in
Chinese patients with non-small cell lung cancer: patterns in
primary tumors and potential diagnostic application in
bronchoalveolar lavage. Clin Cancer Res. 8:3741–3746.
2002.PubMed/NCBI
|
|
25.
|
Fujiwara K, Fujimoto N, Tabata M, Nishii
K, Matsuo K, Hotta K, Kozuki T, Aoe M, Kiura K, Ueoka H and
Tanimoto M: Identification of epigenetic aberrant promoter
methylation in serum DNA is useful for early detection of lung
cancer. Clin Cancer Res. 11:1219–1225. 2005.PubMed/NCBI
|
|
26.
|
Virmani AK, Rathi A, Zöchbauer-Müller S,
Sacchi N, Fukuyama Y, Bryant D, Maitra A, Heda S, Fong KM,
Thunnissen F, Minna JD and Gazdar AF: Promoter methylation and
silencing of the retinoic acid receptor-beta gene in lung
carcinomas. J Natl Cancer Inst. 92:1303–1307. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Zöchbauer-Müller S, Minna JD and Gazdar
AF: Aberrant DNA methylation in lung cancer: biological and
clinical implications. Oncologist. 7:451–457. 2002.PubMed/NCBI
|