Introduction
Helicobacter pylori (H. pylori) is a
gram-negative microaerophilic bacterium which is one of the most
common pathogens in humans and has a worldwide distribution. It is
associated with the development of chronic gastritis, peptic ulcer
and even gastric cancer (1). On
the basis of abundant epidemiological research, H. pylori
was classified as a class I carcinogen in humans by the World
Health Organization International Agency for Research on Cancer
(2).
Several H. pylori virulence genes that may be
associated with the risk of developing diseases have been
identified. The cagA is a marker of genomic pathogenicity
island (cagPAI) encoding the gene product which causes upregulation
of interleukin-8 (IL-8) (3). It is
considered that H. pylori strains possessing cagA are
related to a more severe clinical outcome such as atrophic
gastritis or gastric cancer (4,5). The
vacA exists in all H. pylori strains and encodes
vacuolating cell toxins which cause vacuole degeneration of
epithelial cells. It includes two different parts: the signal (s)
region encoding the signal peptide and the middle (m) region. The
s-region is situated at the 5′ end of the gene and exists as s1 and
s2 alleles. The s1 exists as an s1a, s1b and s1c. The m-region
occurs as m1 or m2 alleles (6).
The mosaic combination of s- and m-region allelic types produces
cytotoxin and is associated with the pathogenicity of the
bacterium. In general, type s1m1 and s1m2 strains produce high and
moderate levels of toxin, respectively, whereas s2m2 strains
produce little or no toxin. (7).
VacA m1 strains are associated with greater gastric
epithelial damage than m2 strains (8). Another virulence gene designated
iceA has two main allelic variants iceA1 and
iceA2 but the function of these variants is unknown.
IceA1 is upregulated upon contact of H. pylori with
the gastric epithelium and has been considered as a marker for
peptic ulcer disease (9).
In Northeast China, there are no data regarding the
pattern of H. pylori genotypes in patients. This study aimed
to investigate the prevalence of the vacA, cagA and
iceA genotypes of H. pylori from patients with upper
gastrointestinal diseases and the relationship with clinical
outcome in Northeast China.
Materials and methods
Study subjects
We evaluated 378 patients with upper
gastrointestinal diseases referred for endoscopy at the Second
Affiliated Hospital of Harbin Medical University in 2007 and 2008.
Gastric mucosal biopsy specimens were obtained from each patient:
one for pathological diagnosis, another for histological detection
of H. pylori and the last for genomic DNA extraction and
polymerase chain reaction (PCR).
The study was approved by the Ethics Committee of
Harbin Medical University. Written informed consent was obtained
from each patient prior to enrolling in the study.
Histological assessment
The biopsy samples were fixated in 10% formalin,
then sliced into 4- to 6-mm pieces, dehydrated in ethanol, embedded
in paraffin wax, sectioned (5-μm thick), and stained with
hematoxylin and eosin (H&E). The presence of H. pylori
in the sections was determined using a modified Gram staining
protocol and taking into consideration its morphological
characteristics which included a curved and spiral form and intense
purple coloring (10).
Pathological diagnoses were evaluated in a blinded manner by two
independent pathologists and were defined as gastritis (active
chronic gastritis or closed-type atrophic gastritis), gastric ulcer
and gastric cancer.
Genomic DNA extraction
DNA was extracted from the biopsy specimens using
the Genomic DNA purification system (Promega, USA) according to the
manufacturer’s instructions and stored at −20°C until analysis.
Diagnosis of H. pylori
infection
H. pylori-positive status was defined as
positive histology and positive 16S-rRNA PCR. A 500-bp region of
16S-rRNA was amplified by PCR using primers CP-1/CP-2 (Table I). Five microlitres of DNA was
added to 50 μl of reaction mixture containing 1X PCR buffer,
0.2 mM dNTPs and 0.3 μM primers as well as 1.25U Taq
polymerase (Takara Bio, Inc., Japan). The incubation conditions
were as follows: a 5-min preincubation at 95°C, followed by 30
cycles of 1 min at 94°C, 1 min at 58°C, 1 min at 72°C, and a final
5-min incubation at 72°C. Positive results were indicative of a
diagnosis of H. pylori infection.
 | Table IPrimer sequences for human HP 16S
rRNA, cagA, vacA and iceA. |
Table I
Primer sequences for human HP 16S
rRNA, cagA, vacA and iceA.
| Gene | Primer | Primer sequence
(5′→3′)a | Product size
(bp) | Reference |
|---|
| 16S rRNA | cp-1 |
GCGCAATCAGCGTCAGGTAATG | 500 | (37) |
| cp-2 |
GCTAAGAGATCAGCCTATGTCC | | |
| cagA | cagA-F |
GATAACAGGCAAGCTTTTGAGG | 349 | (18) |
| cagA-R |
CTGCAAAAGATTGTTTGGCAGA | | |
| s1a | S1a-F |
TCTYGCTTTAGTAGGAGC | 212 | (18) |
| VA1-R |
CTGCTTGAATGCGCCAAAC | | |
| s1b | SS3-R |
AGCGCCATACCGCAAGAG | 187 | (18) |
| VA1-R |
CTGCTTGAATGCGCCAAAC | | |
| s1c | S1c-F |
CTYGCTTTAGTRGGGYTA | 213 | (18) |
| VA1-R |
CTGCTTGAATGCGCCAAAC | | |
| s2 | SS2-F |
GCTAACACGCCAAATGATCC | 199 | (8) |
| VA1-R |
CTGCTTGAATGCGCCAAAC | | |
| m1 | VA3-F |
GGTCAAAATGCGGTCATGG | 290 | (38) |
| VA3-R |
CCATTGGTACCTGTAGAAAC | | |
| m2 | VA4-F |
GGAGCCCCAGGAAACATTG | 352 | (38) |
| VA4-R |
CATAACTAGCGCCTTGCAC | | |
| iceA1 | iceA1-F |
GTGTTTTTAACCAAAGTATC | 247 | (35) |
| iceA1-R |
CTATAGCCASTYTCTTTGCA | | |
| iceA2 | iceA2-F |
GTTGGGTATATCACAATTTAT | 229/334 | (35) |
| iceA2-R |
TTRCCCTATTTTCTAGTAGGT | | |
Genotyping of H. pylori
The systems of PCR were the same as mentioned above
except for the primers. The amplification cycles consisted of an
initial denaturation at 94°C for 5 min and then denaturation at
94°C for 30 sec, primer annealing at 60, 56, 58 and 48°C for
cagA, vacA (s1a, s1b, s1c and s2), vacA (m1,
m2) and iceA, respectively, for one-half minute and
extension at 72°C for 45 sec. All reactions were performed through
35 cycles. The final cycle included an extension step for 5 min.
Primers used for genotyping cagA, vacA and
iceA genes are listed in Table
I. PCR products were analyzed on 1.5% agarose gel
electrophoresis with ethidium bromide. Images were quantified via
the Gene Genius system (Syngene, England, UK). For strains that
were cagA-negative as determined by PCR, Southern blotting
was performed according to the method described by Pan et al
(11).
Statistical analyses
Statistical tests were performed with SPSS software
version 11.5 (SPSS Inc., Chicago, IL, USA). A Chi-square test and
Fisher’s exact test were used to assess the association amongst the
genotypes and between specific genotypes and upper gastrointestinal
diseases. P-values <0.05 were considered to indicate a
statistically significant result.
Results
DNA was successfully extracted from 378 gastric
mucosa tissues of patients with gastrointestinal diseases and 197
were confirmed as H. pylori infection-positive by histology
and PCR amplification. H. pylori-infected patients were
evaluated for the relationship of age and gender with disease as
shown in Table II.
 | Table IIDistribution of 197 patients with
different clinical outcomes, according to age and gender. |
Table II
Distribution of 197 patients with
different clinical outcomes, according to age and gender.
| Clinical status
| |
|---|
| Classification | GUa n=86 (%) | GSb n=58 (%) | GCc n=53 (%) | Total n=197
(%) |
|---|
| Age (years) | | | | |
| 21–30 | 7 (8.1) | 2 (3.5) | 0 (0.0) | 9 (4.6) |
| 31–40 | 12 (14.0) | 11 (19.0) | 4 (7.5) | 27 (13.7) |
| 41–50 | 33 (38.4) | 17 (29.3) | 17 (32.1) | 67 (34.0) |
| 51–60 | 24 (27.9) | 17 (29.3) | 16 (30.2) | 57 (29.0) |
| >60 | 10 (11.6) | 11 (18.9) | 16 (30.2) | 37 (18.7) |
| Gender | | | | |
| Male (M) | 52 (60.5) | 37 (63.8) | 34 (64.2) | 123 (62.4) |
| Female (F) | 34 (39.5) | 21 (36.2) | 19 (35.8) | 74 (37.6) |
| M:F | 1:0.7 | 1:0.6 | 1:0.6 | 1:0.6 |
Detection of H. pylori
genotypes
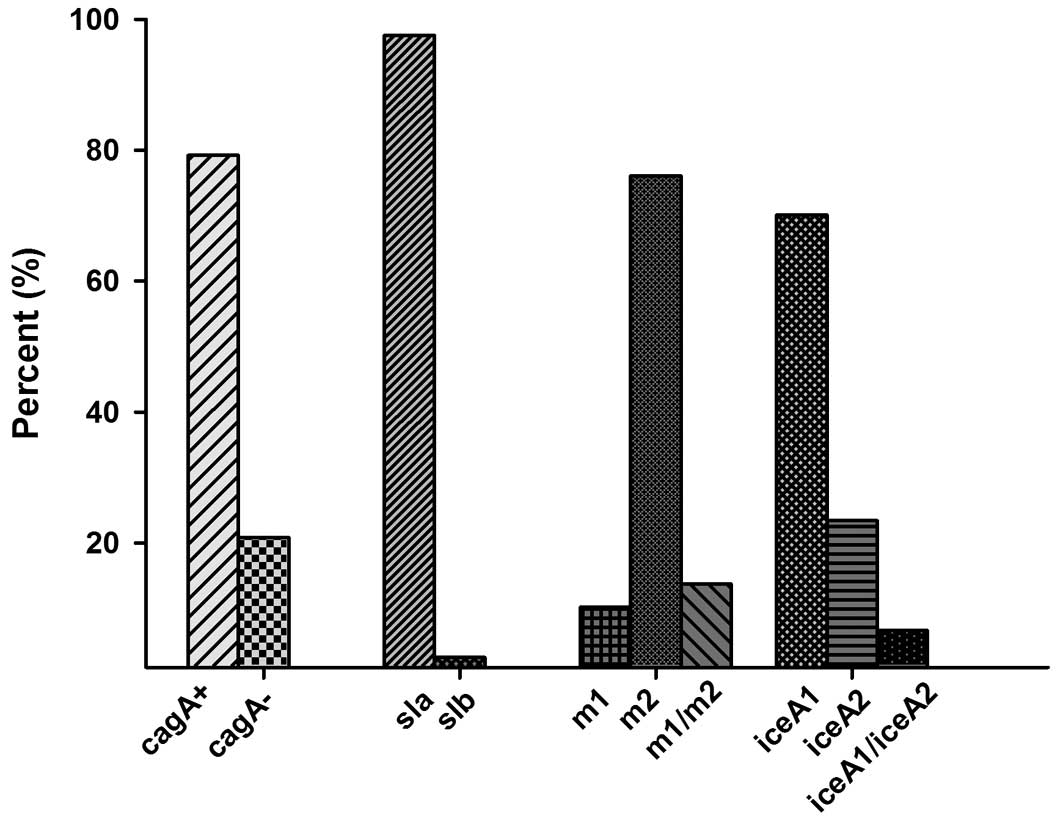
Overall, the presence of the cagA gene was
detected in 176 cases (89.3%). A negative status for the other 21
(10.7%) cases was confirmed by Southern blotting, and the results
were negative as before. All of the samples were positive for
vacA (both the s-region and the m-region). Of the 197 cases,
186 (94.4%) had vacA signal sequence s1c allele, 6 (3%) had
s1a and 5 (2.5%) had s1b. The vacA s2 genotype was not
detected in our study. In the m-region, 27 cases contained both m1
and m2. In these cases the m1 allele was found in 20 (10.2%)
isolates and m2 (76.1%) in 150 cases, which indicating the presence
of mixed infection. The vacA s1am2 genotype was identified
in 6 (3.0%) participants, the vacA s1bm2 was identified in 5
(2.5%) participants, s1cm1 was identified in 20 participants, and
s1cm2 gene was identified in 139 ones. IceA1 was found in
138 (70.1%) and iceA2 was detected in 46 (23.4%) cases. The
iceA2 amplification yielded both the 229-and 334-bp bands
due to the presence of a 105-bp in-frame amplicon present in the
334-bp band that was absent in the 229-bp band. Mixed iceA
(iceA1 + iceA2) genotypes were found in 13 (6.7%) of
our isolates (Fig. 1).
Association among the genotypes
CagA was present in 124 out of 138
iceA1 cases (91.1%) and 44 out of 46 iceA2 cases
(95.5%) (p>0.05) where 29 patients with mixed infection were
excluded. Due to the lack of vacA s2, we could not analyse
the association between cagA status and vacA
genotypes and between iceA and vacA genotypes
(Table III).
 | Table IIIAssociation of vacA with
cagA and iceA genotypes. |
Table III
Association of vacA with
cagA and iceA genotypes.
| vacA | cagA+ n
(%) | cagA− n
(%) | iceA1 n (%) | iceA2 n (%) | iceA1/iceA2 n
(%) |
|---|
| s-region | | | | | |
| s1a | 4 (66.7) | 2 (33.3) | 5 (83.3) | 1 (16.7) | 0 (0.0) |
| s1b | 3 (60.0) | 2 (40.0) | 5 (100.0) | 0 (0.0) | 0 (0.0) |
| s1c | 169 (90.9) | 17 (9.1) | 128 (68.8) | 45 (24.2) | 13 (7.0) |
| m-region | | | | | |
| m1 | 18 (90.0) | 2 (10.0) | 14 (70.0) | 6 (30.0) | 0 (0.0) |
| m2 | 139 (93.0) | 11 (7.0) | 110 (73.4) | 38 (25.3) | 2 (1.3) |
| m1m2 | 19 (84.0) | 8 (16.0) | 14 (70.4) | 2 (22.2) | 11 (7.4) |
| s/m region | | | | | |
| s1am2 | 4 (66.7) | 2 (33.3) | 5 (83.3) | 1 (16.7) | 0 (0.0) |
| s1bm2 | 3 (60.0) | 2 (40.0) | 5 (100.0) | 0 (0.0) | 0 (0.0) |
| s1cm1 | 18 (90.0) | 2 (10.0) | 14 (70.0) | 6 (30.0) | 0 (0.0) |
| s1cm2 | 132 (95.0) | 7 (5.0) | 100 (71.9) | 37 (26.6) | 2 (1.5) |
| s1cm1m2 | 19 (70.4) | 8 (29.6) | 14 (51.9) | 2 (7.4) | 11 (40.7) |
Relationship between genotypes and
gastric diseases
Of the 197 strains studied, 86 were diagnosed with
gastric ulcer, 58 with gastritis and 53 with gastric cancer.
VacA s1cm2 was detected in all the disease conditions, and
it was more significantly associated with the presence of gastric
cancer (p<0.05). S1am2 and s1cm1 were detected in all the
disease except gastric cancer, while s1bm2 was found in gastric
cancer alone (Table IV).
Surprisingly, iceA1 had a statistically significant
association with gastric cancer (p<0.05). Neither cagA
nor iceA2 was associated with various diseases. The most
prevalent combination cagA/s1cm2/iceA1 was present in
56.6% (95 of 168) including 58.0% (40 of 69) of gastric ulcer,
47.0% (23 of 49) of gastritis and 64.0% (32 of 50) of gastric
cancer (Table V). However, no
significant association was found between the combination genotypes
and diseases (p>0.05).
 | Table IVvacA, cagA and
iceA status of H. pylori from 197 patients. |
Table IV
vacA, cagA and
iceA status of H. pylori from 197 patients.
| Clinical status
| |
|---|
| Genotype
status | GUa n=86 (%) | GSb n=58 (%) | GCc n=53 (%) | Total n=197
(%) |
|---|
| vacA | | | | |
| s1am2 | 4 (4.6) | 2 (3.4) | 0 (0.0) | 6 (3.0) |
| s1bm2 | 0 (0.0) | 0 (0.0) | 5 (9.4) | 5 (2.5) |
| s1cm1 | 16 (18.6) | 4 (6.9) | 0 (0.0) | 20 (10.2) |
| s1cm2 | 51 (59.4) | 43 (74.2) | 45 (84.9)d | 139 (70.6) |
| s1cm1m2 | 15 (17.4) | 9 (15.5) | 3 (5.7) | 27 (13.7) |
| cagA | | | | |
|
cagA+ | 78 (90.7) | 53 (91.4) | 45 (84.9) | 176 (89.3) |
|
cagA− | 8 (9.3) | 5 (8.6) | 8 (15.1) | 21 (10.7) |
| iceA | | | | |
| iceA1 | 63 (73.3) | 34 (58.6) | 41 (77.4)d | 138 (70.0) |
| iceA2 | 14 (16.3) | 21 (36.2) | 11 (20.8) | 46 (23.4) |
| iceA1/iceA2 | 9 (10.4) | 3 (5.2) | 1 (1.9) | 13 (6.6) |
 | Table VCombined vacA, cagA,
iceA genotypes. |
Table V
Combined vacA, cagA,
iceA genotypes.
| Clinical status
| |
|---|
| Combination | GUa n (%) | GSb n (%) | GCc n (%) | Total n (%) |
|---|
|
s1am2/cagA+/iceA1 | 1 (1.5) | 2 (4.1) | 0 (0.0) | 3 (1.8) |
|
s1am2/cagA−/iceA1 | 2 (2.9) | 0 (0.0) | 0 (0.0) | 2 (1.2) |
|
s1am2/cagA+/iceA2 | 1 (1.5) | 0 (0.0) | 0 (0.0) | 1 (0.6) |
|
s1bm2/cagA+/iceA1 | 0 (0.0) | 0 (0.0) | 3 (6.0) | 3 (1.8) |
|
s1bm2/cagA−/iceA1 | 0 (0.0) | 0 (0.0) | 2 (4.0) | 2 (1.2) |
|
s1cm1/cagA+/iceA1 | 10 (14.3) | 2 (4.1) | 0 (0.0) | 12 (7.1) |
|
s1cm1/cagA−/iceA1 | 2 (2.9) | 0 (0.0) | 0 (0.0) | 2 (1.2) |
|
s1cm1/cagA+/iceA2 | 4 (5.8) | 2 (4.1) | 0 (0.0) | 6 (3.6) |
|
s1cm2/cagA+/iceA1 | 40 (58.0) | 23 (47.0) | 32 (64.0) | 95 (56.5) |
|
s1cm2/cagA−/iceA1 | 2 (2.9) | 1 (2.0) | 2 (4.0) | 5 (3.0) |
|
s1cm2/cagA+/iceA2 | 6 (8.7) | 18 (36.7) | 11 (22.0) | 35 (20.8) |
|
s1cm2/cagA−/iceA2 | 1 (1.5) | 1 (2.0) | 0 (0.0) | 2 (1.2) |
| Total | 69 (100) | 49 (100) | 50 (100) | 168 (100) |
Discussion
This study was designed to characterize the genotype
of H. pylori from gastric biopsy specimens from patients
with upper gastrointestinal diseases and the relationship with
clinical outcome in Northeast China. H. pylori was analysed
for the presence of the genes for cagA, vacA and
iceA. To our knowledge, this was the first study to analyse
the different proposed virulence genes characterized in H.
pylori and the relationship between the genes and upper
gastrointestinal diseases in Northeast China.
CagA gene, as a major H. pylori
virulence factor, was reported to be strongly associated with
atrophic gastritis and gastric cancer as previously described. This
is probably the main cause of a high incidence of gastric cancer in
the region of East Asia, where the percentage of
cagA-positive strains is above 90% (12). Worldwide, the presence of the
cagA gene varies from 50% in some Middle Eastern countries
to 99% in East Asian countries (13–15).
In this study, cagA was found in 89.3% of H.
pylori-infected patients. The result is similar to data
reported from other districts of China (11,16).
However, we did not find an association between cagA and
clinical results. Notably, of the 29 mixed infection cases, 8 had
cagA-negative and strains with an absence of cagA
appeared to be associated with mixed infection (p=0.004).
The present study demonstrated that all strains of
H. pylori carried the vacA s1 allele. Previous
studies noted that s1c was present exclusively in isolates from
East Asia (16–18). Our report also demonstrated a high
prevalence of type s1c strains in this region, up to 94.4%. The
result was similar to the report of Wang et al (19) and slightly higher than the
prevalence in Beijing and Shanghai, which may result from the fact
that more foreigners from America and Europe live in the two cities
above, as either the s1a or s1b subtype was present in almost all
strains in Central and South America, and in the majority of
strains in Spain and Portugal (20,21),
nevertheless rarely in East Asia (12,16).
The vacA s2 genotype was prominently prevalent in Africa
(9), and consistent to the outcome
reported from China and Korea, s2 failed to be detected in this
study (19,22).
Worldwide prevalence of vacA strains varies
geographically. S1m1 strains were predominant in Japan and Korea
(18,23) while s1m2 was found in Turkey and
Northern and Eastern Europe (20,24).
In Alaskans, H. pylori had either the vacA s1m1
(44.6%) or s2m2 (38.3%) (15). In
China, prevalence of strains documented a greatly distinct pattern,
with s1m1 and s1m2 sharing the same proportion in the Province of
Xi’an (25) and s1m2 strains in
Beijing, Taiwan and Hong Kong (16,19,26).
The latter condition was similar to our study.
Generally, s1m2 forms of vacA bind to and
vacuolate a narrower range of cells than s1m1 forms and induce less
damage, yet they also act as efficient membrane pores and increase
paracellular (27) permeability.
The alleles of s1m1 and s1m2 encode to produce toxin which are
common in patients with gastrosis (27). In Latin America and Germany, s1m1
was found to have a high correlation with gastric ulcer and gastric
carcinoma (21,28). The strains of vacA s2m1 and
s2m2 engender low toxic toxin which rarely correlates with gastric
ulcer and gastric carcinoma (29).
In our study, the vacA gene encoding the s1cm2 was
associated with gastic cancer. Therefore, the s-region should be
responsible for gastrosis other than the m-region.
Another virulent factor is the iceA gene,
with two allelic variants iceA1 and iceA2 having been
identified. The prevalence of the iceA1 genotype is 70.1% in
this study, basically consistent with data reported from China,
Thailand, Korea and Tunisia (9,23,30,31).
Meanwhile, iceA2 is predominant in Brazil, the US, Europe
and South Africa (6,18,32,33).
It was demonstrated that iceA1 was significantly associated
with peptic ulcer disease in Holland (34) and the US (35). However, studies from other
countries such as in Korea, Colombia and India could not confirm
the result (18,36). Some researchers found that the
iceA2 genotype was most frequently found in patients with
duodenal ulcer or gastric carcinoma (18,36).
However, it is difficult to admit that iceA2, a gene that is
considered as a protective factor in some regions and that is
associated with more severe diseases in other places, could be
considered a molecular marker of more virulent H. pylori
strains (33). It was well worth
mentioning that the iceA1 strains, based on this study, have
a significant association with gastric cancer.
In common with other studies, there exists a strong
indication that the presence of multiple H. pylori strains
are detectable in clinical samples. Some studies have claimed
multiple genotypes have a link with duodenal ulcers (9). It may be speculated that multiple
strains contribute to increasing the potential chances of infecting
pathogen. By colonizing a variety of receptors expressed on gastric
epithelial cells, m1 and m2 strains probably tend to bring about
pathological changes. Multi-colonization arising from the
co-existence of more than one strain exert burden to patients under
eradication treatment and furthermore dramatically enhance the risk
of malignant tumors of the digestive tract among adult patients.
However, our data did not indicate that multiple strain infection
increases the risk of developing diseases (p>0.05).
In conclusion, the present study identified the
prevalence of main virulence factor genes cagA, s1cm2 and
iceA1 in Northeast China. The vacA gene encoding
s1cm2 was found to predominate in gastic cancer patients, and the
iceA1 genotype was also associated with gastric cancer. It
may be insufficient to analyse gastrointestinal diseases simply by
genotyping H. pylori, and therefore, we must evaluate the
pathogenesis of diseases by a combination of the analysis of
bacterial factors, genetic factors of the host and environmental
factors.
Acknowledgements
This study was supported by a grant
from the Natural Science Foundation of Heilongjiang Province (grant
no. D2007-72).
References
|
1
|
Blaser MJ: Ecology of Helicobacter
pylori in the human stomach. J Clin Invest. 100:759–762.
1997.
|
|
2
|
Yamazaki S, Yamakawa A, Okuda T, et al:
Distinct diversity of vacA, cagA, and cagE genes of
Helicobacter pylori associated with peptic ulcer in Japan. J
Clin Microbiol. 43:3906–3916. 2005.PubMed/NCBI
|
|
3
|
Jenks PJ, Megraud F and Labigne A:
Clinical outcome after infection with Helicobacter pylori
does not appear to be reliably predicted by the presence of any of
the genes of the cag pathogenicity island. Gut. 43:752–758.
1998.
|
|
4
|
Huang JQ, Zheng GF, Sumanac K, Irvine EJ
and Hunt RH: Meta-analysis of the relationship between cagA
seropositivity and gastric cancer. Gastroenterology. 125:1636–1644.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rathbone M and Rathbone B: Helicobacter
pylori and gastric cancer. Recent Results Cancer Res.
185:83–97. 2011. View Article : Google Scholar
|
|
6
|
Tanih NF, McMillan M, Naidoo N, Ndip LM,
Weaver LT and Ndip RN: Prevalence of Helicobacter pylori
vacA, cagA and iceA genotypes in South African
patients with upper gastrointestinal diseases. Acta Trop.
116:68–73. 2010.
|
|
7
|
Tan HJ, Rizal AM, Rosmadi MY and Goh KL:
Distribution of Helicobacter pylori cagA, cagE and
vacA in different ethnic groups in Kuala Lumpur, Malaysia. J
Gastroenterol Hepatol. 20:589–594. 2005.
|
|
8
|
Atherton JC, Cao P, Peek RM Jr, Tummuru
MK, Blaser MJ and Cover TL: Mosaicism in vacuolating cytotoxin
alleles of Helicobacter pylori Association of specific
vacA types with cytotoxin production and peptic ulceration.
J Biol Chem. 270:17771–17777. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ben Mansour K, Fendri C, Zribi M, et al:
Prevalence of Helicobacter pylori vacA, cagA,
iceA and oipA genotypes in Tunisian patients. Ann Clin
Microbiol Antimicrob. 9:102010.
|
|
10
|
Assumpcao MB, Martins LC, Melo Barbosa HP,
et al: Helicobacter pylori in dental plaque and stomach of
patients from Northern Brazil. World J Gastroenterol. 16:3033–3039.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pan ZJ, van der Hulst RW, Feller M, et al:
Equally high prevalences of infection with cagA-positive
Helicobacter pylori in Chinese patients with peptic ulcer
disease and those with chronic gastritis-associated dyspepsia. J
Clin Microbiol. 35:1344–1347. 1997.PubMed/NCBI
|
|
12
|
Maeda S, Ogura K, Yoshida H, et al: Major
virulence factors, VacA and CagA, are commonly
positive in Helicobacter pylori isolates in Japan. Gut.
42:338–343. 1998.
|
|
13
|
Al Qabandi A, Mustafa AS, Siddique I,
Khajah AK, Madda JP and Junaid TA: Distribution of vacA and
cagA genotypes of Helicobacter pylori in Kuwait. Acta
Trop. 93:283–288. 2005.
|
|
14
|
Lai CH, Kuo CH, Chen YC, et al: High
prevalence of cagA- and babA2-positive Helicobacter
pylori clinical isolates in Taiwan. J Clin Microbiol.
40:3860–3862. 2002.
|
|
15
|
Miernyk K, Morris J, Bruden D, et al:
Characterization of Helicobacter pylori cagA and vacA
genotypes among Alaskans and their correlation with clinical
disease. J Clin Microbiol. 49:3114–3121. 2011.
|
|
16
|
Wong BC, Yin Y, Berg DE, et al:
Distribution of distinct vacA, cagA and iceA
alleles in Helicobacter pylori in Hong Kong. Helicobacter.
6:317–324. 2001.
|
|
17
|
van Doorn LJ, Figueiredo C, Sanna R, et
al: Expanding allelic diversity of Helicobacter pylori vacA.
J Clin Microbiol. 36:2597–2603. 1998.
|
|
18
|
Yamaoka Y, Kodama T, Gutierrez O, Kim JG,
Kashima K and Graham DY: Relationship between Helicobacter
pylori iceA, cagA, and vacA status and clinical
outcome: studies in four different countries. J Clin Microbiol.
37:2274–2279. 1999.
|
|
19
|
Wang J, van Doorn LJ, Robinson PA, et al:
Regional variation among vacA alleles of Helicobacter
pylori in China. J Clin Microbiol. 41:1942–1945. 2003.
View Article : Google Scholar
|
|
20
|
Van Doorn LJ, Figueiredo C, Megraud F, et
al: Geographic distribution of vacA allelic types of
Helicobacter pylori. Gastroenterology. 116:823–830.
1999.
|
|
21
|
Sugimoto M and Yamaoka Y: The association
of vacA genotype and Helicobacter pylori-related
disease in Latin American and African populations. Clin Microbiol
Infect. 15:835–842. 2009.
|
|
22
|
Choe YH, Kim PS, Lee DH, et al: Diverse
vacA allelic types of Helicobacter pylori in Korea
and clinical correlation. Yonsei Med J. 43:351–356. 2002.
|
|
23
|
Kim SY, Woo CW, Lee YM, et al: Genotyping
CagA, VacA subtype, IceA1, and BabA of
Helicobacter pylori isolates from Korean patients, and their
association with gastroduodenal diseases. J Korean Med Sci.
16:579–584. 2001.PubMed/NCBI
|
|
24
|
Erzin Y, Koksal V, Altun S, et al:
Prevalence of Helicobacter pylori vacA, cagA, cagE,
iceA, babA2 genotypes and correlation with clinical outcome
in Turkish patients with dyspepsia. Helicobacter. 11:574–580.
2006.
|
|
25
|
Qiao W, Hu JL, Xiao B, et al: cagA
and vacA genotype of Helicobacter pylori associated
with gastric diseases in Xi’an area. World J Gastroenterol.
9:1762–1766. 2003.
|
|
26
|
Perng CL, Lin HJ, Sun IC and Tseng GY;
Facg: Helicobacter pylori cagA, iceA and vacA
status in Taiwanese patients with peptic ulcer and gastritis. J
Gastroenterol Hepatol. 18:1244–1249. 2003. View Article : Google Scholar
|
|
27
|
Blaser MJ and Atherton JC: Helicobacter
pylori persistence: biology and disease. J Clin Invest.
113:321–333. 2004. View Article : Google Scholar
|
|
28
|
Miehlke S, Kirsch C, Agha-Amiri K, et al:
The Helicobacter pylori vacA s1, m1 genotype and cagA
is associated with gastric carcinoma in Germany. Int J Cancer.
87:322–327. 2000.
|
|
29
|
Bindayna KM and Al Mahmeed A: vacA
genotypes in Helicobacter pylori strains isolated from
patients with and without duodenal ulcer in Bahrain. Indian J
Gastroenterol. 28:175–179. 2009. View Article : Google Scholar
|
|
30
|
Han YH, Liu WZ, Zhu HY and Xiao SD:
Clinical relevance of iceA and babA2 genotypes of
Helicobacter pylori in a Shanghai population. Chin J Dig
Dis. 5:181–185. 2004.
|
|
31
|
Chomvarin C, Namwat W, Chaicumpar K, et
al: Prevalence of Helicobacter pylori vacA, cagA,
cagE, iceA and babA2 genotypes in Thai dyspeptic patients.
Int J Infect Dis. 12:30–36. 2008.
|
|
32
|
Podzorski RP, Podzorski DS, Wuerth A and
Tolia V: Analysis of the vacA, cagA, cagE,
iceA, and babA2 genes in Helicobacter pylori from
sixty-one pediatric patients from the Midwestern United States.
Diagn Microbiol Infect Dis. 46:83–88. 2003.
|
|
33
|
Ashour AA, Collares GB, Mendes EN, et al:
iceA genotypes of Helicobacter pylori strains
isolated from Brazilian children and adults. J Clin Microbiol.
39:1746–1750. 2001. View Article : Google Scholar
|
|
34
|
van Doorn LJ, Figueiredo C, Sanna R, et
al: Clinical relevance of the cagA, vacA, and
iceA status of Helicobacter pylori. Gastroenterology.
115:58–66. 1998.
|
|
35
|
Peek RM Jr, Thompson SA, Donahue JP, et
al: Adherence to gastric epithelial cells induces expression of a
Helicobacter pylori gene, iceA, that is associated
with clinical outcome. Proc Assoc Am Physicians. 110:531–544.
1998.PubMed/NCBI
|
|
36
|
Mukhopadhyay AK, Kersulyte D, Jeong JY, et
al: Distinctiveness of genotypes of Helicobacter pylori in
Calcutta, India. J Bacteriol. 182:3219–3227. 2000.PubMed/NCBI
|
|
37
|
Clayton CL, Kleanthous H, Coates PJ,
Morgan DD and Tabaqchali S: Sensitive detection of Helicobacter
pylori by using polymerase chain reaction. J Clin Microbiol.
30:192–200. 1992.
|
|
38
|
Tummuru MK, Cover TL and Blaser MJ:
Cloning and expression of a high-molecular-mass major antigen of
Helicobacter pylori: evidence of linkage to cytotoxin
production. Infect Immun. 61:1799–1809. 1993.PubMed/NCBI
|