Introduction
Glioma is the most common type of primary brain
tumor in adults. The general outcomes for patients are poor,
particularly for older patients. The etiology of glioma is unclear.
Evidence suggests that exposure to radiation may be a significant
risk factor for glioma, which may explain a small proportion of
glioma since this exposure is generally rare (1). However, only a minority of
individuals exposed to radiation eventually develop glioma,
indicating that host genetic factors may play a critical role in
the carcinogenesis of glioma (2).
Radiation exposure may cause DNA damage and cell
injury. The consequences to the cells may be disastrous, ranging
from single gene mutations to massive chromosomal breakdown and
rearrangements. The instabilities may result in severe human
disorders, including cancer (3).
The repair of various types of DNA damage is vital for the
maintenance of genomic stability and cell survival. Base excision
repair pathways are critical in this process. X-ray repair
cross-complementing gene 1 (XRCC1) is one of the most important DNA
repair genes that plays a key role in the process of base excision
repair. The XRCC1 gene is located on chromosome 19q13.2–13.3 and is
33 kb in length, comprising 17 exons and encoding a 70-kDa protein
(4). A widely studied XRCC1 single
nucleotide polymorphism at codon 194, with a Arg to Trp alteration
(rs1799782), may have a diminished capacity to remove DNA adducts
and oxidized DNA damage (5).
Hence, the Arg194Trp variation has been associated with cancer
susceptibility.
Published data on the association of the XRCC1
Arg194Trp polymorphism with glioma have yielded controversial
results. In the present study, we carried out a quantitative
meta-analysis that increased statistical power to derive a more
precise estimation of this association.
Materials and methods
Literature search strategy
We performed a search of the Medline, Embase, Ovid,
Sciencedirect and Chinese National Knowledge Infrastructure (CNKI)
databases without a language limitation, including all studies
published until May 2012, with a combination of the following
keywords: XRCC1, Arg194Trp, glioma, brain, neoplasm, cancer,
variation and polymorphism. All searched studies were retrieved and
the bibliographies were reviewed for other relevant publications.
Review articles and bibliographies of other relevant studies
identified were searched manually to identify additional eligible
studies.
Inclusion criteria
The following criteria were used for the literature
selection: i) studies should report the association of the XRCC1
Arg194Trp polymorphism with glioma risk; ii) studies are
observational studies (case-control or cohort); iii) studies should
provide the sample size, odds ratios (ORs) and 95% confidence
intervals (CIs), the genetic distribution or the information to
infer the results. After rigorous searching, we reviewed all
studies in accordance with the criteria defined above for further
analysis.
Data extraction
Data were carefully extracted from all eligible
publications independently by two of the authors according to the
inclusion criteria mentioned above. For conflicting evaluations, an
agreement was reached following a discussion. If a consensus could
not be reached, another author was consulted to resolve the dispute
and then a final decision was made by a majority of the votes.
Extracted information was entered into a database.
Statistical analysis
The OR of the XRCC1 Arg194Trp polymorphism and
glioma risk was estimated for each study. The pooled ORs were
assessed for the genetic comparisons of allelic contrast (Trp vs.
Arg), homozygote comparison (Trp/Trp vs. Arg/Arg), dominant model
(Trp/Trp + Trp/Arg vs. Arg/Arg) and recessive model (Trp/Trp vs.
Trp/Arg + Arg/Arg). For the detection of any possible sample size
biases, the OR and 95% CI for each study was plotted against the
number of participants. A Chi-square based Q statistic test was
performed to assess heterogeneity. If the result of the
heterogeneity test was P>0.1, ORs were pooled according to the
fixed-effects model (Mantel-Haenszel); otherwise, the
random-effects model (DerSimonian and Laird) was used. The
significance of the pooled ORs was determined by the Z-test. The
Hardy-Weinberg equilibrium (HWE) was assessed by Fisher’s exact
test.
Publication bias was assessed by visual inspection
of funnel plots (6), in which the
standard error of log (OR) of each study was plotted against its
log (OR). An asymmetric plot indicates a possible publication bias.
The symmetry of the funnel plot was further evaluated by Egger’s
linear regression test (7).
Statistical analysis was undertaken using the program STATA
Results
Study characteristics
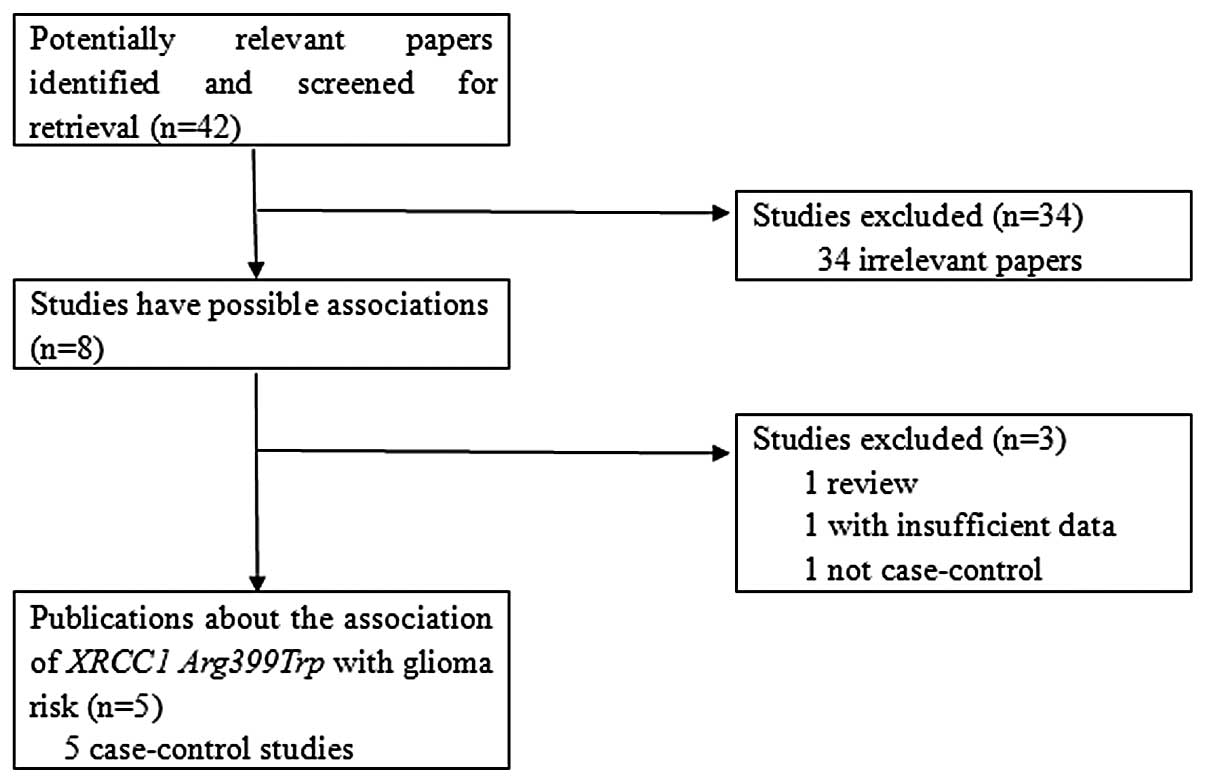
Relevant publications were retrieved and screened.
As shown in Fig. 1, a total of 42
publications were identified, of which 34 irrelevant studies were
excluded. Thus, eight publications were eligible in preliminary
stages, of which one review article (8) and one non-case-control study
(9) were discarded. Subsequently,
one study that did not provide the detailed distributions of the
genotypes (10) was excluded. Five
case-control studies were included for data extraction and analysis
(11–15). However, a study conducted by
Custodio et al (13) was
excluded since it contributed to evident heterogeneity for the
overall data under the four genetic models. Lasty, four
case-control studies were selected for analysis (11,12,14,15).
All the selected publications were written in
English. The relevant information is listed in Table I, including the first author and
the number and characteristics of cases and controls for each study
as well as other relevant information. There were two groups of
Asians (14,15) and two of Caucasians (11,12)
in the present meta-analysis.
 | Table ICharacteristics of studies included in
the meta-analysis. |
Table I
Characteristics of studies included in
the meta-analysis.
| Authors/(Refs.) | Publication year | No. of cases
(male/female) | No. of controls
(male/female) | Type of controls | Median (or mean)age
(range) in years (cases/controls) | Ethnic decent | Country |
|---|
| Kiuru et al
(11) | 2008 | 426 (259/167) | 1560 (705/855) | Healthy controls
(age-, gender-, geographical area-matched; PB) | 48.2 (NA)/63
(NA) | Caucasian | Four countries in
Europe |
| Rajaraman et
al (12) | 2010 | 362 (198/164) | 495 (228/267) | Non-cancer controls
(age-, race-, gender-, hospital-, residence-matched; HB) | 51.2 (18–90)/49.2
(18–90) | Caucasian | USA |
| Hu et al
(14) | 2011 | 127 (87/40) | 249 (166/83) | Non-cancer controls
(age-, gender-matched; HB) | 49.5 (NA)/48.9
(NA) | Asian | China |
| Zhou et al
(15) | 2011 | 271 (168/103) | 289 (180/109) | Healthy controls
(age-matched; PB) | 47.8 (NA)/46.9
(NA) | Asian | China |
The distributions of the XRCC1 Arg194Trp genotypes
as well as the genotyping methods of the included studies are
presented in Table II. The genetic
distributions of the control groups in all studies were consistent
with the HWE, with the exception of one study (14).
 | Table IIDistribution of the XRCC1 Arg194Trp
genotypes among glioma cases and controls included in the
meta-analysis. |
Table II
Distribution of the XRCC1 Arg194Trp
genotypes among glioma cases and controls included in the
meta-analysis.
| | | Cases
| Controls
| |
|---|
| Authors/(Refs.) | Year | Genotyping
method | Trp/Trp | Arg/Trp | Arg/Arg | Trp/Trp | Arg/Trp | Arg/Arg | HWE (control) |
|---|
| Kiuru et al
(11) | 2008 | PCR-RFLP | 3 | 71 | 626 | 2 | 177 | 1,377 | Yes |
| Rajaraman et
al (12) | 2010 | TaqMan | 0 | 38 | 304 | 1 | 73 | 394 | Yes |
| Hu et al
(14) | 2011 | PCR-CTPP | 18 | 38 | 71 | 22 | 64 | 163 | No |
| Zhou et al
(15) | 2011 | TaqMan | 14 | 112 | 145 | 13 | 117 | 159 | Yes |
Test of heterogeneity
As shown in Table
III, we analyzed the heterogeneities for the four genetic
comparisons. Evident heterogeneities for the overall data were
present in the allelic contrast (P=0.030, Q-test) and dominant
model (P=0.061 for Q-test), with the exception of the homozygote
comparison (P=0.561, Q-test) and recessive model (P=0.598, Q-test).
Additionally, the I-square value is another index for a
heterogeneity test (16), with
value less than 25% indicating low, 25% to 50% indicating moderate,
and greater than 50% indicating high heterogeneity. The I-square
values were 66.6 and 59.2% for the overall data of the allelic
contrast and dominant model, respectively, suggesting statistically
significant heterogeneity between the studies; therefore, the
random-effects models were utilized. For the homozygote comparison
and recessive model, the I-square values were both 0.0%, indicating
an absence of the heterogeneity. Thus, the fixed-effects model was
used in these two models.
 | Table IIIMain results of the pooled data from
the meta-analysis. |
Table III
Main results of the pooled data from
the meta-analysis.
| | Trp allele vs. Arg
allele
| Trp/Trp vs. Arg/Arg
| (Trp/Trp + Trp/Arg)
vs. Arg/Arg
| Trp/Trp vs. (Trp/Arg
+ Arg/Arg)
|
|---|
| Analysis | No.
(cases/controls) | OR (95% CI) | P | P (Q-test) | OR (95% CI) | P | P (Q-test) | OR (95% CI) | P | P (Q-test) | OR (95% CI) | P | P (Q-test) |
|---|
| Total | 1440/2562 | 1.01 (0.77–1.33) | 0.933 | 0.030 | 1.56 (0.96–2.54) | 0.073 | 0.561 | 0.98 (0.74–1.31) | 0.909 | 0.061 | 1.48
(0.92–2.38) | 0.107 | 0.598 |
| Ethnicity | | | | | | | | | | | | | |
| Caucasian | 1042/2024 | 0.83
(0.60–1.14) | 0.240 | 0.180 | 1.83
(0.43–7.88) | 0.415 | 0.274 | 0.81
(0.60–1.09) | 0.160 | 0.228 | 1.88
(0.44–8.10) | 0.395 | 0.284 |
| Asian | 398/538 | 1.23
(0.89–1.71) | 0.214 | 0.131 | 1.53
(0.91–2.57) | 0.107 | 0.383 | 1.22
(0.88–1.70) | 0.231 | 0.223 | 1.44
(0.87–2.38) | 0.157 | 0.456 |
| Source of
controls | | | | | | | | | | | | | |
| PB | 971/1845 | 1.00
(0.83–1.21) | 0.998 | 0.551 | 1.39
(0.68–2.85) | 0.368 | 0.303 | 0.97
(0.78–1.21) | 0.797 | 0.487 | 1.36
(0.67–2.76) | 0.391 | 0.286 |
| HB | 469/717 | 1.01
(0.47–2.18) | 0.983 | 0.004 | 1.73
(0.89–3.35) | 0.104 | 0.378 | 0.99
(0.45–2.20) | 0.990 | 0.009 | 1.59
(0.83–3.03) | 0.158 | 0.428 |
Notably, when the overall data were divided for
subgroup analysis, we observed a loss of heterogeneity in the
subgroups in the allelic contrast and dominant models.
Meta-analysis results
The main results of the meta-analysis are listed in
Table III. For the overall data
including 1,440 cases and 2,562 controls, significant associations
of the XRCC1 Arg194Trp polymorphism with glioma risk were not
identified in the four genetic models (allele contrast: OR = 1.01,
95% CI = 0.77–1.33; homozygote comparison: OR = 1.56, 95% CI =
0.96–2.54; dominant model: OR = 0.98, 95% CI = 0.74–1.31; recessive
model: OR = 1.48, 95% CI = 0.92–2.38), indicating that the XRCC1
Arg194Trp polymorphism may not have a correlation with glioma
risk.
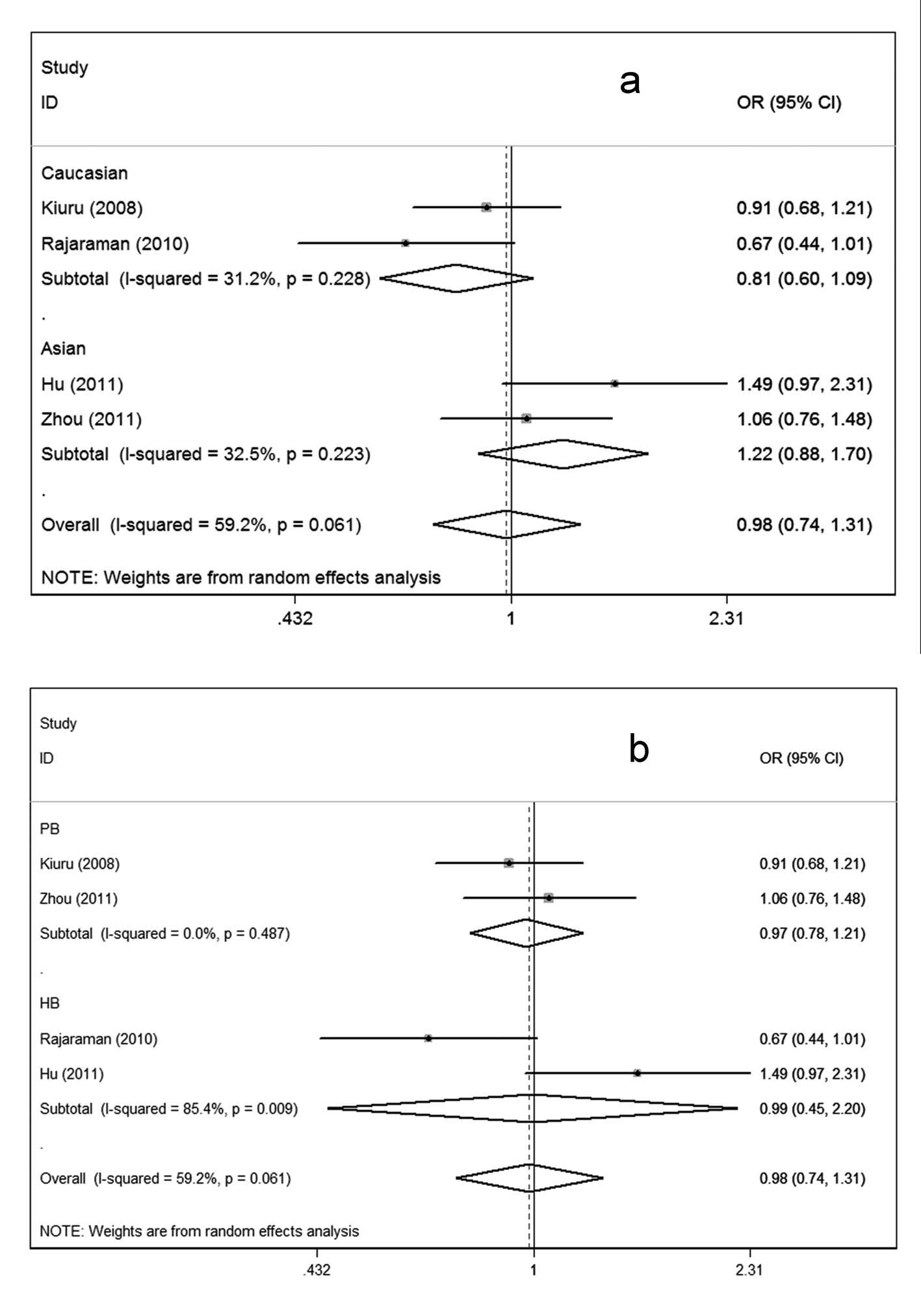
Considering the possible effects of ethnic variation
and selection of controls on the results (Fig. 2), we conducted subgroup analyses.
In the subgroup analysis according to ethnicity, no associations
were observed in either the Asian or Caucasian subgroup (Fig. 2a). Similarly, in the subgroup
analysis by source of controls, significant associations could be
observed in neither the population-based subgroups nor the
hospital-based subgroups under the four genetic comparisons
(Fig. 2b).
Sensitivity analysis
To test the stability of the overall results, we
carried out one-way sensitivity analysis (17). The statistical significance of the
results was not changed when any single study was omitted (data not
shown), indicating the robustness of the results.
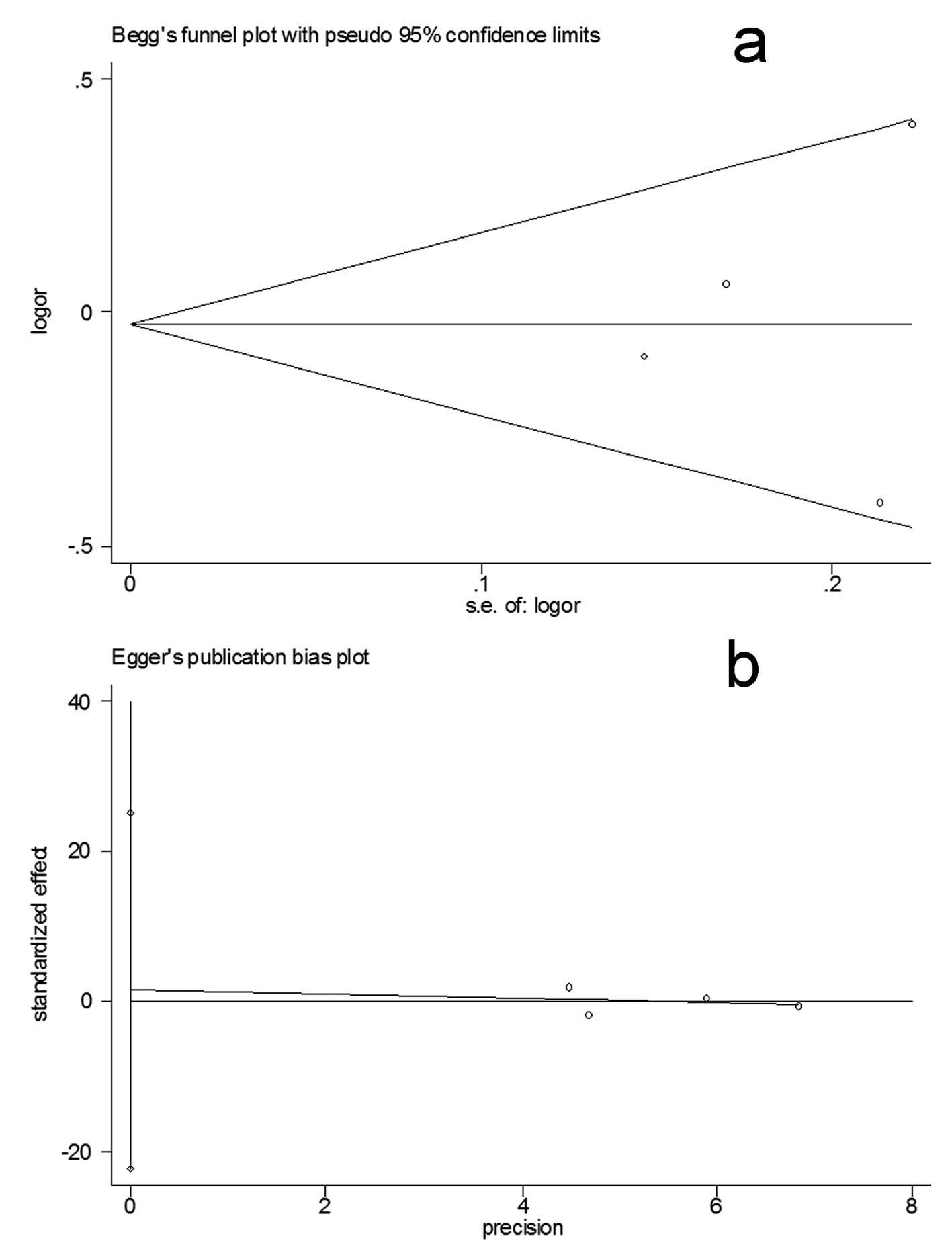
Bias diagnostics
Funnel plots were created for assessment of possible
publication biases (Fig. 3a).
Then, Egger’s linear regression tests were used to assess the
symmetries of the plots. The funnel plots appeared to be
symmetrical for the overall data indicated by the Egger’s tests
(allelic contrast: t=−0.33, P>0.05, homozygote comparison:
t=−0.24, P>0.05; dominant model: t=0.26, P>0.05; recessive
model: t=−0.13, P>0.05; Fig.
3b), suggesting that the publication biases were not
evident.
Discussion
The overall data obtained failed to show a
significant association between the XRCC1 Arg194Trp polymorphism
and glioma risk. Similar results were demonstrated in the subgroup
analysis based on ethnicity and source of controls.
Previous meta-analyses have focused on the
association between the XRCC1 Arg194Trp polymorphism and several
cancer risks and have generated conflicting results. The XRCC1
Arg194Trp variation has been demonstrated to increase risk of lung
and gastric cancer (18,19). However, meta-analyses regarding
skin and esophageal cancer failed to reveal such associations
(20,21). Therefore, the XRCC1 Arg194Trp
polymorphism may exert different effects on different cancers.
In the subgroup analysis based on ethnicity, no
significant associations could be observed in the Asian or
Caucasian subgroup. Evidence indicates that ethnic-specific
variation, different health care and socioeconomic class might
exert an effect on the incidence of glioma (22). The results demonstrated that the
potential effects of ethnic variations on glioma were not evident.
Notably, the results should be interpreted with care since the
limited number of the included studies containing small sample
sizes may result in insufficient statistical power to evaluate a
small effect. Therefore, future investigations regarding different
ethnicities with large sample sizes are required to clarify this
issue.
When the data were stratified based on the source of
controls, no association was observed in either the
population-based or hospital-based subgroup. Since hospital-based
controls may not be representative of the general population, a
selection bias may exist. However, the data of the present
meta-analysis suggest that the potential influence of the selection
bias on the overall results was not evident. However, use of proper
control participants with rigorous matching criteria and large
sample sizes in future studies is important to reduce such possible
selection bias.
In the present meta-analysis, evident between-study
heterogeneities for overall data were observed in the allelic
contrast and dominant models. However, when the subgroup analyses
were conducted, we found that the heterogeneities were removed in
the subgroups regarding Caucasian, Asian and population-based
controls. However, the heterogeneity was still present in the
hospital-based subgroup. Therefore, the heterogeneities might be
multifactorial. In addition to ethnicity and source of controls,
other factors including age, pathology grade and life styles may
also contribute to the heterogeneity.
Several limitations should be addressed. First, in
this meta-analysis, the primary articles only provided data
regarding Caucasians, Asians and mixed races. Other ethnicities
such as African should be investigated in future studies. Second,
subgroup analyses based on age, gender, histological types,
radiation exposure and other factors have not been performed in the
present study since sufficient relevant data were not available in
the primary literature. Third, only studies written in English and
several other languages indexed by the common databases were
searched. Thus, a selection bias might exist. Therefore, the
results should be interpreted with caution. However, the
sensitivity analysis and publication bias analysis indicated the
stability and credibility of the present meta-analysis.
In conclusion, the results of the present
meta-analysis failed to suggest an association of the XRCC1
Arg194Trp polymorphism with glioma risk. Further investigations
with larger sample sizes and rigorous matching criteria in view of
confounding factors are required to confirm the associations.
References
|
1
|
Prasad G and Haas-Kogan DA:
Radiation-induced gliomas. Expert Rev Neurother. 9:1511–1517. 2009.
View Article : Google Scholar
|
|
2
|
Melin B: Genetic causes of glioma: new
leads in the labyrinth. Curr Opin Oncol. 23:643–647. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li GM: A special issue on DNA damage
response and genome stability. Cell Biosci. 2:42012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mutamba JT, Svilar D, Prasongtanakij S, et
al: XRCC1 and base excision repair balance in response to nitric
oxide. DNA Repair (Amst). 10:1282–1293. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ginsberg G, Angle K, Guyton K and Sonawane
B: Polymorphism in the DNA repair enzyme XRCC1: utility of current
database and implications for human health risk assessment. Mutat
Res. 727:1–15. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Munafo MR, Clark TG and Flint J: Assessing
publication bias in genetic association studies: evidence from a
recent meta-analysis. Psychiatry Res. 129:39–44. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Goode EL, Ulrich CM and Potter JD:
Polymorphisms in DNA repair genes and associations with cancer
risk. Cancer Epidemiol Biomarkers Prev. 11:1513–1530.
2002.PubMed/NCBI
|
|
9
|
Liu Y, Shete S, Hosking F, Robertson L,
Houlston R and Bondy M: Genetic advances in glioma: susceptibility
genes and networks. Curr Opin Genet Dev. 20:239–244. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu Y, Scheurer ME, El-Zein R, et al:
Association and interactions between DNA repair gene polymorphisms
and adult glioma. Cancer Epidemiol Biomarkers Prev. 18:204–214.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kiuru A, Lindholm C, Heinavaara S, et al:
XRCC1 and XRCC3 variants and risk of glioma and meningioma. J
Neurooncol. 88:135–142. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rajaraman P, Hutchinson A, Wichner S, et
al: DNA repair gene polymorphisms and risk of adult meningioma,
glioma, and acoustic neuroma. Neuro Oncol. 12:37–48. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Custodio AC, Almeida LO, Pinto GR, et al:
Analysis of the polymorphisms XRCC1Arg194Trp and XRCC1Arg399Gln in
gliomas. Genet Mol Res. 10:1120–1129. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu XB, Feng Z, Fan YC, Xiong ZY and Huang
QW: Polymorphisms in DNA repair gene XRCC1 and increased genetic
susceptibility to glioma. Asian Pac J Cancer Prev. 12:2981–2984.
2011.PubMed/NCBI
|
|
15
|
Zhou LQ, Ma Z, Shi XF, et al:
Polymorphisms of DNA repair gene XRCC1 and risk of glioma: a
case-control study in Southern China. Asian Pac J Cancer Prev.
12:2547–2550. 2011.PubMed/NCBI
|
|
16
|
Higgins JP, Thompson SG, Deeks JJ and
Altman DG: Measuring inconsistency in meta-analyses. BMJ.
327:557–560. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tobias A: Assessing the influence of a
single study in the meta-analysis estimate. Stata Techn Bull.
8:15–17. 1999.
|
|
18
|
Cui Z, Yin Z, Li X, Wu W, Guan P and Zhou
B: Association between polymorphisms in XRCC1 gene and clinical
outcomes of patients with lung cancer: a meta-analysis. BMC Cancer.
12:712012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen B, Zhou Y, Yang P and Wu XT:
Polymorphisms of XRCC1 and gastric cancer susceptibility: a
meta-analysis. Mol Biol Rep. 39:1305–1313. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang H, Li W, Franklin MJ and Dudek AZ:
Polymorphisms in DNA repair gene XRCC1 and skin cancer risk: a
meta-analysis. Anticancer Res. 31:3945–3952. 2011.PubMed/NCBI
|
|
21
|
Yin M, Tan D and Wei Q: Genetic variants
of the XRCC1 gene and susceptibility to esophageal cancer: a
meta-analysis. Int J Clin Exp Med. 2:26–35. 2009.PubMed/NCBI
|
|
22
|
Curry WT Jr and Barker FG II: Racial,
ethnic and socioeconomic disparities in the treatment of brain
tumors. J Neurooncol. 93:25–39. 2009. View Article : Google Scholar : PubMed/NCBI
|