Introduction
Antimicrobial peptides (AMPs) are peptides which are
generated by the innate immune system and confer specific immunity
against microbial invaders. AMPs are able to effectively prevent
pathogen invasion and possess broad-spectrum antibacterial activity
against Gram-positive bacteria, Gram-negative bacteria, fungi,
parasites, viruses and tumor cells. One of their most notable
features is that AMPs rarely induce bacterial resistance which is a
serious problem with conventional antibiotics (1). Therefore, AMPs have emerged as one of
the most promising candidates for a new class of antibiotics.
Cecropins are small molecules with molecular sizes
ranging from 3,500 to 4,000 Da and are considered to be the most
potent AMPs. Structurally, cecropins possess a strong basic amino
(N)-terminus and a long hydrophobic carboxyl (C)-terminus
interrupted by a hinge region composed of a Gly-Pro sequence
(2). Cecropins were first isolated
from Hyalophora cecropia (3,4).
Since then members of the cecropin-family peptides have been
purified from Manduca sexta (5), Bombyx mori (6), Drosophila melanogaster
(7) and Sarcophage
peregrine (8). Cecropin D was
chemically synthesized and its antibacterial activity was analyzed
to allow the development of peptide antibiotics based on the
structure of cecropin (9).
Pichia pastoris serves as a eukaryotic
expression system and has been used successfully to produce
recombinant heterologous proteins of human, animal, plant, fungal,
bacterial and viral origins (10).
The expression of secreted recombinant proteins in P.
pastoris offers several advantages over bacterial expression
systems. These include appropriate folding of molecules and
disulfide bond formation, as well as execution of
post-translational modifications which conserve protein function.
Secretion of recombinant proteins circumvents intracellular
accumulation, a significant aspect in the expression of toxic
proteins. The secretion of recombinant proteins simplifies their
purification by avoiding contamination with intracellular proteins
(11,12). These advantages make secreted
recombinant protein production in P. pastoris popular for
scientific research. For this study, the SMD1168 strain was
selected since it produces low levels of proteases. The present
study demonstrates the successful manipulation of cecropin D for
expression in yeast and the secretion of a functional AMP.
Materials and methods
Strains and plasmid
P. pastoris strain SMD1168 and expression
vector pGAPZαA were purchased from Invitrogen (Carlsbad, CA, USA).
The Escherichia coli strain DH5α and Staphylococcus
aureus strain Cowan I were stored at the College of Veterinary
Medicine, South China Agricultural University (Guangzhou,
Guangdong, China).
Reagents and materials
rTaq DNA polymerase, DNA marker, T4 DNA ligase and
the restriction enzymes XhoI, XbaI and BlnI
were purchased from Takara Bio, Inc. (Shiga, Japan). Zeocin™ was
obtained from Invitrogen. The E.Z.N.A Gel Extraction kit, Plasmid
Mini kit and Cycle-Pure kit were purchased from Promega (Madison,
WI, USA). The routine reagents were domestic and analytically
pure.
Design of primers and synthesis of
cecropin D
The cecropin D gene sequence from GenBank (serial
No. NM_001043368.1) and the optimal codon list for P.
pastoris were employed to design four primers using the Primer
Premier 5.0 software (www.PremierBiosoft.com). The region amplified by the
four primers covered the complete cecropin D coding region. The
length of the amplicon was 108 bp. The four primer sequences were
as follows: P1: 5′-CCATTCAAG GAGTTGGAGAGAG
CTGGTCAAAGAGTCAGAGACGCTATCATCTCCGCT-3′; P2:
5′-CAAAGCGGTAGCTTGAGCGACGGTAGCGACA GCTGGACCAGCGGAGATGATAGCGTC-3′;
P3: 5′-CCCT
CGAGAAAAGATGGAACCCATTCAAGGAGTTGGAG-3′
(containing an XhoI site, underlined; KEX2 signal cleavage
site, bold); P4: 5′-GCTCTAGATTAGTTCTTAGCCAAAGC
GGTAGCTTGAGC-3′ (containing an XbaI site, underlined). The
sequence encoding cecropin D and the proteolytic signal KEX2 fused
to cecropin D was synthesized by PCR using synthetic
oligonucleotides.
The splicing by overlap extension (SOEing) PCR
method was employed to synthesize the cecropin D gene sequence. The
reaction mix for the PCR contained 5 μl 10X PCR Buffer
(Mg2+), 4 μl dNTPs (2.5 mmol/l), 1 μl each
of the primers P1, P2, P3 and P4 (40 pmol/l), 0.25 μl
rTaq DNA polymerase and 34.75 μl distilled water.
Reactions were performed on a GeneAmp PCR System 2400 (Applied
Biosystems, Carlsbad, CA, USA). PCR conditions were as follows: 4
min denaturation at 94°C, then 30 cycles (94°C for 30 sec, 65°C for
45 sec and 72°C for 30 sec) followed by a final 10 min elongation
step at 72°C. The E.Z.N.A Gel Extraction kit was used to purify the
PCR product. Samples (2 μl) were loaded onto a 1% agarose
gel and electrophoresed at 100 V for 15 min. The gel was scanned on
a FluorImager SI (Molecular Dynamics, La Jolla, CA, USA).
The obtained 108-bp fragment was cloned in frame
with the α-mating factor signal peptide of S. cerevisiae
contained in the expression pGAPZαA vector. The insertion of this
DNA fragment into the pGAPZαA vector replaced the original
C-terminal proteolytic recognition sequences (containing a KEX2 and
a STE3 site) with a sequence that contained only the KEX2 protease
site.
Cloning of cecropin D into the yeast
expression vector pGAPZαA and transformation of P. pastoris
SMD1168
The PCR product was digested with the Xbal
and Xhol restriction enzymes and ligated into the linearized
pGAPZαA (Invitrogen) digested with the same enzymes. Following
purification with an E.Z.N.A Cycle-Pure kit, the ligation product
was transformed into competent E. coli DH5α cells.
Restriction endonuclease analysis, PCR and sequencing were
performed to validate the reading frame of the recombinant plasmid
pGAPZαA-cecropin D.
P. pastoris was transformed by
electroporation. Firstly, 5–10 µg of the recombinant plasmid
pGAPZαA-cecropin D was digested with BlnI and a small
aliquot of the digest was used to confirm complete linearization by
agarose gel electrophoresis. Competent yeast cells (80 μl)
were mixed with 5–10 μg of linearized DNA (in 5–10 μl
sterile distilled water). The cell mixture was transferred to an
ice-cold 0.2 cm electroporation cuvette and kept on ice for 5 min,
then pulsed at 305 V for 15 msec. Ice-cold 1 M sorbitol (1 ml) was
added to the cuvette immediately following electroporation.
Aliquots of 10, 25, 50, 100 and 200 μl were spread on
separate yeast extract peptone dextrose (YPD) sorbitol plates
containing 100 μg/ml Zeocin. Plates were incubated for 2 to
3 days at 30°C until colonies had formed. Finally, 6 to 10
Zeocin-resistant P. pichia transformants were analyzed for
the presence of an insert using PCR, or for copy number using
Southern blot analysis.
Expression of cecropin D in P.
pastoris SMD1168
A single colony was inoculated into 10 ml of YPD and
grown at 28–30°C in an agitating incubator (250–300 rpm) overnight.
An aliquot of 0.1 ml of this overnight culture was used to
inoculate 50 ml of YPD in a 250-ml flask which was grown at 28–30°C
in an agitating incubator (250–300 rpm). At the time points
indicated below, 1 ml of the expression culture was transferred
into a 1.5-ml microcentrifuge tube. These samples were used to
analyze expression levels and to determine the optimal time to
harvest the samples. Cultures were centrifuged at maximum speed in
a tabletop microcentrifuge for 2–3 min at room temperature.
Supernatants were transferred into a separate tube and analyzed by
Tricine-SDS-PAGE. Samples were taken at the following time points:
0, 24, 48, 72 and 96 h.
Determination of the protein
concentration
The Bradford Protein Assay kit was used to determine
the protein concentration. Bovine serum albumin (BSA) was used as
standard protein (0.5 mg/ml) and diluted with PBS in a 96-well
plate. Cecropin D supernatant was diluted 1:1 with PBS and 200
μl G250 staining solution was added to each well. A
microplate reader was used to determine A570 which was
used to estimate the total protein in the supernatant and controls.
A light density scanner was used to scan the SDS-PAGE gel to assess
the percentage of the target band in relation to total protein.
These data were used to calculate the expressed level of the AMP
cecropin D.
Antimicrobial assay
The antibacterial activity of cecropin D was tested
using several Gram-positive and Gram-negative bacteria (listed in
Table I) as described by Minagawa
et al (13). A plate
containing 1% agarose in 50 mM phosphate buffer (pH 6.2) and 10
μl Gram-positive or Gram-negative bacteria was prepared.
After solidification, four wells (2 mm in diameter) were carved
into the plate; 30 µl of the supernatant of
SMD1168-pGAPZαA-cecropin D was loaded into one of the wells, while
an equal amount of the supernatant of SMD1168-pGAPZαA and SMD1168
(negative controls) were loaded into two other wells. Ampicillin (2
μl, 100 mg/ml) was loaded into the remaining well as a
positive control. The plate was incubated at 37°C for 6–8 h and the
diameters of the cleared zones were measured.
 | Table IDiameter of cleared zones of cecropin
D exhibiting antibacterial activity towards Gram-positive and
Gram-negative bacteria.a |
Table I
Diameter of cleared zones of cecropin
D exhibiting antibacterial activity towards Gram-positive and
Gram-negative bacteria.a
| Microorganism
|
|---|
| E. coli
DH5α | S. aureus
CowanI | E. coli
K88 |
Streptococcus (SEZ) | S.
choleraesuis | B.
pumilus | E. coli
K99 |
|---|
| Antimicrobial
diameter (mm) | | | | | | | |
| No. 1 | 19 | 17 | 21 | 17 | 16 | 18 | 22 |
| No. 2 | 21 | 16 | 20 | 18 | 18 | 15 | 23 |
| No. 3 | 17 | 15 | 22 | 20 | 16 | 17 | 21 |
| Average
diameter | 19 | 16 | 21 | 18.33 | 16.67 | 16.67 | 22 |
| Standard
deviation | 1.63 | 0.82 | 0.82 | 1.25 | 0.94 | 1.25 | 0.82 |
| MIC
(μg/ml) | 2.17 | 4.55 | 2.05 | 3.13 | 4.62 | 2.12 | 1.98 |
| Ampicillin
(mm) | 30 | 23 | 24 | 27 | 20 | 21 | 31 |
The minimal growth inhibition concentration (MIC)
was determined using a liquid growth inhibition assay (14), with MIC defined as the lowest final
concentration of the peptide at which no growth was observed
(15,16). A stock solution of cecropin D was
serially diluted 10-fold with 0.01% acetic acid and 0.2% BSA.
Aliquots of each dilution were distributed into a 96-well
polypropylene microtiter plate and each well was inoculated with a
100 μl suspension of mid-log phase bacteria in LB broth.
Cultures were grown for 24 h with vigorous agitation at 30°C and
bacterial growth was evaluated by measuring the absorbance of the
bacterial culture at 600 nm using a spectrophotometer.
Tolerance of cecropin D to adverse
conditions (high temperature, acid/base exposure, proteases)
High temperature tolerance
Supernatant from the expression culture of cecropin
D was boiled for 15, 30 and 45 min. The antibacterial activity of
the heat-treated supernatants was measured at the same time and
under the same conditions as untreated supernatants which served as
a control.
Acid and base tolerance
Cecropin D-containing super-natant was treated with
HCl or NaOH (1 M) for 15, 30 and 45 min, after which time the same
volume of NaOH or HCl (1 M) was added in order to neutralize the
solution while using a pH meter. The antibacterial activity of the
treated supernatants was detected as described previously using
untreated control supernatants in parallel.
Tolerance to proteases
Supernatant from the expression culture of cecropin
D was treated with trypsin and pepsin for 15, 30 and 45 min in a
water bath at 37°C. Antibacterial activity in the supernatant was
detected using an agarose diffusion test in the presence of
appropriate controls.
Optimization of fermentation
conditions
In order to improve the expression levels of
cecropin D, the carbon source glucose was compared with glycerol
under identical fermentation conditions. Urea was compared with
tryptone and ammonium sulfate as a nitrogen source. In fed batch
culture studies, carbon and nitrogen sources were supplied
intermittently and compared with a regular supply of the same
source.
Results
Cecropin D characterization and
expression analysis
The sequence of the cloned cecropin D was compared
with the sequence obtained from GenBank (Table II). The base composition was mostly
identical and differed only in one base at position 40 where a
change from Ala to Val had occurred.
 | Table IICecropin D gene sequences obtained
from GenBank and the sequence cloned into expression vector
pGAPZαA-cecropin D. |
Table II
Cecropin D gene sequences obtained
from GenBank and the sequence cloned into expression vector
pGAPZαA-cecropin D.
| Source | Gene sequence |
|---|
| GenBank (ID): |
TGGAACCCATTCAAGGAGTTGGAGAGAGCTGGTCAAAGAGCC(Ala)AGAGACGCT |
|
ATCATCTCCGCTGGTCCAGCTGTCGCTACCGTCGCTCAAGCTACCGCTTTGGCTAAG |
| Cecropin D
Gene: |
TGGAACCCATTCAAGGAGTTGGAGAGAGCTGGTCAAAGAGTC(Val)AGAGACGCT |
|
ATCATCTCCGCTGGTCCAGCTGTCGCTACCGTCGCTCAAGCTACCGCTTTGGCTAAG |
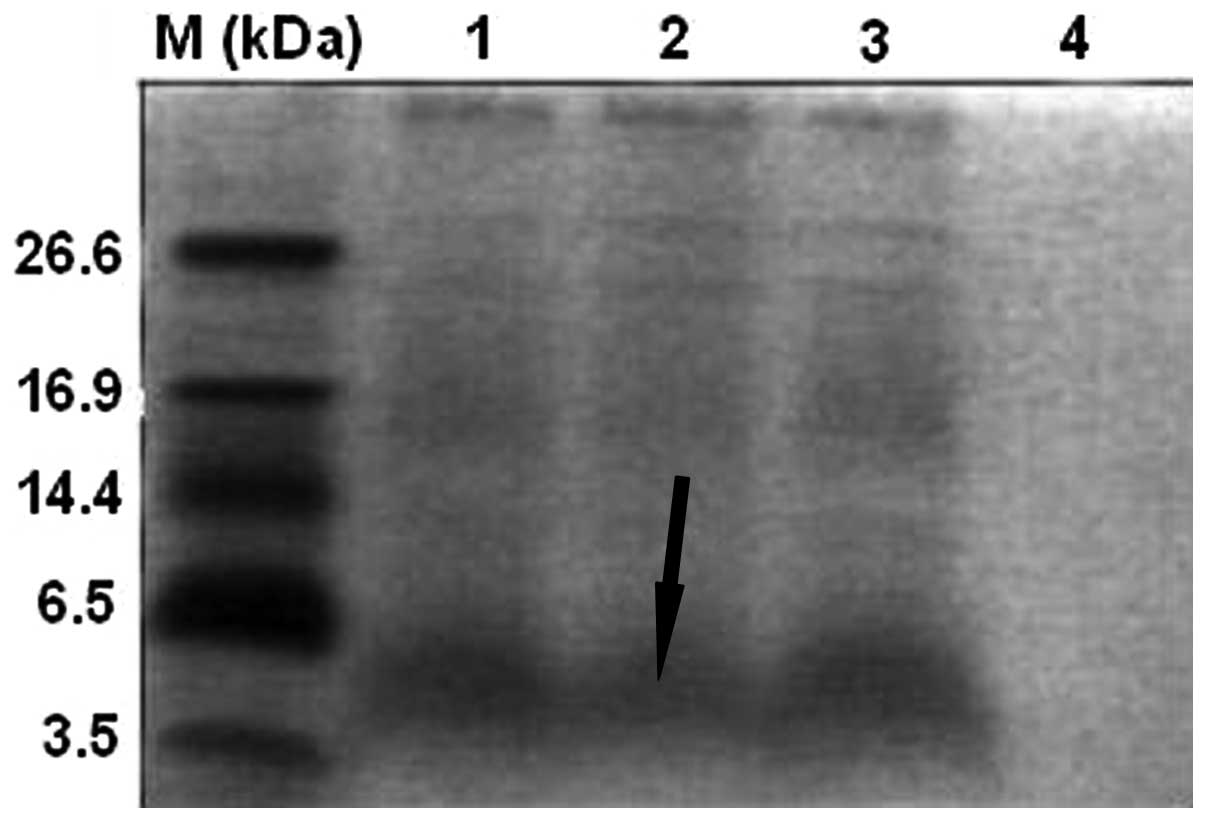
A major band of the polypeptide of interest (3.9
kDa; Fig. 1) was observed by
Tricine-SDS-PAGE in the supernatant of cultures after 3 days of
expression under optimal conditions. This band was absent from the
control cultures. This result suggested that the polypeptide was
successfully expressed in yeast. Secretion of the molecule into the
supernatant further suggested that the signal peptide had been
removed from the N-terminus of cecropin D.
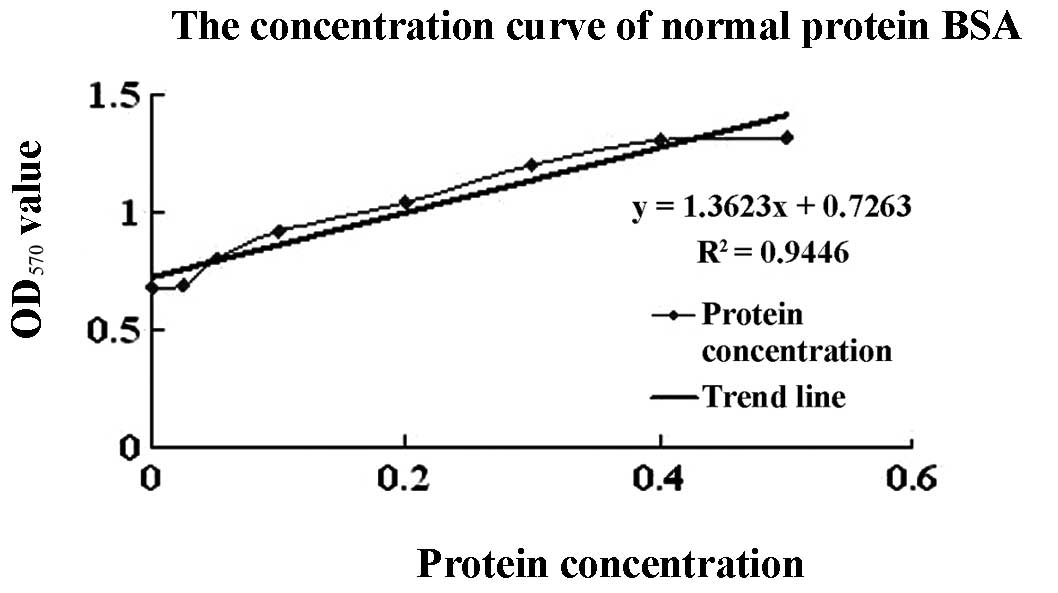
A protein standard concentration curve was generated
using various concentrations of BSA (Fig. 2). The OD570 of samples
expressing cecropin D was 1.0506 and the percentage of the target
protein was 85.34% of total protein determined by the light density
scanner method. Using this information, expression levels of
recombinant cecropin D were calculated to be 485.24 mg/l.
Antibacterial activity of recombinant
cecropin D and MIC
The antimicrobial activity of cecropin D was tested
against Gram-positive and Gram-negative bacteria. The diameters of
the cleared zones were measured and the values are shown in
Table I. The average diameter of
the cleared zones ranged from 16–22 mm for the various bacterial
species included in the experiment, while the cleared zones were
absent from the negative controls. These data suggest that cecropin
D exhibited antibacterial activity to Gram-positive and
Gram-negative bacteria.
Tolerance of recombinant cecropin D to
adverse conditions
Heat treatment of culture supernatants containing
cecropin D had no effect on the antibacterial activity of cecropin
D suggesting that cecropin D is insensitive to high
temperatures.
Similarly, treatment of cecropin D using HCl or NaOH
for 15 or 30 min had no influence on its antibacterial activity.
The clear zones observed in bacteria lawns were similar in diameter
to the samples that were not treated. However, when cecropin D was
treated for 45 min with either HCl or NaOH, the diameters of the
cleared zones decreased to 4 mm for E. coli K99, 5 mm for
Streptococcus equi ssp. zooepidemicus (SEZ), 3 mm for B.
pumilus and 5 mm for S. aureus strain Cowan I, but not
for any of the other bacteria species included.
Cecropin D in culture supernatant was exposed to
trypsin and pepsin for 15, 30 and 45 min. There was no difference
in the diameter of the cleared zones in the antibacterial assay
between protease-treated supernatants and controls. This indicated
that proteases did not have any effect on the antibacterial
activity of cecropin D.
Optimization of fermentation
Initial experiments were performed to explore
whether it was possible to improve the expression level of cecropin
D when using various carbon or nitrogen sources and with various
initial concentrations of glucose. Under the assumption that
improved yeast cell growth would lead to increased expression
levels of the peptide, the change in OD600 of the yeast
cells per 12 h was determined as an indicator of yeast cell growth.
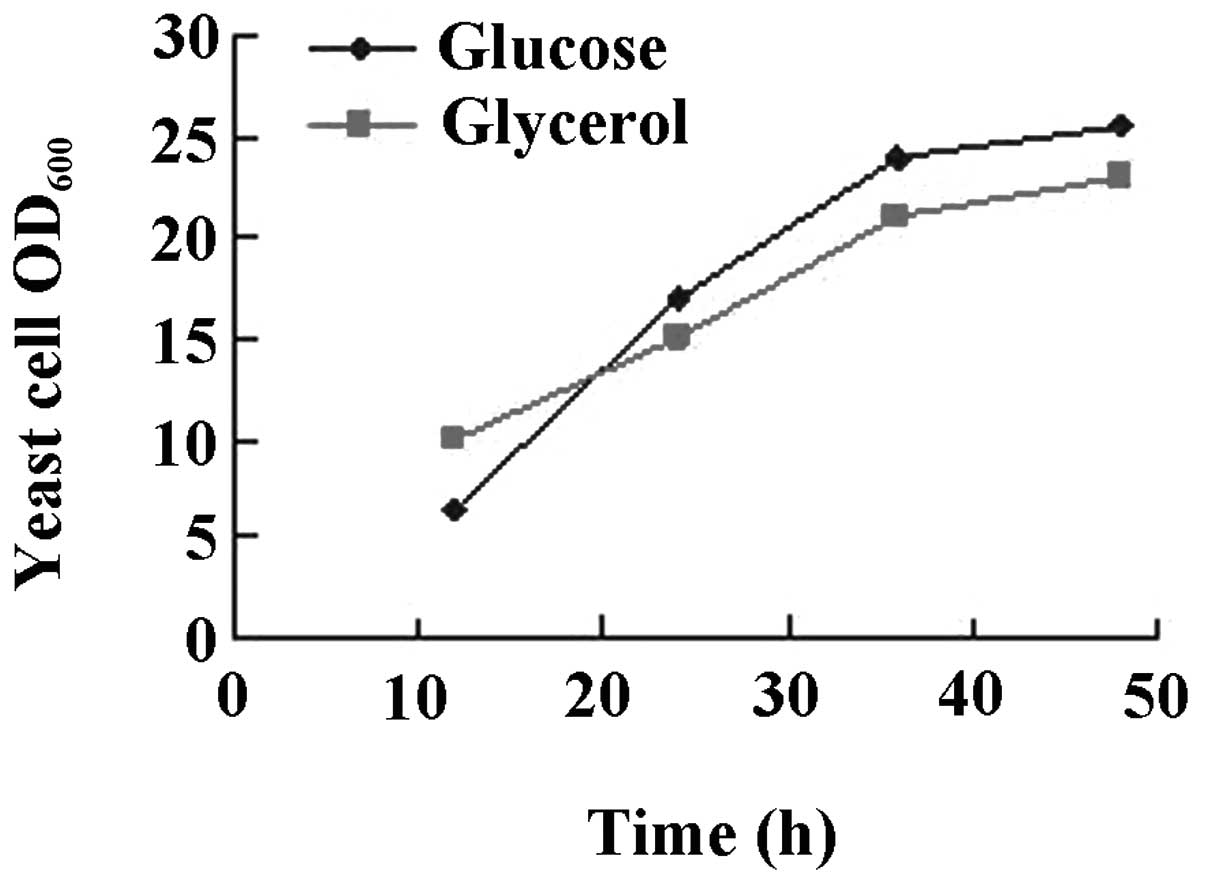
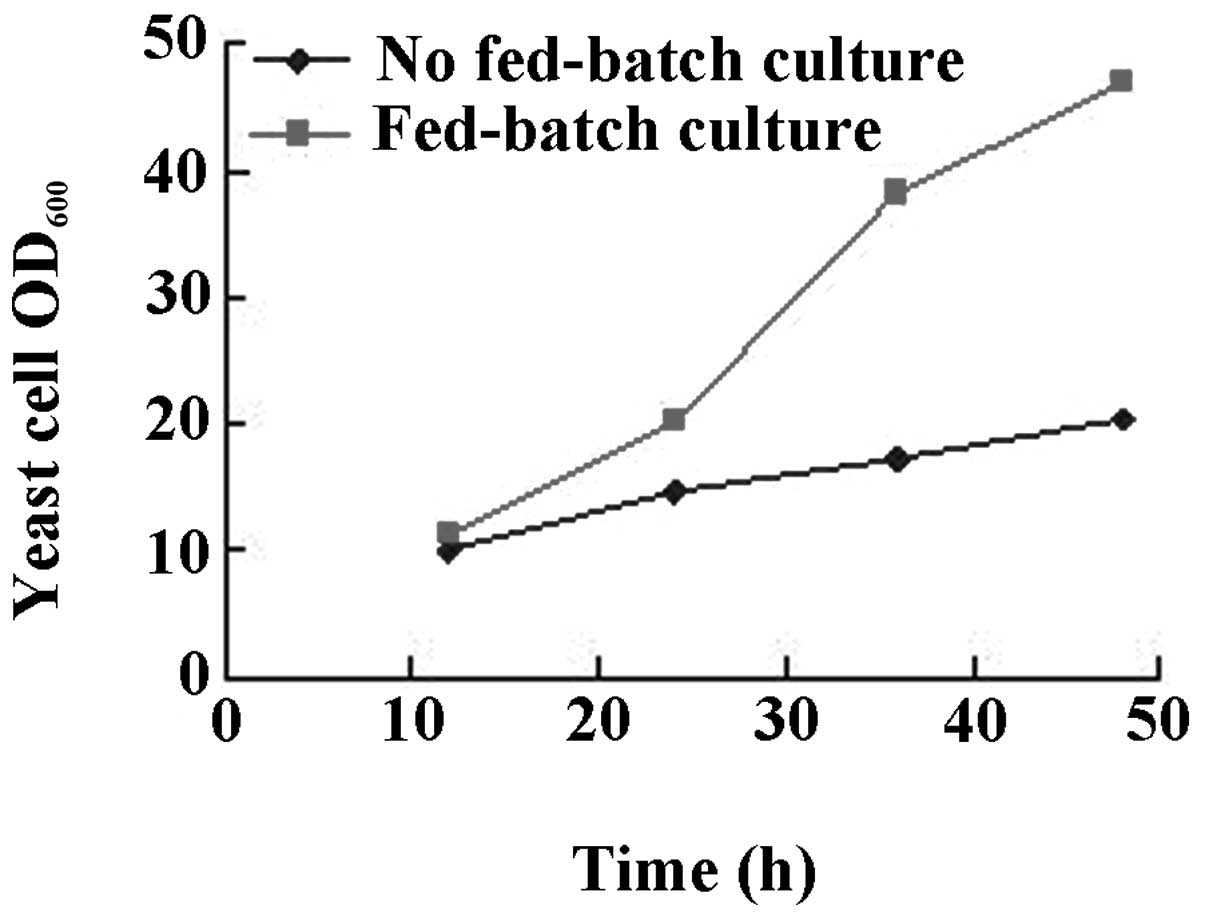
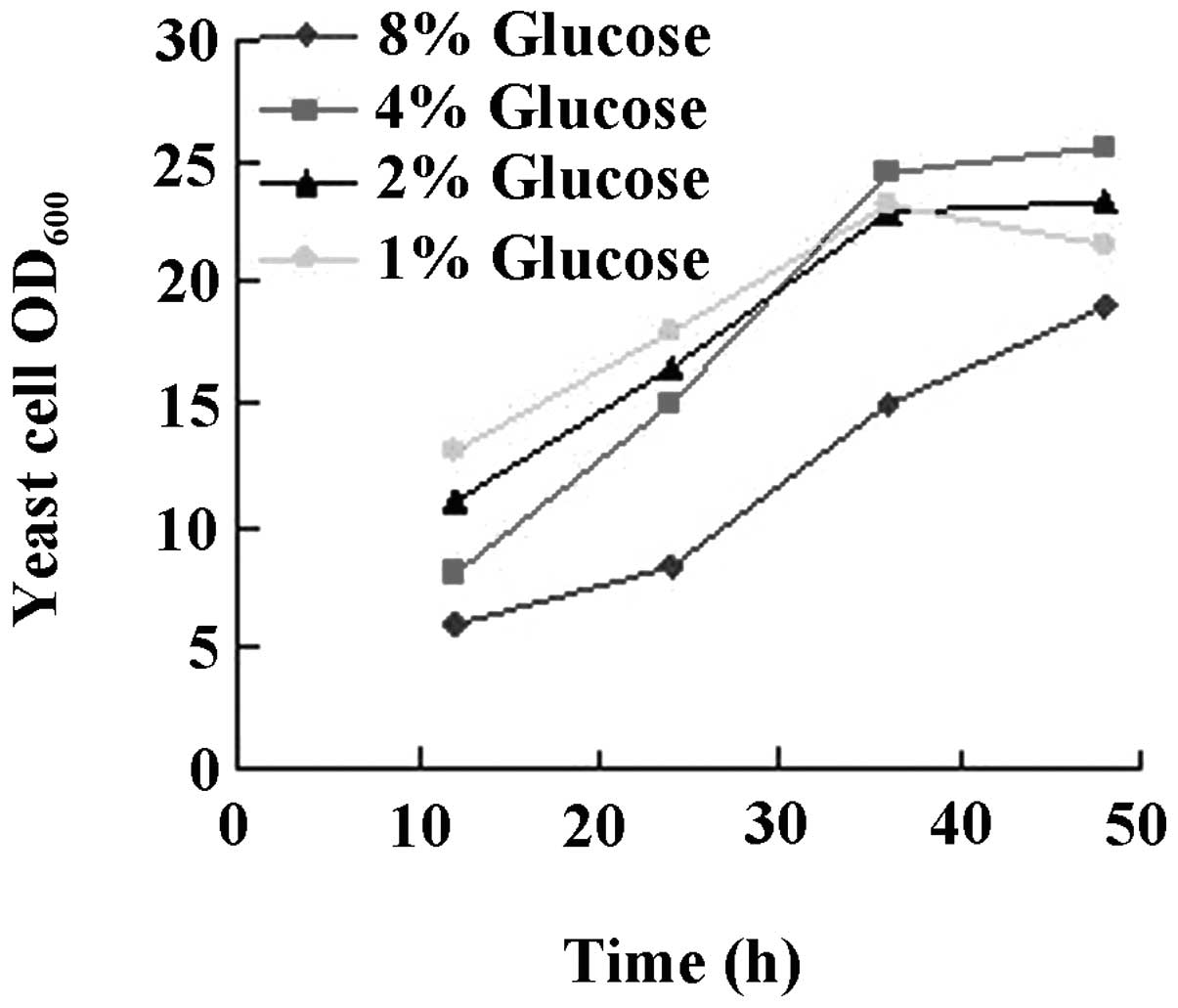
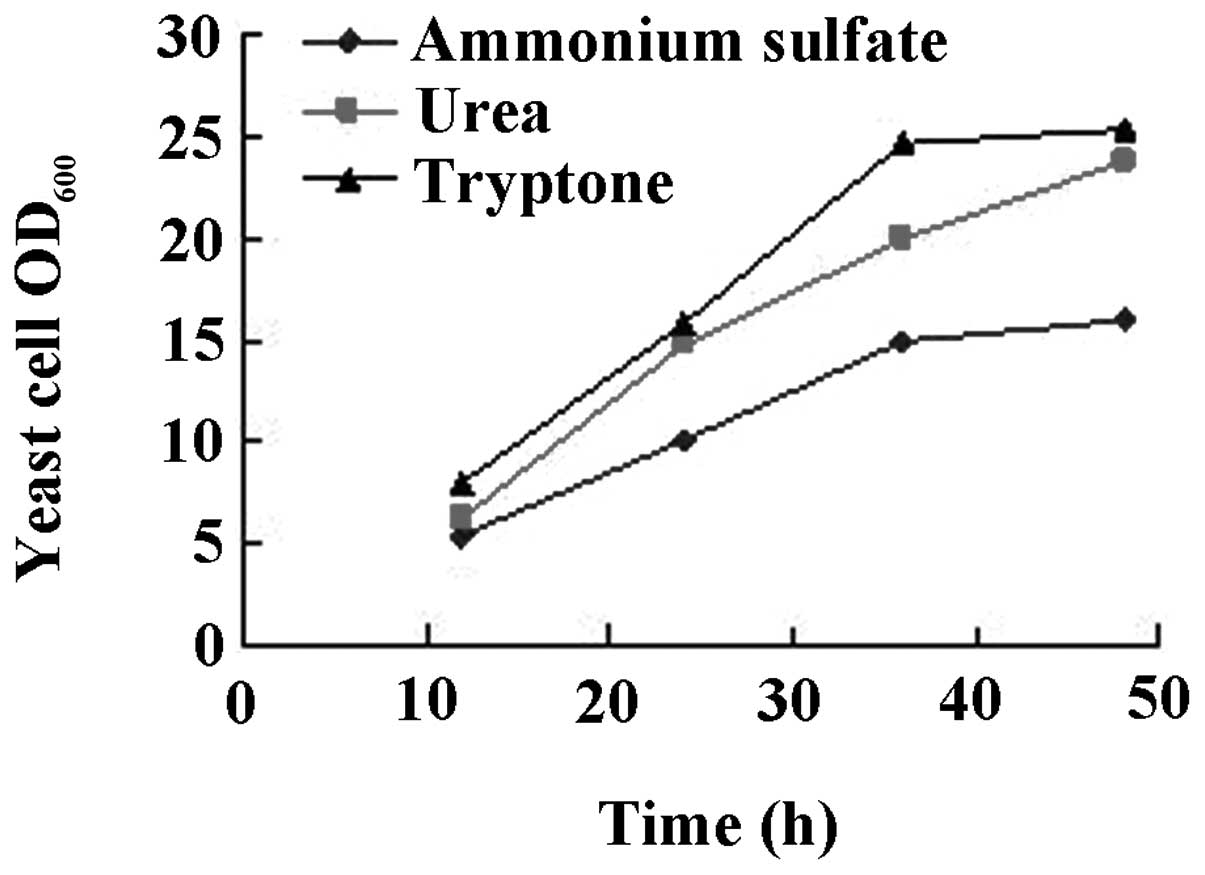
The results are shown in Figs.
3–6. While there was no
difference between glucose or glycerol as the carbon source
(Fig. 3), initiation of the
cultures with 4% glucose had a beneficial effect on yeast growth
(Fig. 4). When urea was used as
the nitrogen source, the level of cell growth was improved compared
with ammonium sulfate. However, there was no significant difference
between urea or tryptone as nitrogen source (Fig. 5). Intermittent batch feeding had a
beneficial effect on cell production as shown in Fig. 6.
Discussion
In the present study the cloning of the cecropin D
gene and its successful expression in P. pastoris were
reported. A base mutation that was found in the cloned cecropin D
gene may have occurred as an error in PCR, sequencing or may be
associated with changes to the codon bias for P. pastoris.
However, optimizing the codons of the gene to the preferred codon
usage of P. pastoris was required to increase expression to
the desired levels. To improve the expression levels of cecropin D,
the protease-deficient P. pastoris strain SMD1168 was
selected, high-copy transformants at a high concentration of Zeocin
were screened for and appropriate concentrations of EDTA
formaldehyde, or protease inhibitors were added to prevent protein
degradation. To optimize the conditions for large-scale
fermentation, initial experiments were performed to explore various
carbon and nitrogen sources and their effects on fermentation
conditions. Considering cost and expressed peptide levels, using
glucose as the carbon source was superior to glycerol, while urea
as a nitrogen source was superior to tryptone or ammonium sulfate.
In addition, the intermittent addition of carbon and nitrogen
(fed-batch culture) was more conducive to the growth of yeast
cells. The amount of dissolved oxygen in the fermentation tank was
vital to the growth of yeast cells and had a positive effect on the
expression levels. Particularly on the third day of fermentation, a
higher dissolved oxygen was required to guarantee a pH between 5.5
and 6.0 in the fermentation broth.
The P. pastoris expression system has been
developed into an excellent tool for the large-scale expression of
proteins from various sources (17). Its advantages are the ability to
perform many of the post-translational modifications of higher
eukaryotes and to secrete high levels of heterologous proteins into
the supernatant under the control of the glyceraldehyde-3-phosphate
dehydrogenase (GAP) promoter. P. pastoris also differs from
bacterial systems in that the vector containing the desired gene is
integrated into the genome during transformation (homologous
recombination) (18). Furthermore,
the vector pGAPZαA does not require methanol for induction, high
levels of which are toxic to cells (19), making pGAPZαA an extremely
convenient expression vector. In the present study cecropin D was
demonstrated to be functionally expressed in P. pastoris.
However, P. pastoris secretes various native proteins and
these must be removed during the purification process of the
desired recombinant protein. To enhance the utility of P.
pastoris, a detailed profile of host-secreted proteins with
regard to their identities and physical properties may provide
critical insights for improving recombinant protein secretion and
purification. D-glucose concentration is a key parameter affecting
protein expression levels in the P. pastoris expression
system. While the expression levels of cecropin D in P.
pastoris were increased with D-glucose concentration, overly
high D-glucose concentrations were unfavorable for protein
expression. This result was similar to that obtained by Johnson
et al (20).
It has been reported that the native N-terminal
segment is a prerequisite for maintaining the activity of
antibacterial peptides (21).
Previously, cecropin1 from M. domestica was expressed in
bacterial cells as a fusion protein with glutathione S-transferase.
Recombinant cecropin1 was obtained following thrombin digestion but
additional amino acid residues at the N-terminus of the recombinant
cecropin1 decreased its activity against microbes (22). Pisum sativum defensin 1
(Psd1) was expressed in P. pastoris, but the recombinant
Psd1 contained 4 additional amino acids (EAEA) at the N-terminus.
In comparison with native Psd1, the anti-fungal activity of the
recombinant Psd1 was decreased by at least 10-fold (23). In order to obtain cecropin D with
the native N-terminus, the gene encoding cecropin D including the
KEX2 cleavage site was cloned. Tricine-SDS-PAGE revealed that
cecropin D was successfully secreted into the culture supernatant
using the α-mating factor signal sequence. Activity assays
demonstrated that cecropin D had a low MIC against Gram-negative
and Gram-positive bacteria. Therefore, the α-factor signal sequence
was efficient at secreting recombinant proteins into the culture
medium and the signal peptide was efficiently processed by the KEX2
protease of P. pastoris.
The large-scale production of peptide antibiotics
such as cecropin provides a significant challenge for developing
commercial products. Bacterial expression systems may not be used
to express cecropin and cecropin-like peptides, such as sarcotoxin
IA, directly (24). Only small
amounts of sarcotoxin IA were obtained when it was expressed in
Bombyx mori cells using a baculovirus expression system (20
μg/450 ml) (25) or in the
yeast Saccharomyces cerevisiae (80 μg/1) (26). In the present study, P.
pastoris was used for the expression of cecropin D for the
following reasons: firstly, it is fast and inexpensive to culture
yeast and secondly, yeast cells are eukaryotes so they have the
machinery for post-translational modifications (27).
In conclusion, the present study demonstrated that
the antibacterial peptide cecropin D may be expressed in a
heterologous expression system, P. pastoris, using the yeast
secretion signal, α-mating factor. Recombinant cecropin D was
expressed at high levels (up to 485.24 mg/l culture medium) and
exhibited antimicrobial activity against Gram-positive and
Gram-negative bacteria. Taken together, the results demonstrate
that P. pastoris is a robust system for expressing the
secreted form of recombinant cecropin D.
References
|
1
|
Lee DG, Park JH, Shin SY, Lee SG, Lee MK,
Kim KL and Hahm KS: Design of novel analogue peptides with potent
fungicidal but low hemolytic activity based on the cecropin
A-melittin hybrid structure. Biochem Mol Biol Int. 43:489–498.
1997.PubMed/NCBI
|
|
2
|
Steiner H, Hultmark D, Engström A, Bennich
H and Boman HG: Sequence and specificity of two antibacterial
proteins involved in insect immunity. Nature. 292:246–248. 1981.
View Article : Google Scholar
|
|
3
|
Hultmark D, Steiner H, Rasmuson T and
Boman HG: Insect immunity: purification and properties of three
inducible bactericidal proteins from hemolymph of immunized pupae
of Hyalophora cecropia. Eur J Biochem. 106:7–16. 1980.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hultmark D, Engström A, Bennich H, Kapur R
and Boman HG: Insect immunity: isolation and structure of cecropin
D and four minor antibacterial components from Cecropia pupae. Eur
J Biochem. 127:207–217. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dickinson L, Russell V and Dunn PE: A
family of bacteria-regulated, cecropin D-like peptides from
Manduca sexta. J Biol Chem. 263:19424–19429. 1988.PubMed/NCBI
|
|
6
|
Morishima I, Suginaka S, Ueno T and Hirano
H: Isolation and structure of cecropins, inducible antibacterial
peptides, from silkworm Bombyx mori. Comp Biochem Physiol B.
95:551–554. 1990.PubMed/NCBI
|
|
7
|
Kylsten P, Samakovlis C and Hultmark D:
The cecropin locus in Drosophila; a compact gene cluster
involved in the response to infection. EMBO J. 9:217–224.
1990.PubMed/NCBI
|
|
8
|
Okada M and Natori S: Primary structure of
sacrotoxin I, an antibacterial protein induced in the hemolymph of
Sarcophaga peregrine (flesh fly) larvae. J Biol Chem.
260:7174–7177. 1985.PubMed/NCBI
|
|
9
|
Kobayashi S and Uchimiya H: Expression and
integration of a foreign gene in orange (Citrus sinensis
Osb.) protoplasts by direct DNA transfer. Jpn J Genet. 64:91–97.
1989. View Article : Google Scholar
|
|
10
|
Cregg JM, Cereghino JL, Shi J and Higgins
DR: Recombinant protein expression in Pichia pastoris. Mol
Biotechnol. 16:23–52. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Daly R and Hearn MT: Expression of
heterologous proteins in Pichia pastoris: a useful
experimental tool in protein engineering and production. J Mol
Recognit. 18:119–138. 2005.PubMed/NCBI
|
|
12
|
Damasceno LM, Pla I, Chang HJ, Cohen L,
Ritter G, Old LJ and Batt CA: An optimized fermentation process for
high-level production of a single-chain Fv antibody fragment in
Pichia pastoris. Protein Expr Purif. 37:18–26. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Minagawa S, Hikima J, Hirono I, Aoki T and
Mori H: Expression of Japanese flounder c-type lysozyme cDNA in
insect cells. Dev Comp Immunol. 25:439–445. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lehrer RI, Rosenman M, Harwig SS, Jackson
R and Eisenhauer P: Ultrasensitive assays for endogenous
antimicrobial polypeptides. J Immunol Methods. 137:167–173. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jin FL, Xu X, Zhang W and Gu D: Expression
and characterization of a housefly cecropin gene in the
methylotrophic yeast, Pichia pastoris. Protein Expr Purif.
49:39–46. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cipáková I, Hostinová E, Ganperik J and
Velebný V: High-level expression and purification of a recombinant
hBD-1 fused to LMM protein in Escherichia coli. Protein Expr
Purif. 37:207–212. 2004.PubMed/NCBI
|
|
17
|
Valore EV and Ganz T: Laboratory
production of antimicrobial peptides in native conformation.
Methods Mol Biol. 78:115–131. 1997.PubMed/NCBI
|
|
18
|
Mølhøj M, Ulvskov P and Dal Degan F:
Characterization of a functional soluble form of a Brassica
napus membrane-anchored endo-1,4-beta-glucanase heterologously
expressed in Pichia pastoris. Plant Physiology. 127:674–684.
2001.
|
|
19
|
Peres MdFS, Silva VC, Valentini SR and
Gattás EAdL: Recombinant expression of glycerol-3-phosphate
dehydrogenase using the Pichia pastoris system. J Mol Catal
B Enzym. 65:128–132. 2010. View Article : Google Scholar
|
|
20
|
Johnson SC, Yang M and Murthy PP:
Heterologous expression and functional characterization of a plant
alkaline phytase in Pichia pastoris. Protein Expr Purif.
74:196–203. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu M and Hancock RE: Interaction of the
cyclic antimicrobial cationic peptide bactenecin with the outer and
cytoplasmic membrane. J Biol Chem. 274:29–35. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liang YL, Wang JX, Zhao XF, Du XJ and Xue
JF: Molecular cloning and characterization of cecropin from the
housefly (Musca domestica), and its expression in
Escherichia coli. Dev Comp Immunol. 30:249–257. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cabral KM, Almeida MS, Valente AP, Almeida
FC and Kurtenbach E: Production of the active antifungal Pisum
sativum defensin 1 (Psd1) in Pichia pastoris: overcoming
the inefficiency of the STE13 protease. Protein Expr Purif.
31:115–122. 2003.PubMed/NCBI
|
|
24
|
Skosyrev VS, Kulesskiy EA, Yakhnin LV,
Temirov YV and Vinokurov LM: Expression of the recombinant
antibacterial peptide sarcotoxin IA in Escherichia coli
cells. Protein Expr Purif. 28:350–356. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Aly R, Granot D, Mahler-Slasky Y, Halpern
N, Nir D and Galun E: Saccharomyces cerevisiae cells
harboring the gene encoding Sarcotoxin IA secrete a peptide that is
toxic to plant pathogenic bacteria. Protein Expr Purif. 16:120–124.
1999. View Article : Google Scholar
|
|
26
|
Yamada K, Nakajima Y and Natori S:
Production of reombinant sarcotoxin IA in Bombyx mori cells.
Biochem J. 272:633–636. 1990.PubMed/NCBI
|
|
27
|
Cereghino JL and Cregg JM: Heterologous
protein expression in the methylotrophic yeast Pichia
pastoris. FEMS Microbiol Rev. 24:45–66. 2000. View Article : Google Scholar
|