Introduction
In China, the hepatitis B surface antigen (HBsAg)
seropositive rate for the general population (between 1 and 59
years of age) is 7.18% (1).
Globally, there are ~93 million individuals with hepatitis B virus
(HBV) infections, 20 million of which are chronic (2). Hepatitis B is the most common risk
factor for liver cirrhosis and hepatocellular carcinoma (HCC), and
the mortality of HBV-induced acute-on-chronic liver failure (ACLF)
may exceed 60%. HBV infections are not easy to cure, and the
mechanisms of the virus are unclear, particularly with regard to
protein expression and regulation function in the pathogenic
process.
Proteomics analysis is a powerful technology used in
a myriad of studies, including those focused on liver diseases
(3–7). Isobaric tags for relative and
absolute quantitation (iTRAQ), as a quantitative method, is a
common tool in proteomics and has been suggested to be as sensitive
as (or more sensitive than) the differential in gel electrophoresis
(DIGE) technique (8).
Specifically, the iTRAQ method has been used to study a variety of
diseases and has been shown to be effective and accurate (4,5,9).
The purpose of this study was to analyze serum
protein levels using iTRAQ in normal controls, as well as patients
with chronic hepatitis B (CHB) and HBV-induced ACLF, and to verify
those results using western blotting. The ultimate aim was to
identify the differences in serum protein levels that were closely
associated with the progression of hepatitis B and HBV-induced
ACLF. The results of this study may provide crucial information
regarding viral mechanisms and the pathogenic process.
Materials and methods
Patients and specimens
Serum samples from healthy individuals and patients
with CHB and HBV-induced ACLF were obtained from the Department of
Infectious Diseases and the Department of Traditional Chinese
Medicine of the Third Affiliated Hospital of Sun Yat-sen University
(Guangzhou, China) (Tables I and
II). HBV-induced ACLF was
diagnosed using the previously described criteria (10,11).
The exclusion criteria included pregnant and lactating females,
patients that had been treated with antivirals or immunomodulatory
therapy within the previous six months, the presence of other
factors causing active liver diseases (e.g. autoimmune,
drug-induced liver, alcoholic liver and inherited metabolic liver
diseases), concomitant human immunodeficiency virus (HIV) infection
or congenital immune deficiency diseases, liver cancer or other
malignancies, severe diabetes, autoimmune diseases, other important
organ dysfunctions (e.g. kidney dysfunction), concomitant infection
(e.g. fever, leukocytosis or neutrophilia; manifestations of
abdominal, biliary tract or lung infection) or other serious
comorbidities (e.g. hepatic encephalopathy and gastrointestinal
bleeding).
 | Table I.Clinical characteristics of the 15
serum samples for iTRAQ testing. |
Table I.
Clinical characteristics of the 15
serum samples for iTRAQ testing.
| No. | Source of serum | Gender | Age (years) | HBsAg | HBsAb | HBeAg | HBeAb | HBcAb | AST (14.5–40.0
U/l) | ALT (3–35 U/l) | TBIL (4.0–23.9
μmol/l) | PT (11.0–14.5
sec) | HBV DNA (IU/ml) |
|---|
| 1 | Healthy control | Male | 34 | − | + | − | − | − | 21.0 | 31 | 13.8 | 12.3 | <100 |
| 2 | Healthy control | Male | 28 | − | + | − | − | − | 31.0 | 30 | 20.1 | 13.1 | <100 |
| 3 | Healthy control | Male | 36 | − | + | − | − | − | 15.0 | 24 | 20.1 | 13.5 | <100 |
| 4 | Healthy control | Male | 27 | − | + | − | − | − | 19.5 | 22 | 22.0 | 11.8 | <100 |
| 5 | Healthy control | Male | 31 | − | + | − | − | − | 17.5 | 20 | 13.3 | 12.7 | <100 |
| 6 | CHB patient | Male | 48 | + | − | + | − | + | 45.0 | 82 | 20.7 | 12.1 |
8.03×107 |
| 7 | CHB patient | Male | 28 | + | − | + | − | + | 38.0 | 84 | 22.4 | 14.1 |
1.76×106 |
| 8 | CHB patient | Male | 32 | + | − | + | − | + | 39.0 | 43 | 15.9 | 13.5 |
8.18×106 |
| 9 | CHB patient | Male | 24 | + | − | + | − | + | 36.0 | 38 | 21.6 | 12.9 |
1.02×106 |
| 10 | CHB patient | Male | 22 | + | − | + | − | + | 49.0 | 57 | 18.7 | 14.2 |
3.21×106 |
| 11 | HBV-induced ACLF
patient | Male | 39 | + | − | − | + | + | 26.0 | 35 | 463.5 | 25.5 |
6.52×104 |
| 12 | HBV-induced ACLF
patient | Male | 51 | + | − | + | − | + | 78.0 | 65 | 312.5 | 35.2 |
1.78×106 |
| 13 | HBV-induced ACLF
patient | Male | 46 | + | − | + | − | + | 101.0 | 46 | 556.7 | 29.1 |
3.55×106 |
| 14 | HBV-induced ACLF
patient | Male | 55 | + | − | + | − | + | 58.0 | 110 | 636.8 | 32.3 |
4.21×105 |
| 15 | HBV-induced ACLF
patient | Male | 32 | + | − | − | + | + | 115.0 | 78 | 482.5 | 28.8 |
6.17×105 |
 | Table II.Clinical characteristics of the nine
serum samples for western blotting verification. |
Table II.
Clinical characteristics of the nine
serum samples for western blotting verification.
| No. | Source of
serum | Gender | Age (years) | HBsAg | HBsAb | HBeAg | HBeAb | HBcAb | AST (14.5–40.0
U/l) | ALT (3–35 U/l) | TBIL (4.0–23.9
μmol/l) | PT (11.0–14.5
sec) | HBV DNA
(IU/ml) |
|---|
| 1 | Healthy
control | Male | 36 | − | + | − | − | − | 21.0 | 31.0 | 13.8 | 12.3 | <100 |
| 2 | Healthy
control | Male | 30 | − | + | − | − | − | 15.0 | 24.0 | 20.1 | 11.1 | <100 |
| 3 | Healthy
control | Male | 30 | − | + | − | − | − | 19.2 | 28.6 | 15.6 | 12.8 | <100 |
| 4 | CHB patient | Male | 41 | + | − | + | − | + | 32.0 | 75.4 | 20.5 | 11.3 |
1.88×106 |
| 5 | CHB patient | Male | 31 | + | − | + | − | + | 45.0 | 47.0 | 19.7 | 14.5 |
7.24×106 |
| 6 | CHB patient | Male | 37 | + | − | + | − | + | 36.0 | 41.0 | 23.6 | 11.9 |
3.22×106 |
| 7 | HBV-induced ACLF
patient | Male | 21 | + | − | + | − | + | 104.5 | 68.0 | 278.9 | 27.3 |
5.66×106 |
| 8 | HBV-induced ACLF
patient | Male | 37 | + | − | + | − | + | 67.0 | 54.0 | 342.1 | 29.7 |
6.58×106 |
| 9 | HBV-induced ACLF
patient | Male | 43 | + | − | − | + | + | 36.0 | 78.0 | 521.4 | 31.5 |
7.33×105 |
The study protocol conformed to the Ethical
Guidelines of the 1975 Declaration of Helsinki and was approved by
the appropriate Institutional Review Committee of Third Affiliated
Hospital of Sun Yat-sen University (Guangzhou, China) and the
Bureau of Health (Guangdong, China). Informed patient consent was
obtained prior to participation in this study.
Sample preparation and protein
extraction
Plasma was fractionated with ProteinMiner Protein
Enrichment Small-Capacity kit, Cat# 163-3006, (ProteinMiner ;
Bio-Rad Laboratories, Hercules, CA, USA) according to the
manufacturer’s instructions. Protein solutions were reduced for 1 h
at 56°C with 10 mM dithiothreitol (DTT) and were cysteine-blocked
with 55 mM iodoacetamide (IAA) at room temperature for 10 min. Each
sample was precipitated with four-times the volume of cold acetone.
The total protein concentration was determined using the Bradford
assay.
Protein digestion and peptide
tagging
Protein solutions [100 μg; 5 mg/ml in 0.5 M
triethyl ammonium bicarbonate (TEAB) containing 0.1% sodium dodecyl
sulfate, pH 8.5]were digested for 24 h with 10 μg
L-1-(4-tosylamido)-2-phenylethyl tosylphenylalanyl chloromethyl
ketone (TPCK)-treated trypsin. Each peptide solution was labeled
for 3 h at room temperature using an iTRAQ reagent that had been
reconstituted in 70 liters of ethanol, in accordance with the iTRAQ
Reagents Multiplex kit instructions (Applied Biosystems, Foster
City, CA, USA). The reaction was terminated by adding MilliQ water,
and the samples were labeled with 114, 115, 116 and 117 mass-tagged
iTRAQ reagents.
Strong cation exchange chromatography
(SCX)
SCX was performed to remove excess iTRAQ reagents
and interfering substances for the mass analysis. The labeled
peptides were then dried in a vacuum centrifuge and resuspended in
200 ml Buffer A, prior to being loaded on a Phenomenex Luna 55
μm SCX 100A column [250×4.60 mm (length × internal
diameter), 5 μm; Phenomenex, Inc., Torrance, CA, USA] on an
Agilent 1100 HPLC unit (Agilent Technologies, Santa Clara, CA,
USA). Buffer A consisted of 10 mM KH2PO4 and
25% acetonitrile at pH 3.0, while Buffer B consisted of 10 mM
KH2PO4, 25% acetonitrile and 2 M KCl at pH
3.0. The 60 min gradient comprised 100% Buffer A for 30 min, 0–5%
Buffer B for 1 min, 5–30% Buffer B for 15 min, 30–50% Buffer B for
5 min, 50% Buffer B for 4 min and 50–100% Buffer A for 5 min.
Thirteen fractions were collected using a Foxy JR Fraction
Collector (Dionex Corp., Sunnyvale, CA, USA). Fractions 2 and 3
were pooled according to the chromatogram profile, based on the
peak intensity. All fractions were then dried in a vacuum
concentrator and stored at −20°C, prior to further analysis using
mass spectrometry.
Nano liquid chromatography (LC) coupled
to quadrupole time-of-flight (Q-TOF) with tandem mass spectrometry
(MS/MS)
Peptides from the SCX fractions were dissolved in
0.3 μl 100% formic acid and diluted to 5 μl in 0.05%
trifluoroacetic acid (TFA). The peptides were loaded onto a
micrOTOF-Q II-nano LC system (Bruker Daltonics, Inc., Billerica,
MA, USA) and mass spectra were acquired in the 250–1,600 m/z range
every second for 60 min.
Database screening
Data were processed using BioTools software (Bruker
Daltonics, Inc.). The files were subsequently submitted to an
in-house Mascot server (Matrix Science Ltd., London, UK) for
database screening. The data were screened against the human
sequence database [National Center for Biotechnology Information
non-redundant (NCBInr)]. The search was performed using trypsin as
a specific enzyme. A maximum of one missed cleavage was permitted
and oxidation (M), iTRAQ 4 plex (K) and iTRAQ 4 plex (N-term) were
selected as variable modifications. The data obtained on the
micrOTOF-Q were searched with a peptide mass tolerance of 10 ppm
and a fragment mass tolerance of 0.8 Da. Protein ratios were
normalized using the overall median ratio for all the peptides in
the sample for each separate ratio in every individual experiment.
The ratio for a given protein was calculated by taking the average
of all the peptide ratios that identified the protein. The final
list of protein ratios was an average of the protein ratios of the
three experiments and consisted only of proteins discovered in ≥2
of the three experiments. Function definitions of the variable
protein contents were searched for using the following two
websites: http://www.uniprot.org/ and http://www.ncbi.nlm.nih.gov/.
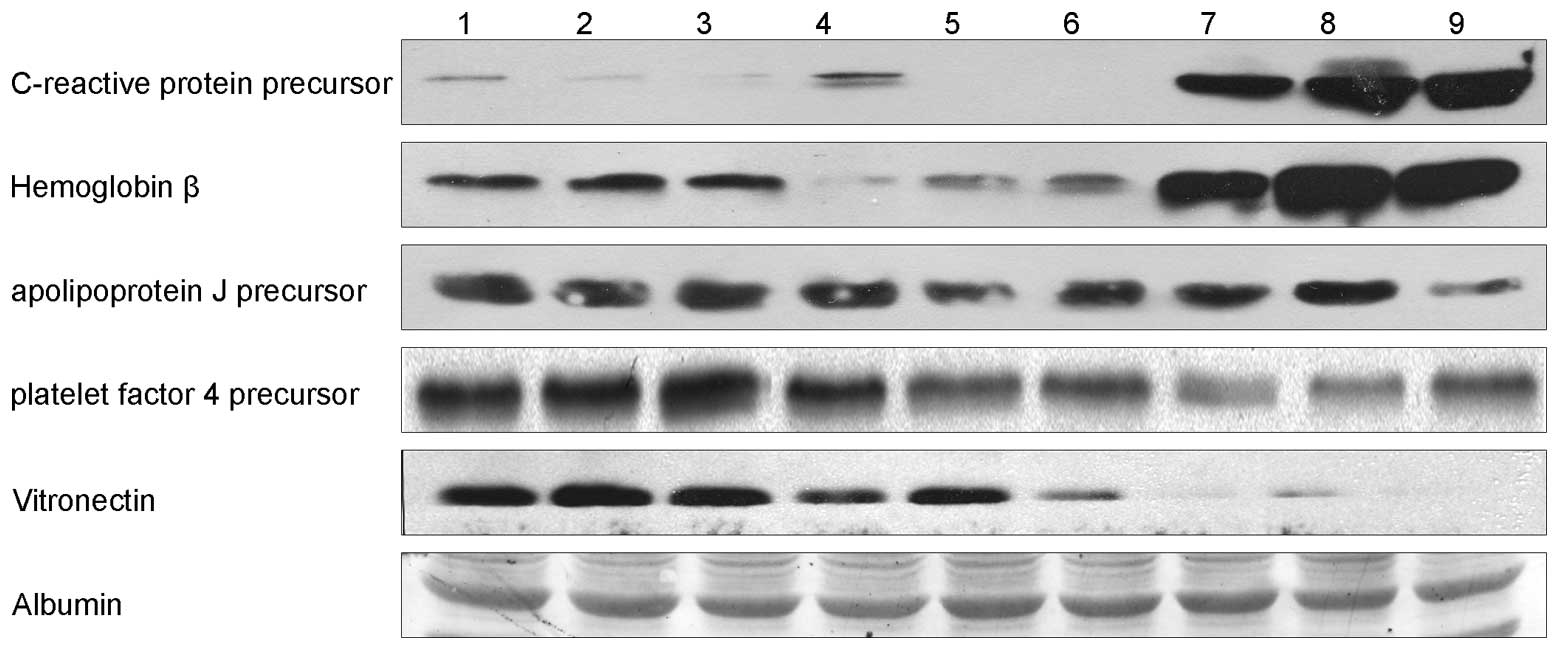
Western blotting
Five proteins [C-reactive protein (CRP) precursor,
hemoglobin β chain variant Hb S-Wake, apolipo-protein J precursor,
platelet factor 4 precursor and vitronectin (VN)], which
demonstrated the greatest differences in their expression levels
and the most significant correlation with liver diseases, were
chosen and verified by western blotting using western blotting kits
(Forevergen, Guangzhou, China). Briefly, the protein lysates were
separated by polyacrylamide gel electrophoresis, transferred to a
polyvinylidene difluoride (PVDF) membrane and subjected to
immunoblotting with the following antibodies: Anti-apolipoprotein J
precursor antibody (ab16077; dilution, 1:2,000; Abcam, Cambridge,
UK), anti-hemoglobin β (sc-21757; dilution, 1:500; Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA), anti-CRP precursor
(ab32412; dilution, 1:1,000; Abcam), anti-platelet factor 4
precursor (AB1488P; dilution, 1:5,000; Millipore, Billerica, MA,
USA) and anti-VN (ab11591; dilution, 1:500; Abcam) at 4°C
overnight. After washing, the membranes were incubated with
horseradish peroxidase-conjugated secondary antibodies and
visualized using an enhanced chemiluminescence system (ECL;
Forevergen, Guangzhou, China).
Results
Tables I and
II summarize the clinical
characteristics of the patients from whom the samples were
collected. iTRAQ identified 16 different proteins that had
≥1.5-fold differences in expression level between the patients with
HBV-induced ACLF and CHB, respectively, and the healthy controls
(Table III). Protein
quantification software, based on the relative content of the
isotopic reporter group and using m/z 114 as a reference, showed
significant results (P≤0.05). Some identified protein was lost in
the corresponding reporter group, giving no quantification
information. The specific information and re-identification will be
published in a future study.
 | Table III.The 16 proteins that were identified
using iTRAQ as having ≥1.5-fold differences in their expression
level between patients with HBV-induced ACLF and CHB, respectively,
and healthy controls. |
Table III.
The 16 proteins that were identified
using iTRAQ as having ≥1.5-fold differences in their expression
level between patients with HBV-induced ACLF and CHB, respectively,
and healthy controls.
| Accession no. | Protein name | Biological
processes | Molecular
function | Protein
function | Levels in patients
with CHB versus controls | Levels in
HBV-induced ACLF patients versus controls |
|---|
| Q7TMA5 | Apolipoprotein
B-100 precursor | Cholesterol and
lipid metabolism, lipid transport | Heparin
binding | Transporter | Upregulated
1.62-fold | Upregulated
1.32-fold |
| P04004 | Vitronectin | Cell adhesion | Heparin and
integrin binding | Cell adhesion
molecule | Downregulated
1.23-fold | Downregulated
2.14-fold |
| P01764 | Monoclonal IgM
antibody heavy chain | Immune
response | Antigen
binding | Defense/immunity
protein | Upregulated
1.41-fold | Upregulated
1.87-fold |
| Unclassified | Unnamed protein
product | Unclassified | Unclassified | Unclassified | Downregulated
1.23-fold | Downregulated
2.14-fold |
| P02768 | Serum albumin
preproprotein | Transport | Binding
capacity | Transporter | Upregulated
1.86-fold | Upregulated
2.83-fold |
| Unclassified | Apolipoprotein J
precursor | Unclassified | Unclassified | Unclassified | Downregulated
1.23-fold | Downregulated
2.00-fold |
| Unclassified | Unnamed protein
product | Unclassified | Unclassified | Unclassified | Upregulated
1.07-fold | Upregulated
2.00-fold |
| D3DUX1 | Serpin peptidase
inhibitor, clade A | Regulatory | Serine-type
endopeptidase inhibitor activity | Regulatory
molecular | Normal | Upregulated
2.64-fold |
| P00450 | Ceruloplasmin
(ferroxidase), isoform CRA_b | Copper and iron
transport | Chaperone binding,
ferroxidase activity | Transporter | Upregulated
2.30-fold | Upregulated
2.64-fold |
| P07203 | Glutathione
peroxidase | UV protection, anti
apoptosis, cell redox homeostasis and glutathione metabolic
process | Oxidoreductase,
peroxidase | Regulatory
molecular | Downregulated
1.07-fold | Upregulated
2.46-fold |
| P68871 | Hemoglobin β chain
variant Hb S-Wake | Oxygen
transport | Hypotensive agent,
vasoactive | Transporter | Downregulated
1.07-fold | Upregulated
9.19-fold |
| B3KUE5 | Phospholipid
transfer protein, isoform CRA_c | Regulatory | Lipid binding | Regulatory
molecular | Upregulated
1.32-fold | Upregulated
4.92-fold |
| Unclassified | Platelet factor 4
precursor | Unclassified | Unclassified | Unclassified | Downregulated
1.15-fold | Downregulated
1.87-fold |
| P19095 | C reactive protein
precursor | Acute phase
response | Sugar binding | Unclassified | Downregulated
2.46-fold | Upregulated
4.59-fold |
| P18428 | Lipopolysaccharide
binding protein, isoform CRA_a | Lipid transport,
transport | Antibiotic,
antimicrobial | Transporter | Upregulated
1.07-fold | Upregulated
1.62-fold |
| P60709 | Actin, β | Cellular component
movement | ATP and kinesin
binding | Extracellular
matrix | Downregulated
1.23-fold | Upregulated
4.29-fold |
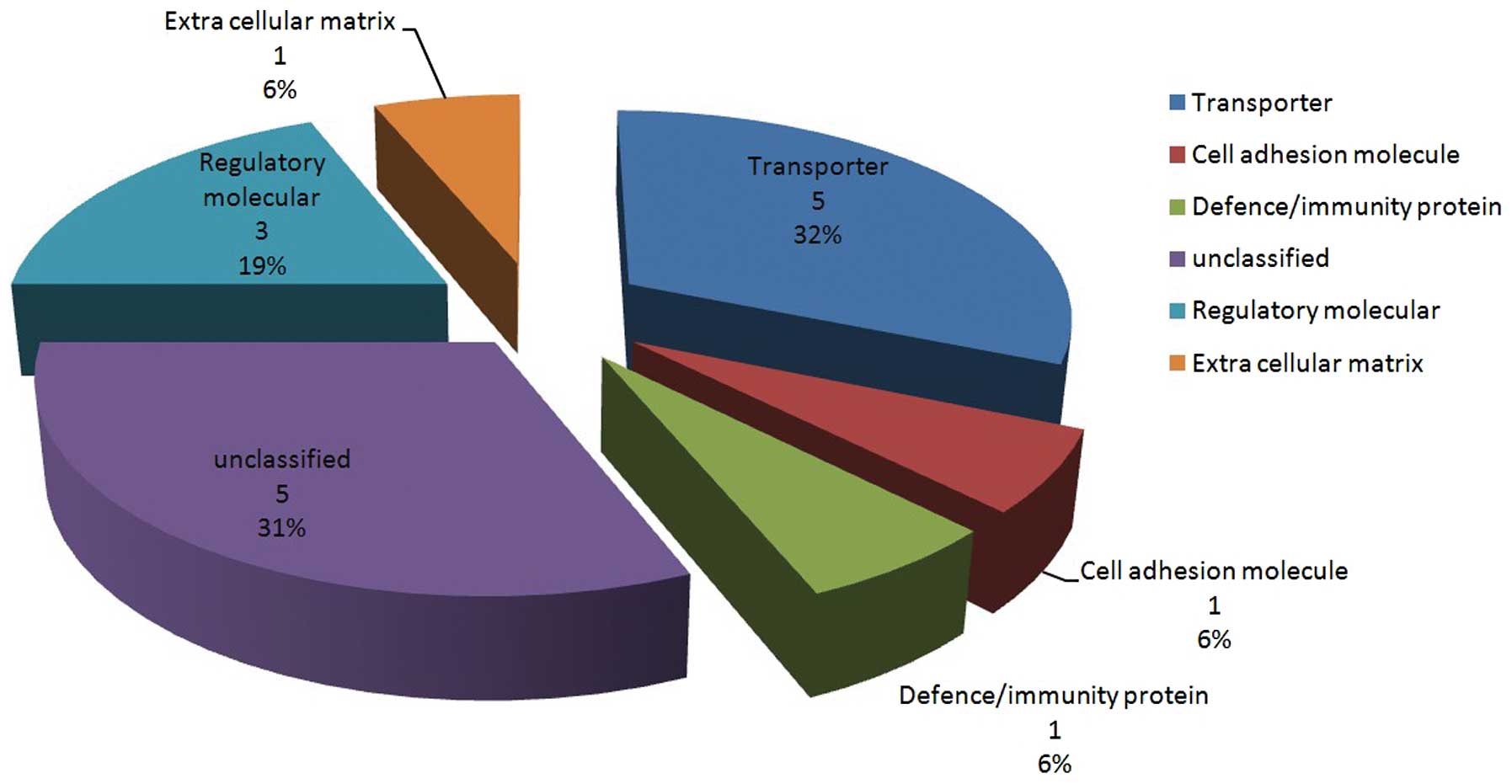
The 16 proteins that were identified using iTRAQ
were classified into six categories, based on protein function
(Fig. 1). The five proteins that
demonstrated the greatest differences in their expression and the
most significant correlation with liver diseases were verified
using western blotting: CRP precursor, hemoglobin β chain variant
Hb S-Wake, apolipoprotein J precursor, platelet factor 4 precursor
and VN (Fig. 2 and Table IV). Two of the five proteins were
not classified according to biological processes (apolipoprotein J
precursor and platelet factor 4 precursor).
 | Table IV.Five serum proteins with the greatest
differences in their expression levels and the most significant
correlation with liver diseases, as verified using western
blotting. |
Table IV.
Five serum proteins with the greatest
differences in their expression levels and the most significant
correlation with liver diseases, as verified using western
blotting.
| Accession no. | Protein name | Biological
processes | Molecular
function | Protein
function | Levels in CHB
patients versus controls | Levels in
HBV-induced ACLF patients versus controls |
|---|
| P19095 | C reactive protein
precursor | Acute phase
response | Sugar binding | Unclassified | Downregulated 2.46
fold | Upregulated 4.59
fold |
| P68871 | Hemoglobin β chain
variant Hb S-Wake | Oxygen
transport | Hypotensive agent,
vasoactive | Transporter | Downregulated
1.07-fold | Upregulated
9.19-fold |
| Unclassified | Apolipoprotein J
precursor | Unclassified | Unclassified | Unclassified | Downregulated
1.23-fold | Downregulated
2.00-fold |
| Unclassified | Platelet factor 4
precursor | Unclassified | Unclassified | Unclassified | Downregulated
1.15-fold | Downregulated
1.87-fold |
| P04004 | Vitronectin | Cell adhesion | Heparin and
integrin binding | Cell adhesion
molecule | Downregulated
1.23-fold | Downregulated
2.14-fold |
Discussion
Proteomics research is a potentially useful and
effective tool for studying pathogenesis, establishing prognosis
and determining treatment outcomes in a variety of diseases. In the
field of CHB, proteomics is not widely performed, presumably due to
the mechanism of CHB being so complicated. The aim of this study
was to describe the changes in serum protein levels in patients
with CHB and HBV-induced ACLF, respectively, compared with healthy
controls using iTRAQ and western blotting. The authors hypothesized
that this approach had the potential to ultimately be beneficial
for the identification of proteins that were important in the
progression of HBV infections.
In the present study, 16 unique proteins were
identified in patients with CHB and those with HBV-induced ACLF
that had ≥1.5-fold differences in expression compared with those in
the healthy controls. Those proteins belonged to six different
categories based on protein function, while two out of the five
proteins that were analyzed using western blotting were not
classified according to biological processes (apolipoprotein J
precursor and platelet factor 4 precursor). The biological
functions of the remaining proteins in HBV infection and
progression that were analyzed have not yet been elucidated;
however, a number of theories are discussed in the following
section.
CRP, synthesized in the liver, is an acute phase
protein that is rapidly identified in the plasma in cases of
infection or tissue damage. CRP is able to activate complement and
enhance phagocytosis to facilitate the removal of pathogenic
microorganisms and injured, necrotic and apoptotic cells.
Therefore, CRP is important in natural immunity (12,13).
Originally, CRP was considered to be a nonspecific biomarker of
inflammation; however, following ~10 years of investigation, it has
been demonstrated that CRP participates in inflammation and
atherosclerosis directly and has been indicated to be a prognostic
factor and a risk factor (14). In
this study, it was revealed that levels of the precursor of CRP
were decreased in the patients with CHB and markedly increased in
the patients with ACLF. Nearly identical changes were observed for
the hemoglobin β chain variant Hb S-Wake, which has been implicated
in in orphan diseases (15),
although not in liver diseases.
The apolipoprotein J precursor is important in cell
aging and tumorigenesis with apolipoprotein J, lipid transfer
inhibitor protein, complement C3d, corticosteroid-binding globulin
and apolipoprotein L1 (16,17).
These latter five markers of fibrosis are secreted in the blood and
show consistent changes with the increasing stage of fibrosis when
compared with the markers used in the FibroTest and enhanced liver
fibrosis (ELF), Hepascore and FIBROSpect tests. In the present
study, the levels of apolipoprotein J precursor decreased in the
progression of HBV from CHB to ACLF. Therefore, further studies,
with a larger number of samples, are required in order to better
establish the role of apolipoprotein J precursor as a potential
biomarker for HBV progression.
Platelet factor 4 is chemotactic for numerous cell
types. It also functions as an inhibitor of hematopoiesis,
angiogenesis and T-cell function, which may be used as a biomarker
for establishing a prognosis in severe acute respiratory syndrome
(18). Changes in platelet
morphology, with accompanying increases in megathrombocyte
fraction, may occur in chronic liver diseases, and thrombocytes are
activated in chronic liver diseases and liver cirrhosis. Platelet
sensitivity to stimuli in patients with liver cirrhosis has been
demonstrated to be higher than in healthy controls (19) and it was indicated that platelet
factor 4 was activated and upregulated in chronic hepatitis and
liver cirrhosis. In the present study, levels of platelet factor 4
precursor decreased in the progression of hepatitis B to liver
failure. This result suggests that further studies are required to
investigate the potential of platelet factor 4 precursor as a
biomarker in the progression of CHB.
VN is another protein that commonly features in
studies on liver disease. It is a multifunctional plasma
glycoprotein produced by hepatocytes that is detectable in plasma
and the extracellular matrix. Glycosylation of VN modulates
multi-merization and collagen binding in liver regeneration
(20) and alterations in VN
glycosylation modulate substrate adhesion to rat hepatic stellate
cells (HSCs), which is responsible for matrix restructuring
(21). Immunoblotting data
identified increases in VN levels in cirrhotic livers, and VN
immunoreactivity has been shown to be markedly increased in the
cirrhotic liver matrix, irrespective of the apparent reduction in
plasma VN. VN immunoreactivity in the liver may be considered to be
a marker of fibrosis, particularly for chronic/mature fibrosis,
paralleling previous observations concerning the enhanced orcein
staining of cirrhotic septa (22).
These results suggest that VN may be important in the progression
of liver disease and/or in hepatic fibrosis through its
collagen-binding domain I, and that VN levels are likely to be
higher in patients with chronic hepatitis, liver cirrhosis and
liver cancer than in controls (20–23).
In the present study, VN levels decreased in the progression of
hepatitis B (from CHB to HBV-induced ACLF). The observation of
decreased VN levels was consistent with two other studies (24,25).
In one of these studies, it was demonstrated that, in chronic liver
disease, the concentration of plasma VN was significantly lower
than in healthy controls and was associated with the severity of
liver disease. Notably, levels of VN in the liver tissue were
significantly increased in patients with chronic liver disease
compared with those in normal controls. These results suggested
that VN was deposited in injured tissue during the repair process
and functioned as an adhesive protein (24). In the other study, it was
demonstrated that there were lower levels of plasma VN in chronic
liver disease, which was possibly due to a decreased synthesis,
deposition in injured tissues or a combination of the two (25). The same group of authors also
revealed that decreased levels of plasma VN and fibronectin (FN)
and increased levels of serum laminin (LM) P1 in patients with
chronic liver diseases were associated with hepatic dysfunction,
and that changes in the levels of the glycoproteins involved in
cell attachment were significant in the development of hepatic
fibrosis in patients with chronic liver diseases (25). Based on the accumulating
information on VN and liver fibrosis, it appears that VN requires
further study.
In the present study, the authors aimed to describe
changes in serum protein levels in patients with liver dysfunction
induced by hepatitis B and patients with severe liver damage
(ACLF). The purpose of this was to specifically identify one or
more proteins that are able serve as biomarkers for predicting a
pathogenic condition and for prognostic purposes, similar to
α-fetoprotein (AFP) for hepatocellular carcinoma (HCC). It was not
possible in this study to obtain a series of serum samples from
individual patients as they progressed through the various stages
of hepatitis B infection to severe liver damage (ACLF). However,
using iTRAQ and western blotting, several candidate proteins were
identified (namely apolipoprotein J precursor, platelet factor 4
precursor and VN) that warrant further study using a larger number
of samples.
Acknowledgements
The authors would like to thank Miss.
Qin Zhang and Mr. Bing-Quan Lai for their commitment to this study.
This study was supported by the National Natural Science Foundation
of China (grant no. 81101256), National Science and Technology
Major Projects (grant nos. 2012ZX10002004 and 2012ZX10002007), the
Fundamental Research Funds for the Central Universities (grant no.
12ykpy35) and the New Teacher Fund of the Ministry of Education
(grant no. 20090171120079).
References
|
1.
|
Chinese Society of Hepatology and Chinese
Society of Infectious Diseases, Chinese Medical Association: The
guideline of prevention and treatment for chronic hepatitis B (2010
version). Zhonghua Gan Zang Bing Za Zhi. 19:13–24. 201l.(In
Chinese).
|
|
2.
|
Lu FM and Zhuang H: Management of
hepatitis B in China. Chin Med J (Engl). 122:3–4. 2009.
|
|
3.
|
Ren F, Chen Y, Wang Y, Yan Y, Zhao J, Ding
M, Zhang J, Jiang Y, Zhai Y and Duan Z: Comparative serum proteomic
analysis of patients with acute-on-chronic liver failure:
alpha-1-acid glycoprotein maybe a candidate marker for prognosis of
hepatitis B virus infection. J Viral Hepat. 17:816–824. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Yang L, Rudser KD, Higgins L, Rosen HR,
Zaman A, Corless CL, David L and Gourley GR: Novel biomarker
candidates to predict hepatic fibrosis in hepatitis C identified by
serum proteomics. Dig Dis Sci. 56:3305–3315. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Jin GZ, Li Y, Cong WM, Yu H, Dong H, Shu
H, Liu XH, Yan GQ, Zhang L, Zhang Y, et al: iTRAQ-2DLC-ESI-MS/MS
based identification of a new set of immunohistochemical biomarkers
for classification of dysplastic nodules and small hepatocellular
carcinoma. J Proteome Res. 10:3418–3428. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Lee HJ, Na K, Choi EY, Kim KS, Kim H and
Paik YK: Simple method for quantitative analysis of N-linked
glycoproteins in hepatocellular carcinoma specimens. J Proteome
Res. 9:308–318. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Goh WW, Lee YH, Zubaidah RM, Jin J, Dong
D, Lin Q, Chung MC and Wong L: Network-based pipeline for analyzing
MS data: an application toward liver cancer. J Proteome Res.
10:2261–2272. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Wu WW, Wang G, Baek SJ and Shen RF:
Comparative study of three proteomic quantitative methods, DIGE,
cICAT, and iTRAQ, using 2D gel- or LC-MALDI TOF/TOF. J Proteome
Res. 5:651–658. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Kolla V, Jenö P, Moes S, Tercanli S,
Lapaire O, Choolani M and Hahn S: Quantitative proteomics analysis
of maternal plasma in Down syndrome pregnancies using isobaric
tagging reagent (iTRAQ). J Biomed Biotechnol. 2010:9520472010.
View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Lok AS and McMahon BJ: Chronic hepatitis
B: update 2009. Hepatology. 50:661–662. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Sarin SK, Kumar A, Almeida JA, Chawla YK,
Fan ST, Garg H, et al: Acute-on-chronic liver failure: consensus
recommendations of the Asian Pacific Association for the Study of
the Liver (APASL). Hepatol Int. 3:269–282. 2009. View Article : Google Scholar
|
|
12.
|
Nahum E, Livni G, Schiller O, Bitan S,
Ashkenazi S and Dagan O: Role of C-reactive protein velocity in the
diagnosis of early bacterial infections in children after cardiac
surgery. J Intensive Care Med. 27:191–196. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Sklavou R, Karavanaki K, Critselis E,
Kossiva L, Giannaki M, Tsolia M, Papadakis V, Papargyri S, Vlachou
A, Karantonis F, Gourgiotis D and Polychronopoulou S: Variation of
serum C-reactive protein (CRP) over time in pediatric cancer
patients with febrile illness and its relevance to identified
pathogen. Clin Biochem. 45:1178–1182. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Kashiwagi M, Tanaka A, Kitabata H,
Tsujioka H, Matsumoto H, Arita Y, Ookochi K, Kuroi A, Kataiwa H,
Tanimoto T, et al: Relationship between coronary arterial
remodeling, fibrous cap thickness and high-sensitivity C-reactive
protein levels in patients with acute coronary syndrome. Circ J.
73:1291–1295. 2009. View Article : Google Scholar
|
|
15.
|
Kutlar F, Redding-Lallinger R, Meiler SE,
Bakanay SM, Borders L and Kutlar A: A new sickling variant ‘Hb
S-Wake β [(Glu6Val-Asn139 Ser)]’ found in a compound heterozygote
with Hb S β (Glu6Val) coinherited with homozygous α-thalassemia-2:
phenotype and molecular characteristics. Acta Haematol.
124:120–124. 2010.
|
|
16.
|
Trougakos IP and Gonos ES:
Clusterin/apolipoprotein J in human aging and cancer. Int J Biochem
Cell Biol. 34:1430–1448. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Gangadharan B, Bapat M, Rossa J, Antrobus
R, Chittenden D, Kampa B, Barnes E, Klenerman P, Dwek RA and
Zitzmann N: Discovery of novel biomarker candidates for liver
fibrosis in hepatitis C patients: a preliminary study. PLoS One.
7:e396032012. View Article : Google Scholar
|
|
18.
|
Poon TC, Pang RT, Chan KC, Lee NL, Chiu
RW, Tong YK, Chim SS, Ngai SM, Sung JJ and Lo YM: Proteomic
analysis reveals platelet factor 4 and beta-thromboglobulin as
prognostic markers in severe acute respiratory syndrome.
Electrophoresis. 33:1894–1900. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Panasiuk A, Prokopowicz D, Zak J,
Matowicka-Karna J, Osada J and Wysocka J: Activation of blood
platelets in chronic hepatitis and liver cirrhosis P-selectin
expression on blood platelets and secretory activity of
beta-thromboglobulin and platelet factor-4. Hepatogastroenterology.
48:818–822. 2001.PubMed/NCBI
|
|
20.
|
Sano K, Asanuma-Date K, Arisaka F, Hattori
S and Ogawa H: Changes in glycosylation of vitronectin modulate
multimerization and collagen binding during liver regeneration.
Glycobiology. 17:784–794. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Sano K, Miyamoto Y, Kawasaki N, Hashii N,
Itoh S, Murase M, Date K, Yokoyama M, Sato C, Kitajima K and Ogawa
H: Survival signals of hepatic stellate cells in liver regeneration
are regulated by glycosylation changes in rat vitronectin,
especially decreased sialylation. J Biol Chem. 285:17301–17309.
2010. View Article : Google Scholar
|
|
22.
|
Koukoulis GK, Shen J, Virtanen I and Gould
VE: Vitronectin in the cirrhotic liver: an immunomarker of mature
fibrosis. Hum Pathol. 32:1356–1362. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Yamada S, Kobayashi J, Murawaki Y, Suou T
and Kawasaki H: Collagen-binding activity of plasma vitronectin in
chronic liver disease. Clin Chim Acta. 252:95–103. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Kobayashi J, Yamada S and Kawasaki H:
Distribution of vitronectin in plasma and liver tissue:
relationship to chronic liver disease. Hepatology. 20:1412–1417.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Tomihira M: Changes in plasma vitronectin,
fibronectin, and serum laminin P1 levels and immunohistochemical
study of vitronectin in the liver of patients with chronic liver
diseases. Fukuoka Igaku Zasshi. 82:21–30. 1991.(In Japanese).
|