Introduction
Disc degenerative disease (DDD) is a common and
frequently occurring orthopedic disease. The main clinical
manifestations are disc herniation, vertebral instability and
spinal stenosis. The intervertebral disc (IVD) is an immune-free
organ that is completely closed, without vessels and nerves. Its
immune privilege provides a potential environment for treatment
with gene therapy. However, the etiology and pathophysiological
mechanisms of DDD require further investigation (1).
Gene therapy is a novel therapeutic strategy that is
capable of consistently changing and affecting the physiological
function of cells. It has received considerable attention in the
field of biological therapy, and promising developments have been
made in this area (2). With regard
to gene therapy for DDDs, depending on use of the appropriate dose
and on the treatment area in the body, gene therapy is capable of
producing positive therapeutic effects. However, devastating side
effects are likely to arise if the therapeutic gene is not correct,
if leakage occurs when the therapeutic genes are injected or if the
dose is inappropriate. Therefore, an important aspect of gene
therapy is determining the correct target gene (3).
Genes work in synergy with other genes to perform
biological functions. They simultaneously interact with multiple
genes and trigger a variety of changes that lead to diverse
reactions. The functional annotation of regulated genes, using Gene
Ontology, has enabled the identification of severely affected
groups of genes that correlate with the disease phenotypes
(4). Genes involved in mutation of
the IVD are enriched in the Gene Ontology terms ‘multicellular
organism development’ and ‘pattern specification’ (5).
A phenotype is the result of a series of complex
molecular reactions. A protein interaction network embodies the
complex interactions between genes (6–7). An
abundant and organized network of elastic fibers has been
previously observed in the non-degenerated human disc using
immunohistochemical staining (8).
In addition, Yu et al(9)
revealed that an abundant and organized network of elastic fibers
was present in adolescent (12- and 17-year-olds) human IVDs, and
suggested that the elastic fiber network had a significant
biomechanical function (9). Trp2
and Trp3 allelic products have been incorporated into the
cross-linked fibrillar network to study the process of developing
human cartilage (10).
Once the disease-related genes have been identified,
the active binding site may provide another perspective for the
treatment of the DDD. It has been demonstrated that the deletion of
transforming growth factor β receptor 2 (Tgfβr2) in
Col2a-expressing mouse tissue results in alterations in the
development of the IVD annulus fibrosus (5). Based on the genes associated with the
disease phenotype, drugs to repress these genes are screened. The
Connectivity Map (CMAP) database is a collection of genome-wide
transcriptional expression data that have been obtained from
cultured human cells treated with bioactive small molecules and
simple pattern-matching algorithms. These data enable the discovery
of functional connections between drugs, genes and diseases through
the transitory feature of common changes in gene expression
(11). Yeh et al (12)
screened drugs that targeted cancer stem cells (CSCs) to improve
current treatment and overcome drug resistance. Gene signatures
between embryonic and CSCs were used to identify potential drugs
that were capable of reversing the gene expression profile of CSCs,
based on CMAP.
The present study identified differentially
expressed genes (DEGs) by comparing the gene expression profiles of
non-degenerative and degenerative discs. The Gene Ontology terms
enriched among the DEGs were screened. Following the construction
of the interaction network of the DEGs, hub genes were considered
as targets for gene therapy, and small molecules that were
associated with DDD were studied to facilitate the treatment of
DDD.
Materials and methods
Gene expression profiles of DDD
The gene expression profiles of DDD were assessed
using the Affymetrix Human Genome U133A Array (HG-U133A) at
platform GPL96 (Affymetrix, Santa Clara, CA, USA), which covered
22,283 probes in the human genome. The profiles were submitted by
Zhou et al to the Gene Expression Omnibus (13). The samples included three
degenerative and three non-degenerative IVDs from elderly patients
and younger patients, respectively. The annotation information for
the gene expression profile was downloaded, as well as the raw
data.
Data preprocessing and differential gene
analysis
The original expression profile in CEL format was
transformed into a matrix using affy package in R language
(14–15). The median method was used for
normalizing the expression matrix. Subsequently, the Limma package
(16) was utilized to identify the
differential genes between the degenerative and non-degenerative
IVDs. The threshold for the P-value was set to 0.05 and |logFC| was
set to 1.
Enrichment analysis of differential
genes
The WEB-based GEne SeT AnaLysis Toolkit (WebGestalt)
was designed for the enrichment analysis of differential genes, and
comprises a set of analytical tools for biological analysis.
WebGestalt contains 59,278 functional categories derived from
centrally and publicly curated databases, as well as computational
analyses, including National Center for Biotechnology Information,
Ensemble, Kyoto Encyclopedia of Genes and Genomes and Gene Ontology
(17–18), for eight species, such as humans,
rats and mice. In this study, the functional enrichment analysis
was based on the hypergeometric test [false discovery rate (FDR),
<0.05] for the DEGs.
Constructing an interaction network of
differential genes
Since one gene always acts in synergy with other
partners, the interactive protein should also be studied when
exploring the function of one gene and its protein (19). The Osprey platform (20) facilitates the study of
protein-protein interaction networks and protein complexes by
integrating the interaction information from the Biomolecular
Interaction Network Database (21)
and the General Repository for Interaction Datasets (22), which includes >50,000 of the
interactions.
Therefore, Osprey software was used to analyze the
interactions between differential genes and build the interaction
network. Following topological analysis of the interaction network
of differential genes, the genes with high degrees were screened as
a hub. Hubs are to key maintenance of the integrity and robustness
of the network and if specific hub genes are knocked out, the
network is likely to be divided into fragments or even undergo
paralysis (23).
Functional enrichment analysis for
differential genes in the interaction network
In gene enrichment analysis, a set of genes with
similar or related functions are considered as a whole. By
calculating the global and significant gene expression changes for
the genes in the set, it is possible to determine whether the
biological function has altered. This strategy greatly reduces the
dimensions of the data; however, it makes the analysis much closer
to the actual biological problems. Thus, gene enrichment analysis
is popular in gene expression profiles (24). To date, there are numerous tools
providing gene function enrichment analysis. In the present study,
the Database for Annotation, Visualization and Integrated Discovery
(DAVID) was used (25) for the
functional enrichment analysis of the differential gene hubs in the
interaction network. The FDR threshold was set to 0.05.
Screening of drug-like small
molecules
The DEGs in the interaction network were divided
into two groups of upregulated and downregulated genes. By
comparing the expression pattern similarities of the differential
genes and genes perturbed by small molecules in the CMAP, small
molecules involved in the disease were able to be identified
(11,26). Small molecules with a score >0.8
were considered to be associated with the disease.
Active site screening for hub genes
Proteins are the basis of life and are composed of
~20 amino acids. The structure of proteins is maintained by
numerous interactions, including hydrogen bonds and ionic
interaction forces. Due to the influence of these forces, protein
molecules fold, forming a complex 3-D structure (27). In the molecular structure of the
protein, the selective interaction region, which binds with other
molecules, is referred to as the active site. The selective
interaction between proteins is determined by the functional groups
of the amino acids. The accurate prediction of a selective
interactive region provides an enhanced understanding of specific
protein interactions, which is of great significance for biologists
(28). The Universal Protein
(UniProt) (29) database stores
comprehensive information regarding proteins, and is the most
popular database for protein research. UniProt is integrated from
Swiss-Prot, TrEMBL and the Protein Information Resource-Protein
Sequence Database. In the current study, the active sites were
screened for hub differential genes based on the UniProt
database.
Results
Differential gene screening
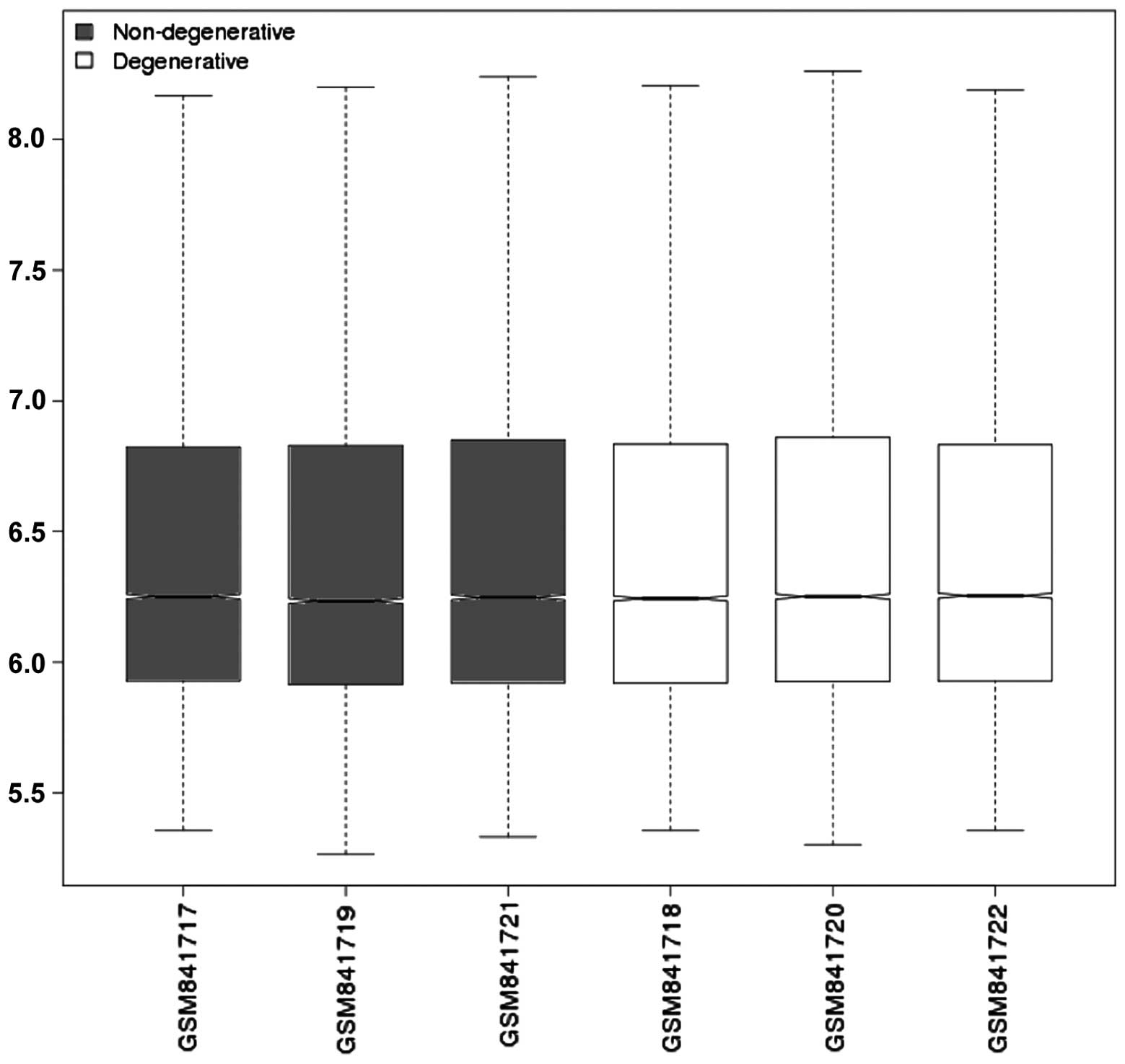
The gene expression profiles were normalized, as
shown in Fig. 1. The median of
each sample was the same, which showed that the data were well
normalized (Fig. 1). When
comparing the degenerative and non-degenerative IVDs, 53 DEGs were
identified under the threshold of FDR <0.05 and |logFC| >1. A
total of 16 genes were downregulated and 37 genes were upregulated.
Matrix metalloproteinase 2 (MMP2) was upregulated in degenerative
IVDs with its corresponding probe of 218468_s_at and the adjusted
P-value of 0.04.
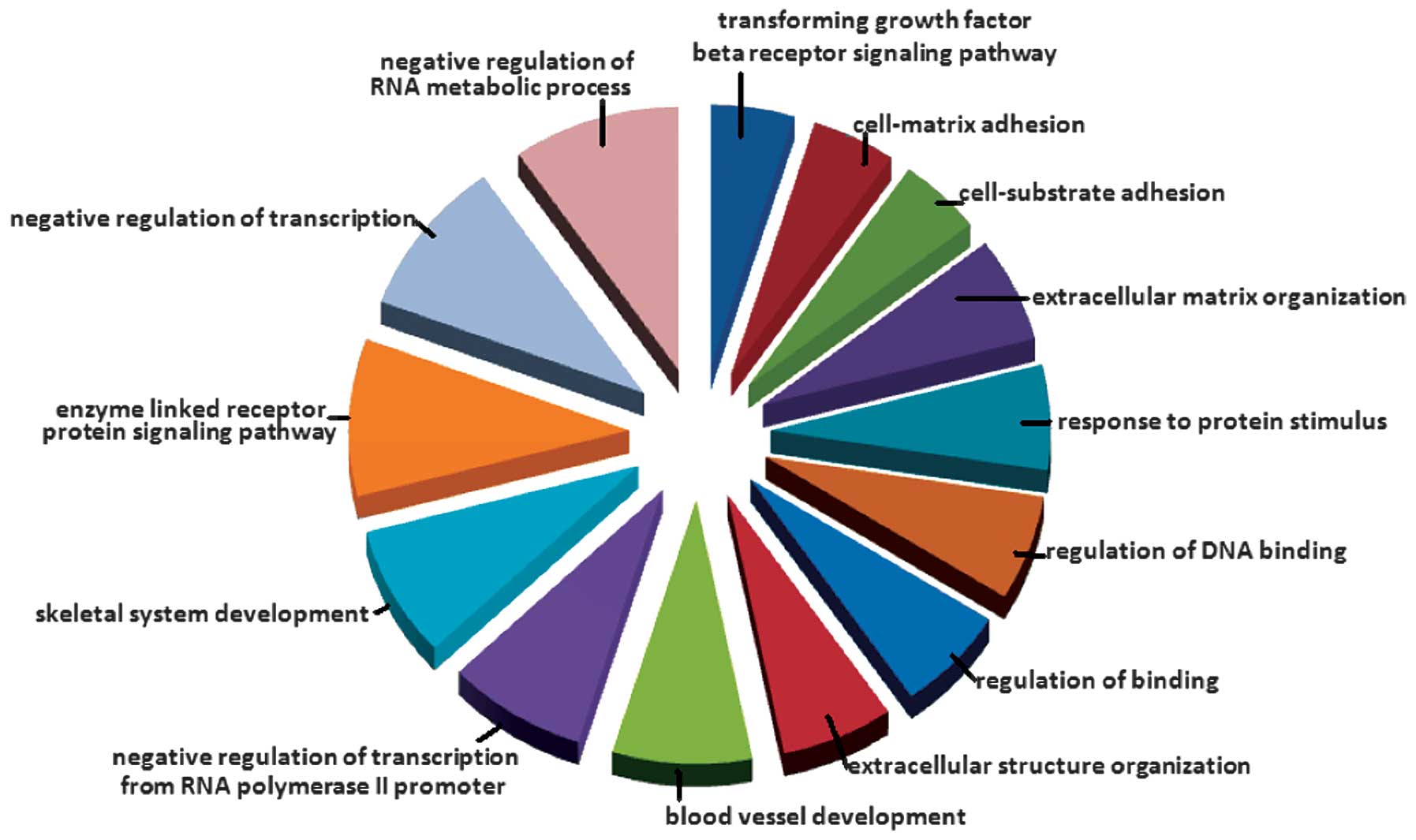
Enriched terms among the DEGs
The WebGestalt web tool was used for the enrichment
analysis. As shown in Fig. 2, 14
Gene Ontology terms, including extracellular matrix, were enriched
among the DEGs (Fig. 2).
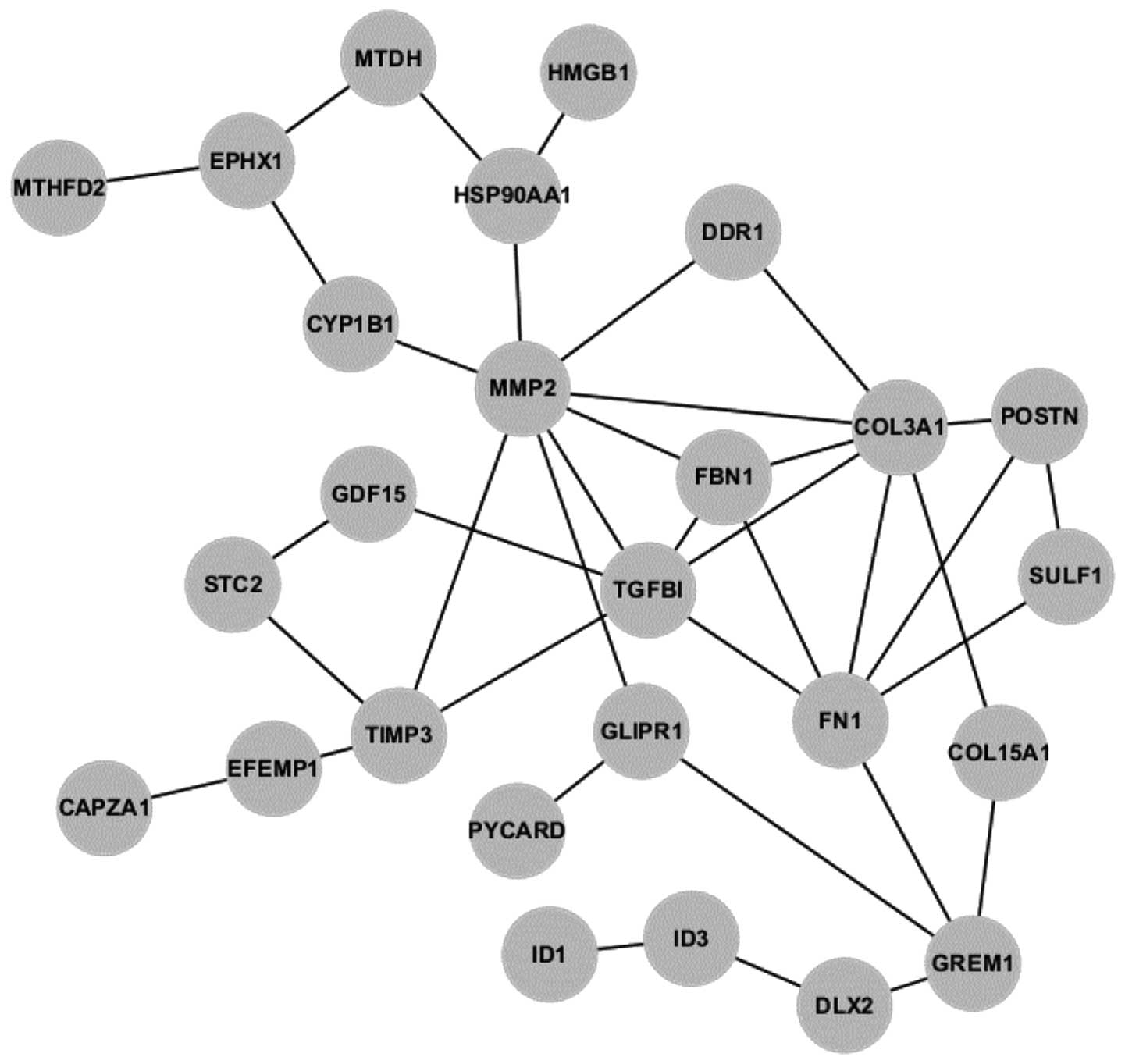
DEG interaction network
Osprey software was utilized to explore the
interactions among the DEGs. Fig.
3 shows the interaction network, which consists of 19 DEGs.
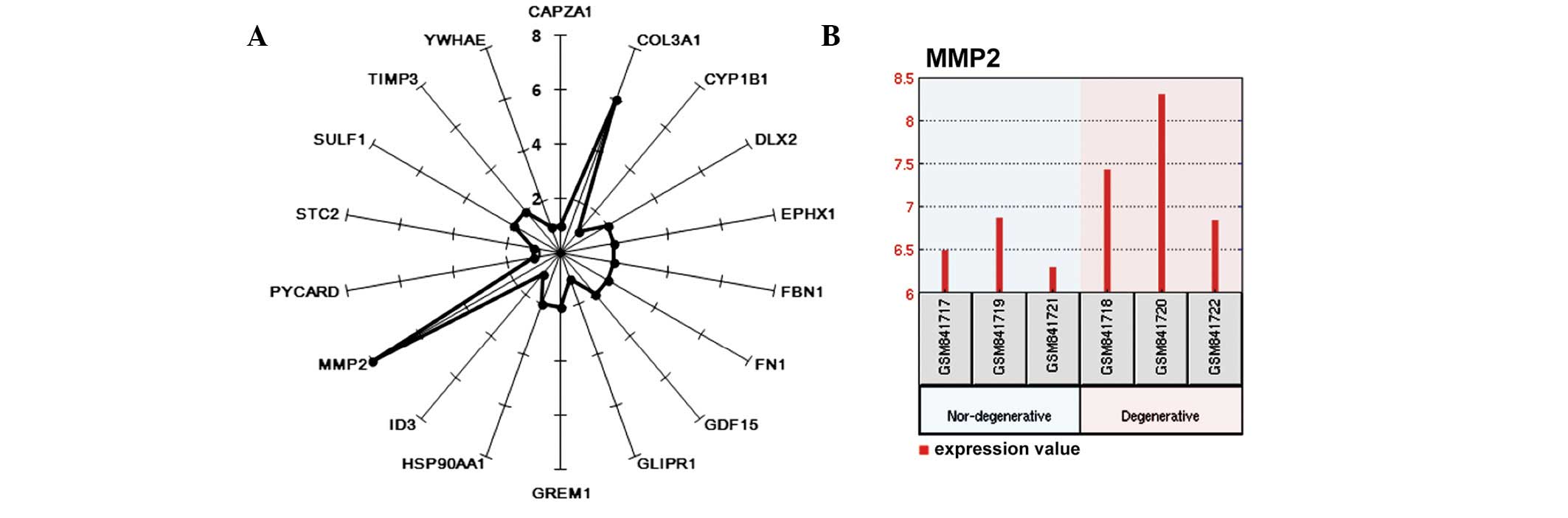
Following construction of the network, the degree of each gene in
the network was calculated. As shown in Fig. 4A, the gene MMP2 was connected to
eight other genes, and was, therefore, the hub of the network
(Fig. 4A). MMP2 was upregulated in
degenerative IVDs (Fig. 4B).
Gene Ontology terms enriched among the
genes in the interaction network
In this study, DAVID was used to identify the Gene
Ontology terms enriched among the genes in the interaction network.
As shown in Table I, nine terms
were significantly enriched. The most significant term was organ
development, which was the only term associated with MMP2.
 | Table IDEGs in the interaction network. |
Table I
DEGs in the interaction network.
| GO-ID | P-value | x | Description |
|---|
| 48513 | 0.004904 | 11 | Organ
development |
| 48731 | 0.010540 | 12 | System
development |
| 9653 | 0.015218 | 8 | Anatomical structure
morphogenesis |
| 48856 | 0.015218 | 12 | Anatomical structure
development |
| 7275 | 0.029394 | 12 | Multicellular
organismal development |
| 32501 | 0.029394 | 15 | Multicellular
organismal process |
| 48523 | 0.033435 | 9 | Negative regulation
of cellular process |
| 9892 | 0.035170 | 6 | Negative regulation
of metabolic process |
| 32502 | 0.040190 | 12 | Developmental
process |
Small molecules involved in degenerative
IVDs
The DEGs were divided into two groups of upregulated
and downregulated genes. Using the two gene groups as inputs for
the CMAP database, the small molecules that were associated with
the degenerative IVD were determined. The small molecules with a
score of >0.8 were focused on for further analysis. As shown in
Table II, the small molecule,
MG-262, had the highest score (score=0.896).
 | Table IISignificant small molecules revealed
by CMAP. |
Table II
Significant small molecules revealed
by CMAP.
| CMAP name | Score | P-value |
|---|
| Ioxaglic acid | −0.828 | 0.01022 |
| Trazodone | −0.826 | 0.01052 |
| Cortisone | −0.805 | 0.01462 |
| Diperodon | 0.817 | 0.01208 |
| Procainamide | 0.85 | 0.00074 |
| MG-262 | 0.896 | 0.00218 |
Hub gene active sites
For the hub protein in the interaction network, the
UniProt database was searched for active binding sites of the MMP2
ligand. As shown in Table III,
22 metal ligand binding sites were revealed, with binding sites for
metal ions, such as calcium and zinc.
 | Table IIISearch results for active binding
sites. |
Table III
Search results for active binding
sites.
| Type | Binding site | Metal type |
|---|
| Metal binding | 102 | Zinc 2; in
inhibited form |
| Metal binding | 134 | Calcium |
| Metal binding | 168 | Calcium 2 |
| Metal binding | 178 | Zinc |
| Metal binding | 180 | Zinc |
| Metal binding | 185 | Calcium 3 |
| Metal binding | 186 | Calcium 3; via
carbonyl oxygen |
| Metal binding | 193 | Zinc |
| Metal binding | 200 | Calcium 2; via
carbonyl oxygen |
| Metal binding | 202 | Calcium 2; via
carbonyl oxygen |
| Metal binding | 204 | Calcium 2 |
| Metal binding | 206 | Zinc |
| Metal binding | 208 | Calcium 3 |
| Metal binding | 209 | Calcium |
| Metal binding | 211 | Calcium 3 |
| Metal binding | 403 | Zinc 2;
catalytic |
| Metal binding | 407 | Zinc 2;
catalytic |
| Metal binding | 413 | Zinc 2;
catalytic |
| Metal binding | 476 | Calcium 4; via
carbonyl oxygen |
| Metal binding | 521 | Calcium 4; via
carbonyl oxygen |
| Metal binding | 569 | Calcium 4; via
carbonyl oxygen |
| Metal binding | 618 | Calcium 4; via
carbonyl oxygen |
Discussion
In the present study, the gene expression microarray
of non-degenerative and degenerative disc tissue samples was
compared. The differential genes were screened out and their
functions were further analyzed. The proposed method provides basis
for the identification of candidate targets for the clinical
treatment of DDD. It also facilitates clinical medication.
The results revealed that the DEGs were associated
with TGF-β and the extracellular matrix, which was the fourth most
enriched term. Degeneration of the IVD is predominantly a chronic
process, involving the excessive destruction of the extracellular
matrix (30). IVD degeneration has
been demonstrated to be a progressive disease, mainly characterized
by the alteration of the extracellular matrix composition (31). Factors involved in these processes
have been suggested to be important for the identification of
target genes in degenerative discs (32). In the interaction network of the
DEGs, MMP2 was shown to have the highest degree. The MMPs are a
class of proteolytic enzymes containing zinc and calcium, which are
predominantly involved in the metabolism of the extracellular
matrix. Roberts et al(33)
suggested that MMPs were one of the primary factors leading to the
disorder of the matrix structure and the degradation of the matrix
in IVDs. The expression of MMPs has been shown to be significantly
increased in the degenerative IVD, disrupting the balance between
the MMPs and their endogenous inhibitor. In addition, it has been
observed that MMPs are produced by IVD autologous cells and cells
involved in invasive neovascularization in the IVDs. Therefore,
Roberts et al(33)
suggested that inhibiting the activity of MMPs may be an effective
method of treating disc degeneration (33). Based on the downregulated and
upregulated genes, six small molecules, which were most relevant to
the degenerative IVD, were screened out. Hexabrix (ioxaglate), the
most significant negatively-associated small molecule, has been
introduced as a novel low-osmolality contrast agent for lumbar
epidural double-catheter venography (34). Cortisone, which has been predicted
to cure DDD, has been shown to result in satisfactory remission of
articular facets syndrome, leaving the patient free from pain.
Epidural cortisone injections are able to efficiently and safely
release anterior and anteroposterior fusion for lumbar disc pain
(34).
Further studies on activity sites may aid the
inhibition and promotion of the normal expression of DEGs. Zinc
metalloproteinases exert particularly important effects, directly
and indirectly through the promotion of neovascularization. The
zinc metalloproteinases closely interact with other metabolic
factors to produce disc disorders (35). Calcium has been used as an
indicator of calcification potential in human IVD degeneration and
scoliosis (36).
At present, biological treatment must focus on the
biological changes occurring in IVD degeneration, promote the
synthesis of extracellular matrix and inhibit the reduction in
levels of the extracellular matrix. In this manner, the metabolism
of the extracellular matrix in the IVD is likely to return to
normal. The mechanism underlying IVD degeneration is complex and
has been suggested to correlate with changes at the molecular
level. With an enhanced understanding of the disease from a
molecular biology perspective, novel strategies are likely to
emerge to prevent and treat degenerative IVDs. Although significant
efforts have been committed to further elucidate the degenerative
mechanism at the molecular and genetic levels, studies must be
performed to evaluate the pathological roles of a number of factors
and their interactions in the process of disc degeneration.
References
|
1
|
Sambrook PN, MacGregor AJ and Spector TD:
Genetic influences on cervical and lumbar disc degeneration: a
magnetic resonance imaging study in twins. Arthritis Rheum.
42:366–372. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sobajima S, Kim JS, Gilbertson LG and Kang
JD: Gene therapy for degenerative disc disease. Gene Ther.
11:390–401. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wallach CJ, Kim JS, Sobajima S, et al:
Safety assessment of intradiscal gene transfer: a pilot study.
Spine J. 6:107–112. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Beltran S, Angulo M, Pignatelli M, Serras
F and Corominas M: Functional dissection of the ash2 and ash1
transcriptomes provides insights into the transcriptional basis of
wing phenotypes and reveals conserved protein interactions. Genome
Biol. 8:R672007. View Article : Google Scholar
|
|
5
|
Sohn P, Cox M, Chen D and Serra R:
Molecular profiling of the developing mouse axial skeleton: a role
for Tgfbr2 in the development of the intervertebral disc. BMC Dev
Biol. 10:292010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pache RA, Zanzoni A, Naval J, Mas JM and
Aloy P: Towards a molecular characterisation of pathological
pathways. FEBS Lett. 582:1259–1265. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thorn CF, Whirl-Carrillo M, Klein TE and
Altman RB: Pathway-based approaches to pharmacogenomics. Current
Pharmacogenomics. 5:79–86. 2007. View Article : Google Scholar
|
|
8
|
Cloyd JM and Elliott DM: Elastin content
correlates with human disc degeneration in the anulus fibrosus and
nucleus pulposus. Spine (Phila Pa 1976). 32:1826–1831. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yu J, Fairbank JC, Roberts S and Urban JP:
The elastic fiber network of the anulus fibrosus of the normal and
scoliotic human intervertebral disc. Spine (Phila Pa 1976).
30:1815–1820. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Matsui Y, Wu JJ, Weis MA, Pietka T and
Eyre DR: Matrix deposition of tryptophan-containing allelic
variants of type IX collagen in developing human cartilage. Matrix
Biol. 22:123–129. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lamb J, Crawford ED, Peck D, et al: The
Connectivity Map: using gene-expression signatures to connect small
molecules, genes, and disease. Science. 313:1929–1935. 2006.
View Article : Google Scholar
|
|
12
|
Yeh CT, Wu AT, Chang PM, et al:
Trifluoperazine, an antipsychotic agent, inhibits cancer stem cell
growth and overcomes drug resistance of lung cancer. Am J Respir
Crit Care Med. 186:1180–1188
|
|
13
|
Zhou Y, Jiang S, Chen J, Wang T, Jiang D,
Chen H and Yu H: 32k Da protein improve ovariectomy-induced bone
loss in rats. Prion. 7:335–340. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Troyanskaya O, Cantor M, Sherlock G, et
al: Missing value estimation methods for DNA microarrays.
Bioinformatics. 17:520–525. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fujita A, Sato JR, de Rodrigues LO,
Ferreira CE and Sogayar MC: Evaluating different methods of
microarray data normalization. BMC Bioinformatics. 7:4692006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Smyth GK, Michaud J and Scott HS: Use of
within-array replicate spots for assessing differential expression
in microarray experiments. Bioinformatics. 21:2067–2075. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang B, Kirov S and Snoddy J: WebGestalt:
an integrated system for exploring gene sets in various biological
contexts. Nucleic Acids Res. 33:W741–W748. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Duncan DT, Prodduturi N and Zhang B:
WebGestalt2: an updated and expanded version of the Web-based Gene
Set Analysis Toolkit. BMC Bioinformatics. 11:2010. View Article : Google Scholar
|
|
19
|
Patil A and Nakamura H: Filtering
high-throughput protein-protein interaction data using a
combination of genomic features. BMC Bioinformatics. 6:1002005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Breitkreutz BJ, Stark C and Tyers M:
Osprey: a network visualization system. Genome Biol. 4:R222003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Willis RC and Hogue CW: Searching,
viewing, and visualizing data in the Biomolecular Interaction
Network Database (BIND). Curr Protoc Bioinformatics. 8.9.1–8.9.30.
2006.PubMed/NCBI
|
|
22
|
Breitkreutz BJ, Stark C and Tyers M: The
GRID: The General Repository for Interaction Datasets. Genome Biol.
4:R232003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Albert R, Jeong H and Barabasi AL: Error
and attack tolerance of complex networks. Nature. 406:378–382.
2000. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009.PubMed/NCBI
|
|
25
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
26
|
Wen Z, Wang Z, Wang S, et al: Discovery of
molecular mechanisms of traditional Chinese medicinal formula
Si-Wu-Tang using gene expression microarray and connectivity map.
PLoS One. 6:e182782011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Leland WE, Taqqu MS, Willinger W and
Wilson DV: On the self-similar nature of Ethernet traffic. IEEE/ACM
Transactions on Networking (TON). 2:1–15. 1994. View Article : Google Scholar
|
|
28
|
Papagiannaki K, Taft N, Zhang ZL and Diot
C: Long-term forecasting of internet backbone traffic. IEEE Trans
Neural Netw. 16:1110–1124. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Suzek BE, Huang H, McGarvey P, Mazumder R
and Wu CH: UniRef: comprehensive and non-redundant UniProt
reference clusters. Bioinformatics. 23:1282–1288. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ding F, Shao ZW and Xiong LM: Cell death
in intervertebral disc degeneration. Apoptosis. 18:777–785. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Vadalà G, Russo F, Di Martino A and Denaro
V: Intervertebral disc regeneration: from the degenerative cascade
to molecular therapy and tissue engineering. J Tissue Eng Regen
Med. Mar 20–2013.(Epub ahead of print).
|
|
32
|
Haro H, Crawford HC, Fingleton B, et al:
Matrix metalloproteinase-3-dependent generation of a macrophage
chemoattractant in a model of herniated disc resorption. J Clin
Invest. 105:133–141. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Roberts S, Caterson B, Menage J, Evans EH,
Jaffray DC and Eisenstein SM: Matrix metalloproteinases and
aggrecanase: their role in disorders of the human intervertebral
disc. Spine (Phila Pa 1976). 25:3005–3013. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Meijenhorst GC and de Bruin JN: Hexabrix
(ioxaglate), a new low osmolality contrast agent for lumbar
epidural double-catheter venography. Neuroradiology. 20:29–32.
1980. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Grang L, Gaudin P, Trocme C, Phelip X,
Morel F and Juvin R: Intervertebral disk degeneration and
herniation: the role of metalloproteinases and cytokines. Joint
Bone Spine. 68:547–553. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hristova GI, Jarzem P, Ouellet JA, et al:
Calcification in human intervertebral disc degeneration and
scoliosis. J Orthop Res. 29:1888–1895. 2011. View Article : Google Scholar : PubMed/NCBI
|