Introduction
Thyroid cancer is the most common malignant tumor of
the endocrine organs and its incidence has been steadily increasing
over the past few decades (1–3).
Consistent with the majority of malignant neoplasms, thyroid
cancers are usually associated with specific genetic abnormalities,
as well as environmental factors (4,5).
Genome-wide association studies (GWAS) determined the
predisposition to papillary thyroid cancer (PTC), out of which two
single nucleotide polymorphisms (SNPs; rs965513 and rs944289)
located on 9q22.33 and 14q13.3, respectively, were shown to have a
significant association with PTC (6–9). A
long non-coding RNA gene (lncRNA) named papillary thyroid carcinoma
susceptibility candidate 3 (PTCSC3) is located 3.2 kb downstream of
rs944289 at 14q.13.3 (10). PTCSC3
expression is strictly thyroid-specific and is dramatically
downregulated in thyroid tumor tissues and thyroid cell lines.
LncRNAs are involved in a number of regulatory
functions, including modulation of apoptosis and invasion,
reprogramming of induced pluripotent stem cells, acting as a marker
of cell fate and parental imprinting. Previously, studies
identified that lncRNA functions as a competing endogenous RNA
(ceRNA) for shared miRNAs (11,12).
ceRNAs demonstrate a post-transcriptional regulatory role in miRNA
molecule distribution on the targets. In this study, PTCSC3, as a
target of miRNAs involved in thyroid cancer, was investigated using
in silico and biological analyses.
Materials and methods
Construction of the expression
plasmid
The cDNA of PTCSC3 was amplified from normal human
thyroid tissues (surgical specimen from benign thyroid lesion). The
sequences of the forward and reverse primers were 5′-GTACGGTAC
CCTCCTTCAGACTTCTCAGTACTC-3′ and 5′-CGACTC
GAGATTGCTACTGTGAGCATAACCTAC-3′, respectively. Subsequently, PTCSC3
was subcloned into a pcDNA3 vector (Invitrogen Life Technologies,
Carlsbad, CA, USA) to create the expression plasmid for PTCSC3 and
the products were confirmed by polymerase chain reaction (PCR) and
sequencing.
Cell lines and transfection
Thyroid tumor cell lines BCPAP, FTC133 and 8505C
were cultured in RPMI-1640 medium supplemented with 10% calf serum,
0.1 mM non-essential amino acids, 1 mM sodium-pyruvate and 1%
penicillin-streptomycin in a 37°C humidified incubator with 5%
CO2. The thyroid cancer cell lines were assessed for
PTCSC3 expression and no endogenous expression was observed. BCPAP,
FTC133 and 8505 cells, which are of papillary, follicular and
anaplastic cancer origin, respectively, were transfected with the
PTCSC3 expression construct and the empty vector (pcDNA3) as the
control, respectively, using 2 μl Lipofectamine 2000 reagent
(Invitrogen Life Technologies). At 24 h post-transfection, the
cells were harvested and total RNA was extracted using TRIzol
reagent and Ambion® DNase I (Invitrogen Life
Technologies). An Agilent 2100 BioAnalyzer (Agilent Technologies,
Santa Clara, CA, USA) was used to assess its integrity and a
high-capacity reverse transcriptase kit (Applied Biosystems, Foster
City, CA, USA) was applied to produce cDNA. To confirm successful
transfection, reverse transcription (RT)-PCR was performed to
detect PTCSC3 expression (forward primer,
5′-TCAAACTCCAGGGCTTGAAC-3′; reverse primer,
5′-ATTACGGCTGGGTCTACCT-3′). The study was approved by the ethics
committee of Xiangya Hospital, Central South University, Changsha,
China.
MTT assay and flow cytometry
To study the changes in the biological
characteristics of the thyroid cancer cell lines following PTCSC3
transfection, cell proliferation was analyzed with the MTT assay
and apoptosis and cell cycle with flow cytometry, as previously
described (13). For each group,
cells in the logarithmic phase were used in all experiments and
incubated at 37°C in 5% CO2. For the MTT assay, cells
were incubated with 0.5 mg/ml MTT for 4 h. The formazan crystals
produced by the living cells in the culture were dissolved with 100
ml dimethyl sulfoxide and the absorbance [optical density (OD)
value] was measured at 570 nm using a 96-well plate reader at
various time points (0, 24, 48, 72 and 96 h). For flow cytometry,
the prepared cells were collected and digested into single-cell
suspensions using 0.25% trypsin. Then, the cells were centrifuged
at 500 × g for 5 min and washed with phosphate-buffered saline
(PBS; 0.01 M, pH 7.4) twice. Seventy percent ethanol stored at 4°C
was used to fix the cells for 24 h before they were fully shaken
and dispensed. The plates were incubated with 0.5% Triton X-100
(Sigma, St. Louis, MO, USA) and 0.05% RNase (Sigma) in 1 ml PBS at
37°C for 30 min and then centrifuged at 1,500 × g for 5 min. The
cells were stained with 50 mg/ml propidium iodide (Sigma) at room
temperature for 30 min and the cell number was adjusted to
1×106/ml. Samples were immediately analyzed using a
FACSCalibur flow cytometer (Becton-Dickinson, Mountain View, CA,
USA).
In silico analysis of miRNAs that match
PTCSC3
Bioinformatic analysis was carried out based on the
online software, PITA (http://genie.weizmann.ac.il/pubs/mir07/mir07_prediction.html).
Briefly, the PTCSC3 sequence, NR_049735.2, was obtained from PubMed
and entered into the PITA system. As PTCSC3 is a non-protein coding
RNA, it is unlikely that the open reading frame (ORF) is occupied
by ribosomes. We uploaded the full sequence of PTCSC3, including
the 5-untranslated region (UTR), ORF and 3-UTR, into the PITA
system. Only the top 20 miRNAs presenting targeting sites in PTCSC3
were selected for further analysis.
Quantitative RT-PCR analysis of PTCSC3
and miR-574-5p expression
To investigate the expression of miR-574-5p in
thyroid cancer cells following transfection with PTCSC3, a
quantitative RT-PCR technique was used. Briefly, total RNA was
isolated from tissues using the TRIzol reagent (Invitrogen Life
Technologies) according to the manufacturer’s instructions.
Quantitative real-time PCR was performed on an ABI 7300 real-time
PCR system (Applied Biosystems) using SYBR-Green mix (Applied
Biosystems). Relative gene expression was calculated using the ΔΔCt
method, following the manufacturer’s instructions. All reactions
were carried out in triplicate. The primer sequences were:
glyceraldehye 3-phosphate dehydrogenase (GAPDH), 5′-
GGTGATGCTGGTGCTGAGTATGT-3′ and 5′-AAGAATGGGAGTTGCTGTTGAAGTC-3′;
PTCSC3, 5′-TCAAACTCCAGGGCTTGAAC-3′ and 5′-ATT ACGGCTGGGTCTACCT-3′;
miR-574-5p, 5′-GGGGTG AGTGTGTGTGTG-3′ and 5′-TGCGTGTCGTGGAGTC-3′.
For each plate, a dissociation curve was obtained to monitor any
additional double stranded DNA. GAPDH was used as an internal
control and the formula ΔΔCt, where ΔΔCt = Ct(gene) - Ct(GAPDH),
was used to calculate the relative mRNA level.
Statistical analysis
Statistical analysis was performed using SPSS 13.0
software (SPSS Inc., Chicago, IL, USA). Comparisons were performed
using Student’s t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
PTCSC3 inhibits cell growth and induces
cell cycle arrest in thyroid cancer cells
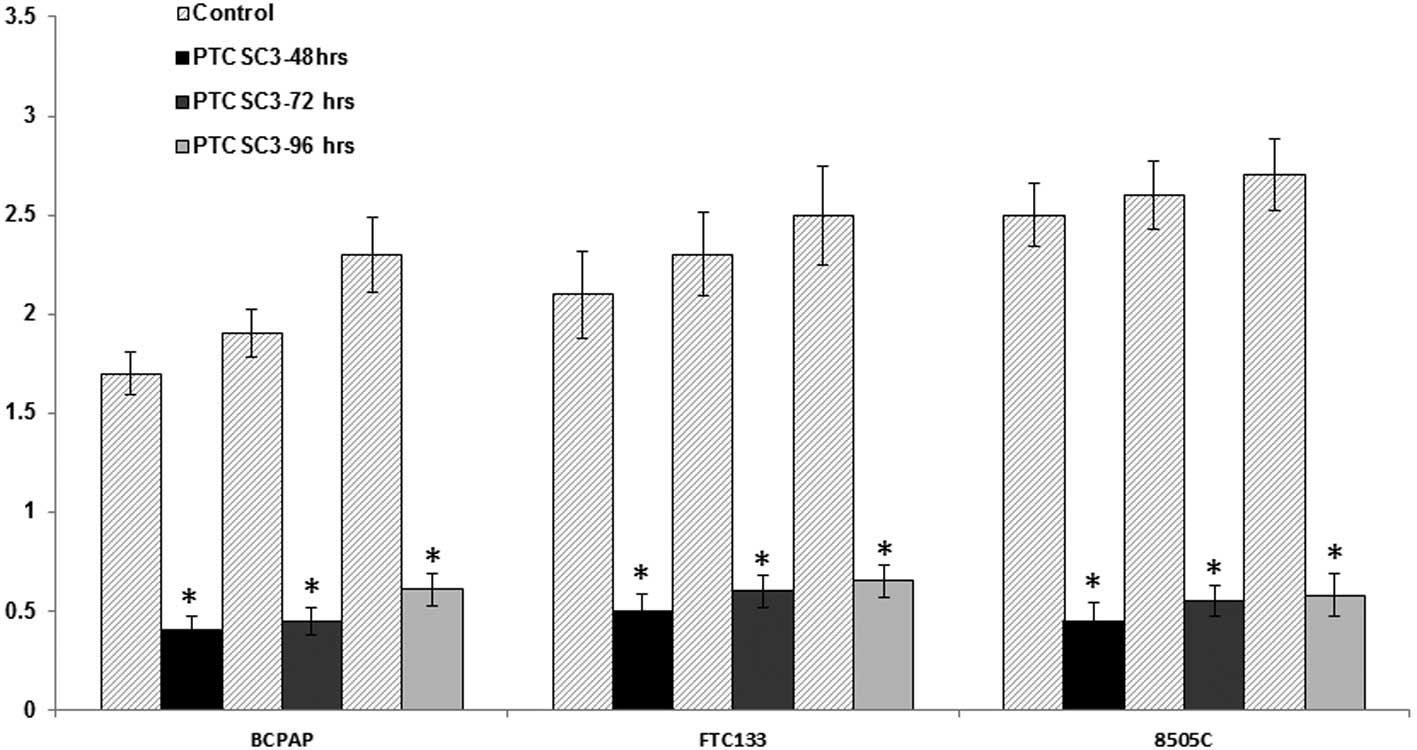
In order to assess the inhibitory effect of PTCSC3
on thyroid cancer cells, a cell growth assay was performed. The MTT
assay revealed that absorptance values of thyroid cells (BCPAP,
FTC133, 8505C) transfected with PTCSC3 at 48, 72 and 96 h were
significantly lower than those transfected with the empty plasmid
(P<0.01; Fig. 1). Additionally,
cell cytometry revealed significant cell cycle arrest at G1/S and
M/G2 phases and an increased rate of apoptosis in thyroid cancer
cells (BCPAP, FTC133, 8505C) transfected with PTCSC3 compared with
those transfected with the empty plasmid (P<0.01; Table I).
 | Table IG0/G1, S, G2/M and apoptosis rate in
thyroid cancer cell lines transfected with PTCSC3. |
Table I
G0/G1, S, G2/M and apoptosis rate in
thyroid cancer cell lines transfected with PTCSC3.
| Group | G0/G1 | S | G2/M | Apoptosis |
|---|
| BCPAP | | | | |
| Control | 26.2±2.1 | 46.6±5.1 | 27.2±3.6 | 3.6±1.0 |
| PTCSC3 | 46.8±5.2a | 35.9±4.1a | 17.3±2.2a | 6.9±1.9a |
| FTC133 | | | | |
| Control | 27.7±3.4 | 45.6±4.4 | 26.7±2.5 | 2.9±1.2 |
| PTCSC3 | 46.4±6.7a | 34.2±3.6a | 19.4±1.8a | 5.8±1.5a |
| 8505C | | | | |
| Control | 25.7±3.2 | 54.2±5.3 | 14.2±2.1 | 2.1±1.1 |
| PTCSC3 | 45.6±5.7a | 40.2±4.3a | 20.1±2.6a | 6.3±2.3a |
Bioinformatic analysis of PTCSC3
targets
As the majority of lncRNAs share the same biogenesis
mechanisms and have a similar structure with mRNAs, including the
5-UTR, ORF and 3-UTR, it is logical to assume that PTCSC3, a
lncRNA, may be targeted by certain miRNAs that are upregulated in
thyroid cancers. We carried out bioinformatic analyses aiming to
screen miRNAs that match the 3-UTR of PTCSC3 using the online
software, PITA (14). Since the
ORF of mRNA is occupied by ribosomes, the majority of miRNAs target
the 3-UTR of mRNAs. However, certain miRNAs target ORF of mRNAs,
including the targeting of c-Myc by miR-85-3p (15). Therefore, we uploaded the full
sequence of PTCSC3, including 5-UTR, ORF and 3-UTR, into the PITA
system, to identify miRNAs that may target PTCSC3. As shown in
Table II, twenty miRNAs have
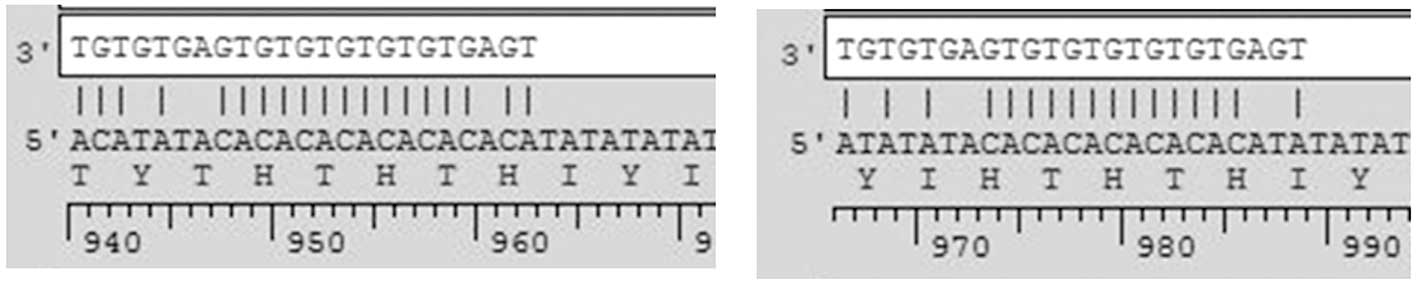
putative targeting sites in PTCSC3. Among these miRNAs, miR-574-5p
is the top candidate, since it has the highest score (−30.64)
according to the PITA results(14). Additionally, the matching between
miR-574-5p and PTCSC3 (Fig. 2)
appears perfect according to the results of miRNA-mRNA
matching.
 | Table IITop 20 microRNAs as candidates for
PTCSC3 targets. |
Table II
Top 20 microRNAs as candidates for
PTCSC3 targets.
| Gene | microRNA | Sites | Score |
|---|
| PTCSC3 | hsa-miR-574-5p | 17 | −30.64 |
| PTCSC3 | hsa-miR-1207-5p | 2 | −21.84 |
| PTCSC3 | hsa-miR-939 | 4 | −21.15 |
| PTCSC3 | hsa-miR-637 | 4 | −18.05 |
| PTCSC3 | hsa-miR-1260 | 3 | −17.18 |
| PTCSC3 | hsa-miR-297 | 22 | −16.66 |
| PTCSC3 | hsa-miR-1229 | 4 | −16.04 |
| PTCSC3 | hsa-miR-920 | 4 | −15.82 |
| PTCSC3 | hsa-miR-212 | 4 | −15.49 |
| PTCSC3 | hsa-miR-574-3p | 2 | −15.48 |
| PTCSC3 | hsa-miR-1182 | 4 | −15.4 |
| PTCSC3 | hsa-miR-1280 | 1 | −15.04 |
| PTCSC3 | hsa-miR-885-5p | 4 | −14.98 |
| PTCSC3 | hsa-miR-326 | 3 | −14.96 |
| PTCSC3 | hsa-miR-453 | 2 | −14.2 |
| PTCSC3 | hsa-miR-511 | 10 | −14.13 |
| PTCSC3 | hsa-miR-34c-3p | 1 | −13.96 |
| PTCSC3 | hsa-miR-612 | 3 | −13.9 |
| PTCSC3 | hsa-miR-188-3p | 2 | −13.49 |
| PTCSC3 | hsa-miR-210 | 17 | −13.45 |
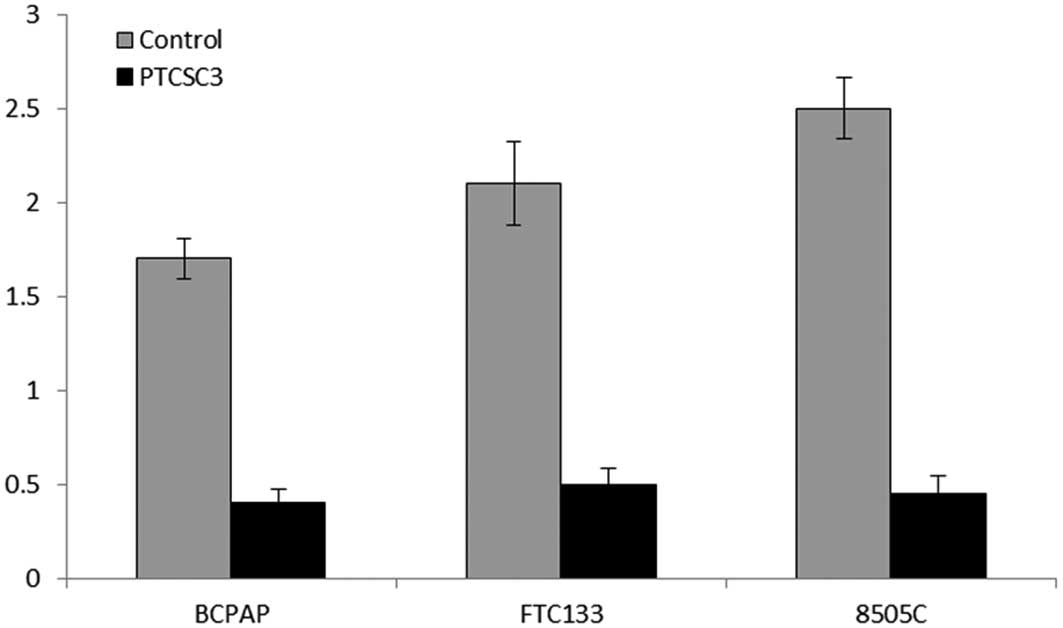
Downregulation of miR-574-5p expression
in thyroid cancer cells transfected with lnc-PTCSC3
Based on the bioinformatics study, we next confirmed
this targeting in thyroid cancer cells. To investigate the possible
binding of miR-574-5p with PTCSC3, miR-574-5p expression in thyroid
cancer cells was determined by quantitative RT-PCR. As shown in
Fig. 3, a dramatic decrease of
miR-574-5p expression in all three thyroid cancer cell lines
following transfection with PTCSC3 was detected (P<0.01). These
data demonstrate that overexpression of PTCSC3 significantly
downregulates miR-574-5p expression in thyroid cancer cells that
are of papillary, follicular and anaplastic origin.
Discussion
The newly termed PTCSC3 gene was reported to be
involved in the predisposition to PTC. However, as an lncRNA, the
presence and significance of PTCSC3 in thyroid cancer is
undetermined. The function of individual lncRNAs may be related to
epigenetic changes, action as antisense transcripts or decoys for
splicing factors, as well as a competing endogeous RNA for miRNA
binding (16,17). Aberrant expression of lncRNAs has a
potential role in the occurrence and development of various human
cancers and highlights the requirement for improved understanding
of the mechanisms involved (18).
In the present study, the effect of PTCSC3 on cell growth and
apoptosis in thyroid cancer cells was evaluated and the possible
binding of PTCSC3 with specific miRNAs was studied by bioinformatic
and biological analyses.
The highly thyroid-specific expression of PTCSC3 and
downregulated expression in PTC tissues and cell lines implicated
its potential role in thyroid cancer. To date, only one study has
investigated the biological feature changes of PTC cells following
transfection with PTCSC3 (10). In
the present study, three thyroid cancer cell lines of various
histo-pathological origin (papillary, follicular and anaplastic
thyroid cancer) were selected to study the effect of PTCSC3 on
thyroid cancer originating from follicular epithelial cells.
Collectively, our data demonstrate that overexpression of PTCSC3 in
thyroid cancer cells inhibits cellular proliferation and induces
cell cycle arrest and apoptosis, suggesting that dysfunction of
PTCSC3 in thyroid cancer may be a common molecular event. It is
essential to develop in vivo animal models with PTCSC3
expression in the thyroid to demonstrate the role of PTCSC3 in
tumorigenesis in the future.
According to the ceRNA hypothesis, lncRNAs may
elicit their biological activity through their ability to act as
endogenous decoys for miRNAs; such activity in turn affects the
distribution of miRNAs on their targets (11). Bioinformatic analysis of lncRNAs is
an important method for discovering the relevant functions
(19). We searched for miRNA
recognition motifs in the PTCSC3 sequence and the presence of
recognition sites for possible miRNAs. As shown in Table II, out of the top 20 miRNAs that
were identified, miR-574-5p was selected to validate the
interaction in thyroid cancer cells. To further comfirm the binding
possibility of PTCSC3 with miR-574-5p, the miR-574-5p expression in
various thyroid cell lines transfected with PTCSC3 was detected
using quantitative RT-PCR. The results revealed that miR-574-5p
expression was dramatically reduced due to overexpression of
PTCSC3. The significant inverse correlation between PTCSC3 and
miR-574-5p suggests that PTCSC3 acts as a competing endogenous RNA
to target miRNAs and in turn regulate cell growth and apoptosis in
thyroid cancer. Site-directed mutagenesis assay was performed to
confirm the binding site. Although there are no expression studies
regarding miR-574-5p in thyroid cancer cells, miR-574-5p was
previously identified to be associated with various human cancers
as oncogenic miRNA. As far as miR-574-5p is concerned, the
discovered functions include proliferation and
anchorage-independent growth of cancer cells in head and neck
squamous cell carcinoma, chemoresistance and poor survival in
patients with small-cell lung cancer, as well as colorectal cancer
tumorigenesis and progression at the early stages (20–22).
The present study provides information concerning the function of
PTCSC3; however, further investigations are required on lncRNA
PTCSC3 and its association with miRNAs in thyroid cancer. Moreover,
understanding the molecular mechanisms of PTCSC3 in thyroid cancer
is fundamentally important in developing new molecular markers for
earlier diagnosis and novel therapeutic targets.
Acknowledgements
This study was supported by the
National Natural Science Foundation of China (grant no. 30600601).
The authors would like to thank Junming Liao from Tulane
University, School of Medicine, USA for excellent technical support
and critical review of the manuscript.
References
|
1
|
Wang Y and Wang W: Increasing incidence of
thyroid cancer in Shanghai, China, 1983–2007. Asia Pac J Public
Health. Mar 16–2012.(Epub ahead of print).
|
|
2
|
Chen AY, Jemal A and Ward EM: Increasing
incidence of differentiated thyroid cancer in the United States,
1988–2005. Cancer. 115:3801–3807. 2009.
|
|
3
|
Davies L and Welch HG: Increasing
incidence of thyroid cancer in the United States, 1973–2002. JAMA.
295:2164–2167. 2006.
|
|
4
|
Li X, Abdel-Mageed AB and Kandil E: BRAF
mutation in papillary thyroid carcinoma. Int J Clin Exp Med.
5:310–315. 2012.
|
|
5
|
Li X, Abdel-Mageed AB, Mondal D and Kandil
E: The nuclear factor kappa-B signaling pathway as a therapeutic
target against thyroid cancers. Thyroid. Aug 13–2012.(Epub ahead of
print).
|
|
6
|
Gudmundsson J, Sulem P, Gudbjartsson DF,
et al: Common variants on 9q22.33 and 14q13.3 predispose to thyroid
cancer in European populations. Nat Genet. 41:460–464. 2009.
View Article : Google Scholar
|
|
7
|
Gudmundsson J, Sulem P, Gudbjartsson DF,
et al: Discovery of common variants associated with low TSH levels
and thyroid cancer risk. Nat Genet. 44:319–322. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Takahashi M, Saenko VA, Rogounovitch TI,
et al: The FOXE1 locus is a major genetic determinant for
radiation-related thyroid carcinoma in Chernobyl. Hum Mol Genet.
19:2516–2523. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
He H, Nagy R, Liyanarachchi S, et al: A
susceptibility locus for papillary thyroid carcinoma on chromosome
8q24. Cancer Res. 69:625–631. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jendrzejewski J, He H, Radomska HS, et al:
The polymorphism rs944289 predisposes to papillary thyroid
carcinoma through a large intergenic noncoding RNA gene of tumor
suppressor type. Proc Natl Acad Sci U S A. 109:8646–8651. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: the Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
McCarthy N: Regulatory RNA: layer by
layer. Nat Rev Genet. 12:8042011.PubMed/NCBI
|
|
13
|
Li X, Wang Z, Liu J, Tang C, Duan C and Li
C: Proteomic analysis of differentially expressed proteins in
normal human thyroid cells transfected with PPFP. Endocr Relat
Cancer. 19:681–694. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kertesz M, Iovino N, Unnerstall U, Gaul U
and Segal E: The role of site accessibility in microRNA target
recognition. Nat Genet. 39:1278–1284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liao JM and Lu H: Autoregulatory
suppression of c-Myc by miR-185-3p. J Biol Chem. 286:33901–33909.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mohammad F, Pandey GK, Mondal T, et al:
Long noncoding RNA-mediated maintenance of DNA methylation and
transcriptional gene silencing. Development. 139:2792–2803. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cesana M, Cacchiarelli D, Legnini I, et
al: A long noncoding RNA controls muscle differentiation by
functioning as a competing endogenous RNA. Cell. 147:358–369. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kogo R, Shimamura T, Mimori K, et al: Long
noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Krzyzanowski PM, Muro EM and
Andrade-Navarro MA: Computational approaches to discovering
noncoding RNA. Wiley Interdiscip Rev RNA. 3:567–579. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Meyers-Needham M, Ponnusamy S, Gencer S,
et al: Concerted functions of HDAC1 and microRNA-574-5p repress
alternatively spliced ceramide synthase 1 expression in human
cancer cells. EMBO Mol Med. 4:78–92. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ranade AR, Cherba D, Sridhar S, et al:
MicroRNA 92a-2*: a biomarker predictive for chemoresistance and
prognostic for survival in patients with small cell lung cancer. J
Thorac Oncol. 5:1273–1278. 2010.
|
|
22
|
Ji S, Ye G, Zhang J, et al: miR-574-5p
negatively regulates Qki6/7 to impact β-catenin/Wnt signalling and
the development of colorectal cancer. Gut. April 5–2012.(Epub ahead
of print).
|