Introduction
The Epstein-Barr virus (EBV) was the first described
onco-virus. Previously, research mainly focused on the role of
Burkitt lymphoma, nasopharyngeal carcinoma, Hodgkin’s disease and B
lymphoma in patients with acquired immuno-deficiency syndrome
(AIDS) (1). It has been identified
that 7–16% gastric cancer cases are EBV-positive (2,3).
Therefore, the role of EBV in gastric cancer is becoming
increasingly acknowledged. In this study, using
immunohistochemistry, we examined the expression of PCNA and c-myc
in 13 cases of EBV-associated gastric carcinoma (EBVaGC) and 45
cases of EBV-negative gastric carcinoma (EBVnGC) as the control.
Additionally, the pericancer tissues were also examined. The
Epstein-Barr nuclear antigen (EBNA)-1, EBNA-2 and latent membrane
protein 1 (LMP1) genes, which are expressed in the virus, were
assayed by reverse transcription-polymerase chain reaction (RT-PCR)
and the early genes BARF1 and BHRF2 were also explored. The aim of
this study was to establish the correlation between EBV-related
genes and PCNA and c-myc gene expression and to identify the role
of EBV-related genes in the pathophysiological process of
EBVaGC.
Materials and methods
Subjects
The samples of gastric cancer and the pericancer
tissues were obtained from inpatients who received surgical
treatment in the Affiliated Hospital of Qingdao University Medical
College, Qingdao Municipal Hospital and Yantai Yuhuangding
Hospital, China, from January 2008 to December 2009. Of the 185
specimens, 13 EBV-positive cases were identified by PCR and
southern blotting. Additionally, 45 cases that were paired with the
13 cases in age, gender, pathological type and clinical phase
served as the controls. In the 58 cases, there were 48 males and 10
females, with an average age of 58 years (range, 31–81 years). All
cases were confirmed by pathological examination. The study was
approved by the ethics committee of Yantai Yuhuangding Hospital,
Yantai, China. Informed consent was obtained from each patient.
Reagents and materials
A reverse transcription kit (Promega Corporation,
Madison, WI, USA), TRIzol (Gibco, Carlsbad, CA, USA),
Hybond-N+ membrane (Amersham Pharmacia Biotech,
Piscataway, NJ, USA), digoxigenin (DIG) oligonucleotide 3′-end
labeling kit, CSPD, DNA molecular weight marker VIII (Roche
Diagnostics, Mannheim, Germany), Taq polymerase and
deoxyribonucleotide triphosphate (dNTP; Sangong Biotech, Shanghai,
China) were used in this study. The monoclonal antibody for c-myc,
9E10, PCNA, PC10-vascular endothelial growth factor (VEGF) and
SP-type immunohistochemistry kits were purchased from Zhongshan
Golden Bridge Biotechnology Co., Ltd. (Beijing, China). All primers
and probes were synthesized by Beijing Saibaisheng Biotech Co.,
Ltd. (Beijing, China).
RNA extraction
The total RNA was extracted with TRIzol following
the manufacturer’s instructions for the reverse transcription kit.
cDNA was synthesized and served as the template for the PCR
reaction. The specific primers and probes for the latent and the
early phase gene were designed according to previous studies
(4–7). All primers and probes are listed in
Table I. The oligo probe was
marked with the DIG oligonucleotide 3′-end labeling kit, following
the manufacturer’s instructions.
 | Table IOligonucleotide primers and probes of
EBV related genes used for RT-PCR analysis. |
Table I
Oligonucleotide primers and probes of
EBV related genes used for RT-PCR analysis.
| Transcripts | Primer sequences
(5′-3′) | Product size
(bp) |
|---|
| EBNA1 | | |
| 5′ primer |
GATGAGCGTTTGGGAGAGCTGATTCTGCA | 273 |
| 3′ primer |
TCCTCGTCCATGGTTATCAC | |
| Probe |
AGACCTGGGAGCAGATTCAC | |
| EBNA2 | | |
| 5′ primer |
GCTGCTACGCATTAGAGACC | 339 |
| 3′ primer |
TCCTGGTAGGGATTCGAGGG | |
| Probe |
CAGCACTGGCGTGTGACGTGGTGTAAGTT | |
| LMP1 | | |
| 5′ primer |
TCCTCCTCTTGGCGCTACTG | 490 |
| 3′ primer |
TCATCACTGTGTCGTTGTCC | |
| Probe |
GAACAGCACAATTCCAAGGAACAATGCCTG | |
| BARF1 | | |
| 5′ primer |
GGCTGTCACCGCTTTCTTGG | 203 |
| 3′ primer |
AGGTGTTGGCACTTCTGTGG | |
| Probe |
CTGGTTTAAACTGGGCCCAGGAGAGGAGCA | |
| BHRF1 | | |
| 5′ primer |
GTCAAGGTTTCGTCTGTGTG | 211 |
| 3′ primer |
TTCTCTTGCTGCTAGCTCCA | |
| Probe |
ATGCACACGACTGTCCCGTATACAC | |
PCR
A 30 μl PCR system was employed with 1.0 IU
Taq DNA polymerase, 3 μl 10X buffer, 1.5 mmol/l
MgCl2, 0.1 mmol/l deoxyribonucleotide triphosphate
(dNTP), 0.5 μmmol/l each upper and lower primer and 3
μl cDNA product. The PCR system was denatured at 94°C for 5
min, followed by 35 cycles with the following conditions: 45 sec at
94°C, 45 sec at 58°C, 1 min at 72°C and 10 min at 72°C. For
glyceraldehyde 3-phosphate dehydrogenase (GAPDH), 25 cycles were
completed.
The cDNA from the lymphoblastoid cell line (LCL) was
employed as the negative control and the Ramos cell line was used
as the positive control for EBV. The 10 μl PCR product was
detected by 2% agarose gel containing 0.5 μg/ml ethidium
bromide under ultraviolent radiation. The gel was transferred to a
Hybond-N+ membrane and assessed by southern
blotting.
Immunohistochemistry (IHC) for c-myc and
PCNA
The antibody was diluted 150-fold for c-myc,
200-fold for PCNA and 100-fold for VEGF. Phosphate-buffered saline
(PBS) was employed as the negative control and a known breast
carcinoma specimen was used for the positive control. All IHC
experiments followed the manufacturer’s instructions.
The scoring criteria for immunohistochemistry were
as follows: i) c-myc, the positive signal for c-myc existed in the
nucleus as brown granular shapes. Five visual fields were randomly
selected. The samples were regarded as positive when the positive
area was >25% in the nucleus. ii) PCNA, the positive signal for
PCNA existed in the nucleus as yellow particles. PCNA labeling
index (PCNA LI) was calculated as the percentage of positive cells
in 1,000 tumor cells.
Statistical analysis
All data were analyzed with SPSS software (SPSS
Inc., Chicago, IL, USA). The quantitative data were expressed as
mean ± standard deviation (SD). The difference between the groups
was compared with t-test or χ2 test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of virus-related genes in the
EBVaGC tissues
The internal marker gene GAPDH was assayed in all 13
samples, which demonstrated the the success of PCR amplification.
The expression of EBNA1 mRNA was identified in all cases; however,
the expression of EBNA2 and LMP1 was negative in all cases. The
expression of BARF1 was identified in 6 cases and BHRF1 in 2 cases.
The RT-PCR and southern blotting results are shown in Fig. 1.
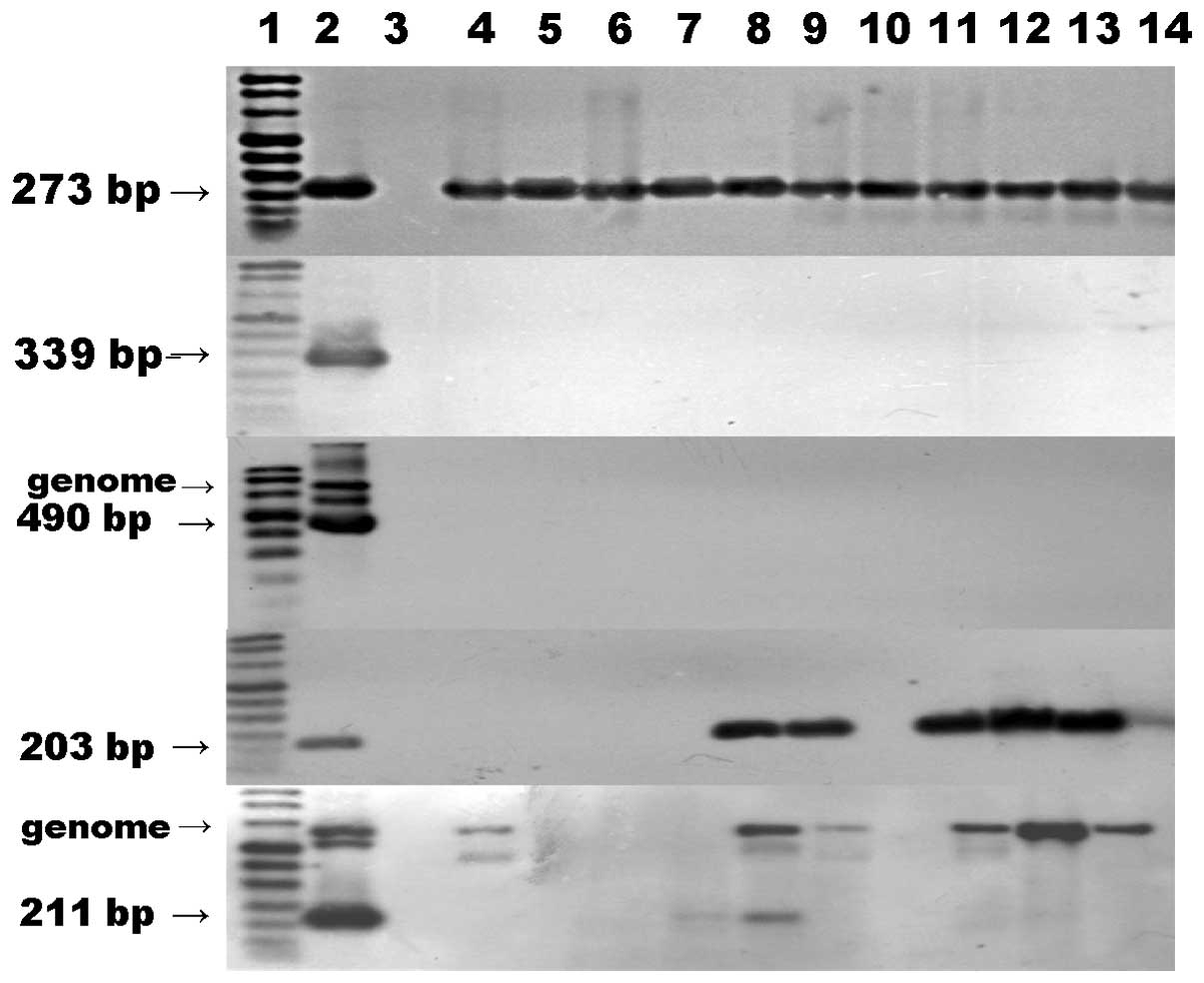 | Figure 1RT-PCR-southern blot analysis for
EBNA1, EBNA2, LMP1, BARF1 and BHRF1 gene expression. Lane 1, DNA
molecular weight marker VIII Digoxigenin-labeled; lane 2, LCL was
used as a positive control for the detection of EBNA1; lane 3,
Ramos cells (negative control); other lanes, EBVaGC specimens.
RT-PCR, reverse transcription-polymerase chain reaction; EBNA,
Epstein-Barr nuclear antigen; LMP1 latent membrane protein 1; LCL,
lymphoblastoid cell line; EBVaGC, Epstein-Barr virus-associated
gastric carcinoma. |
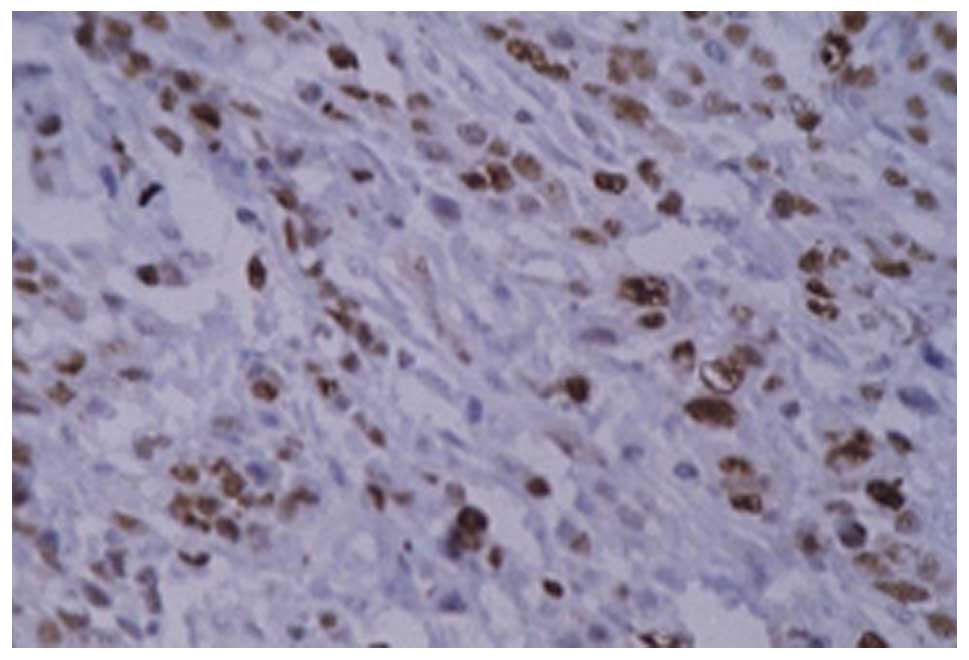
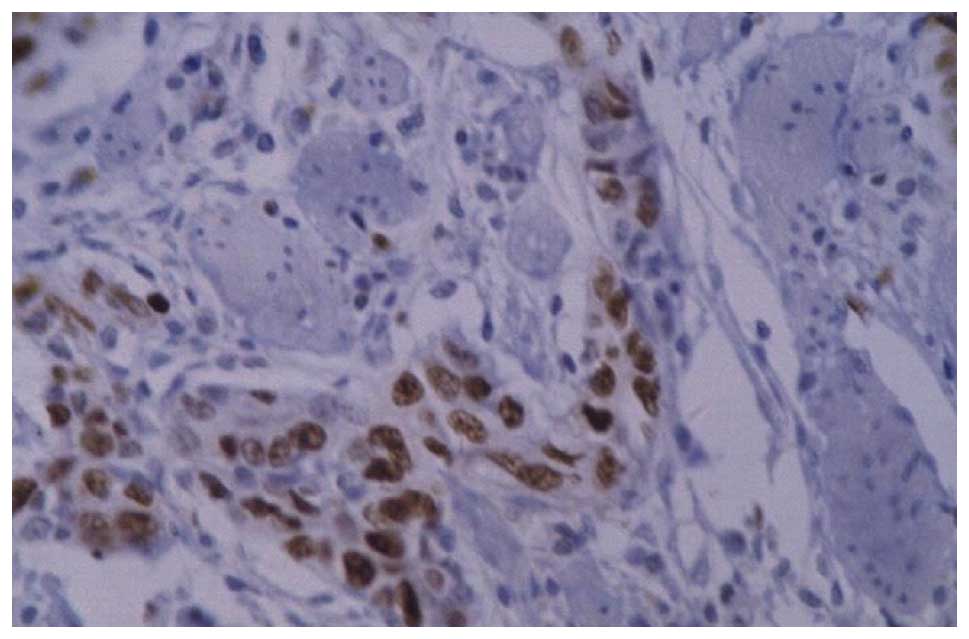
PCNA immunohistochemistry
The positive signal was observed as brown granules
in the nucleus (Figs. 2 and
3). The PCNA LI were
49.3768±12.1823, 14.8396±7.1847, 35.6139±8.3831 and 24.2735±10.1332
in the EBVaGC, the EBVaGC neighboring tissue, EBVnGC and the EBVnGC
neighboring tissue, respectively. The difference between EBVaGC and
EBVnGC was significant (t=4.686, P<0.01) and the difference
between EBVaGC and the neighboring tissue was also significant
(t=8.805, P<0.01). In all 58 cases, PCNA LI was significantly
higher in the c-myc-positive group than in the c-myc-negative group
(t=9.687, P<0.01). The results are listed in Table II.
 | Table IICorrelation between c-myc expression
and PCNA LI in 58 cases of gastric carcinoma. |
Table II
Correlation between c-myc expression
and PCNA LI in 58 cases of gastric carcinoma.
| N | LI |
|---|
| c-myc (+) | 33 | 51.645±9.172 |
| c-myc (−) | 25 | 32.013±4.916 |
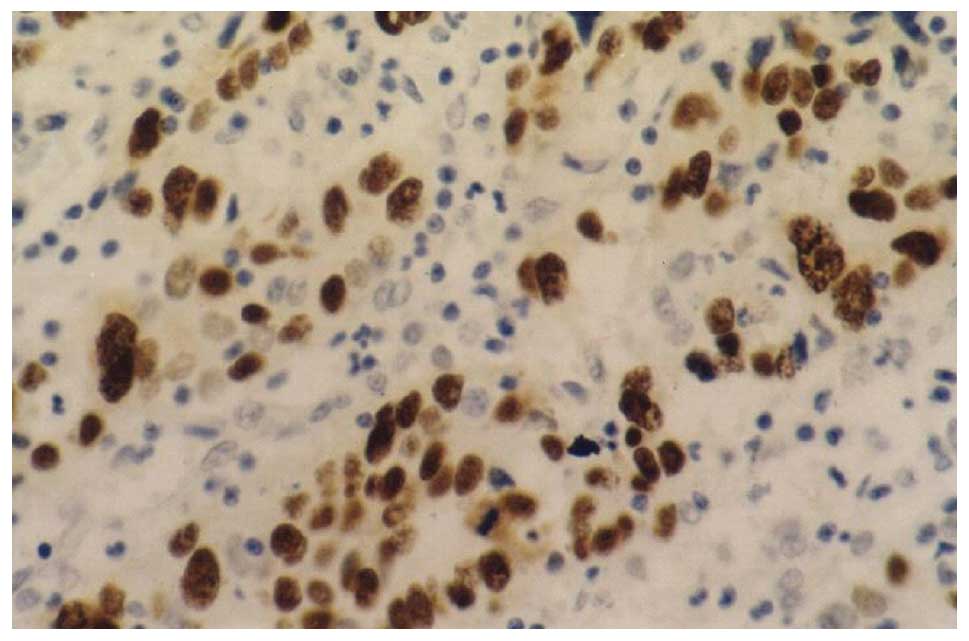
c-myc immunohistochemistry
c-myc was observed in the cytoplasm, as shown in
Figs. 4–6. The results demonstrated that the
c-myc-positive rate in the 58 gastric cancer cases was 55.93%
(33/58) and was 36.21% (21/58) in the neighboring tissues
(χ2=4.989, P<0.05). The c-myc-positive rate in EBVaGC
was 61.54% and in EBVnGC it was 55.56% (25/45). No significant
difference was identified between these two groups
(χ2=0.147, P>0.05). All results are listed in
Tables III and IV.
 | Table IIIComparison of c-myc-positive rate
between the gastric carcinoma tissues and corresponding
paracarcinoma tissues. |
Table III
Comparison of c-myc-positive rate
between the gastric carcinoma tissues and corresponding
paracarcinoma tissues.
| Gastric
carcinoma | Corresponding
paracarcinoma tissue
| Total |
|---|
| c-myc (+) | c-myc (−) |
|---|
| c-myc (+) | 15 | 18 | 33 |
| c-myc (−) | 6 | 19 | 25 |
 | Table IVComparison of c-myc protein expression
between EBVaGC and EBVnGC. |
Table IV
Comparison of c-myc protein expression
between EBVaGC and EBVnGC.
| N | c-myc (+) | c-myc (−) |
|---|
| EBVaGC | 13 | 8 | 5 |
| EBVnGC | 45 | 25 | 20 |
Discussion
In normal-dividing cells, the proliferation is
partially regulated by c-myc, which plays an essential role in the
regulation of cell proliferation and apoptosis. Besides, serving as
a pro-oncogene, c-myc is closely related with tumor cell
proliferation, transformation and the induction of apoptosis. c-myc
is markedly increased in cells with fast division and low
differentiation. Ishii et al (7) identified that in patients with
EBVnGC, the expression of c-myc was significantly lower in the
early phase than that in the proceeding phase. However, in EBVaGC
patients, the level of c-myc is slightly higher in the early phase
than that in the developing phase, although no significance was
identified. It is considered that EBV affects the expression of
c-myc. In the early phase of EBVaGC, EBV induces the expression of
c-myc and inhibits the over-expression of p53, promoting cell
proliferation. However, in the developing phase, EBV inhibits the
expression of c-myc and the apoptosis of tumor cells, which makes
therapy more difficult for late-phase gastric cancer. In our study,
we did not identify a significant difference between the positive
rate of c-myc in EBVaGC and EBVnGC. This suggests that there is no
clear correlation between c-myc gene expression and the infection
of EBV. The underlying reason may be due to sample bias. In our
study, the samples were obtained from developing and later-stage
patients. In these cases, the inducing effect of EBV on c-myc is
lower than that in the early phase.
PCNA plays an important role in the initiation of
cell proliferation. This molecule is observed in proliferating
cells or cells that demonstrate a proliferating tendency.
Therefore, PCNA reflects the proliferating condition of the whole
cell population. In our study, the content of PCNA LI was
significantly higher in EBVaGC cases than in EBVnGC cases, which
suggests that there is a correlation between the expression of PCNA
and EBV infection. EBV infection promotes the expression of PCNA.
We also identified that the level of PCNA LI was clearly higher in
the c-myc-positive group than that in the c-myc-negative group.
This indicates that c-myc expression is closely related to the
level of PCNA.
The viral proteins expressed in each phase during
EBV replication have shown the potential effect on cell
proliferation. A study has demonstrated that EBV-encoded LMP1 and
BHRF1 are the major genes closely conneted with cell proliferation
and c-myc levels. Yang et al (8) identified that EBV LMP1 induced the
expression of endogenous c-myc. Despite this fact, it remains
elusive whether LMP1 regulates the expression of c-myc directly or
via the NF-κB pathway or other signal transduction pathways.
By analyzing 55 nasopharyngeal carcinoma specimens,
Li and Zong (9) identified that
the index of PCNA in the EBV LMP1-positive samples was
significantly higher than that in the EBV LMP1-negative samples.
This indicates that EBV LMP1 may promote the proliferation of tumor
cells. Another study determined that EBNA2 promotes the expression
of c-myc. In our study, neither LMP1 nor EBNA2 were expressed in
the 13 samples, which indicates that cell proliferation and the
expression of c-myc is independent of LMP1 and EBNA2.
The EBV early gene BARF-1 is regarded as the second
main viral oncogene after LMP1. Studies have identified that BARF1
activates the expression of c-myc and B cell lymphoma 2 (Bcl-2)
(10,11). zur Hausen et al (5) identified transcriptional BARF1 mRNA
in 90% of the EBV-related gastriointestinal cancer cases. Despite
the LMP1 mRNA-negative results, the BARF1 protein still exists in
the epithelial cells. Therefore, BARF1, surrogating LMP1, plays a
role in tumorigenesis. In our study, 6 EBVaGCs samples were
BARF1-positive, which provides new evidence for its role in cancer.
The correlation between the BARF1 gene, cell proliferation and
c-myc gene expression requires further investigation with a greater
number of cases.
BHRF1 and Bcl-2 share a highly homologous gene
sequence. Similar to Bcl-2, BHRF1 inhibits cell apoptosis, promotes
cell growth and transforms and elongates cell lifespan. Huang et
al (12) observed a
high-efficiency BHRF1-expressing clone in the CNE2 cell line and
examined the biological activity following 60Co
irradiaton. The authors identified that BHRF1 inhibits the
expression of PCNA, ameliorates the sensitivity to the radiation
and promotes the survival of tumor cells under nutrient-deficient
conditions and contributes to the genesis and development of tumor
cells. In our study, only two EBVaGCs samples demonstrated positive
expression of BHRF1 and the other 11 cases demonstrated negative
expresion. Combined with results from the study by Huang et
al, we consider that BHRF1 inhibits cell proliferation and
promotes the advancement of tumor cells in specific circumstances,
including malnutrition, despite the fact that it does not
contribute to cell proliferation in physiological conditions.
References
|
1
|
Shah KM and Young LS: Epstein-Barr virus
and carcinogenesis: beyond Burkitt’s lymphoma. Clin Microbiol
Infect. 15:982–988. 2009.
|
|
2
|
Wu MS, Shun CT, Wu CC, Hsu TY, Lin MT,
Chang MC, Wang HP and Lin JT: Epstein-Barr virus-associated gastric
carcinomas: relation to H. pylori infection and genetic
alterations. Gastroenterology. 118:1031–1038. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Akiba S, Koriyama C, Herrera-Goepfert R
and Eizuru Y: Epstein-Barr virus associated gastric carcinoma:
epidemiological and clinicopathological features. Cancer Sci.
99:195–201. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sugiura M, Imai S, Tokunaga M, et al:
Transcriptional analysis of Epstein-Barr virus gene expression in
EBV-positive gastric carcinoma: unique viral latency in the tumor
cells. Br J Cancer. 74:625–631. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
zur Hausen A, Brink AA, Craanen ME, et al:
Unique transcription pattern of Epstein-Barr virus (EBV) in
EBV-carrying gastric carcinomas: expression of the transforming
BARF1 gene. Cancer Res. 60:2745–2748. 2000.PubMed/NCBI
|
|
6
|
Oudejans JJ, van den Brule AJ, Jiwa NM, et
al: BHRF1, the Epstein-Barr virus (EBV) homologue of the bcl-2
protooncogene, is transcribed in EBV associated B-cell lymphomas
and in reactive lymphocytes. Blood. 86:1893–1902. 1995.PubMed/NCBI
|
|
7
|
Ishii H, Gobe G, Kawakubo Y, et al:
Interrelationship between Epstein-Barr virus infection in gastric
carcinomas and the expression of apoptosis-associated proteins.
Histopathology. 38:111–119. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang J, Deng X, Deng L, et al:
Epstein-Barr virus LMP1 activates human telomerase transcription
via c-myc expression. Chinese Journal of Virology. 19:240–244.
2003.
|
|
9
|
Li Z and Zong Y: Effects of LMP-1
expression on neoplastic cell proliferation and apoptosis in
nasopharyngeal carcinoma. Zhonghua Bing Li Xue Za Zhi. 28:340–343.
1999.(In Chinese).
|
|
10
|
Sheng W, Decaussin G, Sumner S, et al:
N-terminal domain of BARF1 gene encoded by Epstein-Barr virus is
essential for malignant transformation of rodent fibroblasts and
activation of BCL-2. Oncogene. 20:1176–1185. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sheng W, Decaussin G, Ligout A, et al:
Malignant transformation of Epstein-Barr virus-negative Akata cells
by introduction of the BARF1 gene carried by Epstein-Barr virus. J
Virol. 77:3859–3865. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang H, Pan X and Yu L: The effect of
Epstein-Barr virus gene BHRF1 expression on the apoptotic
resistance of nasopharyngeal carcinoma cells. Zhonghua Zhong Liu Za
Zhi. 20:245–247. 1998.(In Chinese).
|