Introduction
Gliomas are the most common primary malignant tumor
of the adult central nervous system, accounting for ~40% of
intracranial tumors (1). Gliomas
have the worst prognosis due to chemoresistance and
radioresistance. Despite multimodality treatments with extensive
surgical resection, radiotherapy or chemotherapy, the median
survival time of glioma patients is ~1 year (2,3).
Therefore, novel strategies are urgently required to increase the
survival rate of glioma patients.
The C6 cell line is a rat glioma cell line that is
induced by N-nitrosomethylurea. This cell line is widely adopted
for glioma study due to the histocompatibility in various
categories of rats and the infiltrative growth pattern that is
similar to human glioma (4).
Therefore, C6 cells were employed in the present study as the cell
model for glioma.
Doxorubicin (DOX) is an anticancer agent with a wide
spectrum, including breast cancer, lung cancer, leukemia, lymphoma
and glioma (5). DOX can penetrate
through the cell membrane into cells and combine with chromosomes.
The antitumor effects of DOX are exerted mainly through forming a
complex with double stranded DNA and strongly interfering with the
synthesis of DNA, RNA and also proteins (6). A previous study demonstrated that DOX
is able to insert into DNA and cause the splitting of DNA by
topoisomerase II. An additional antitumor mechanism of DOX is
associated with redox (7). A
series of NADPH-dependent cytoreductases reoxidize DOX into
semiquinone radicals, which react with oxygen to produce cytotoxic
chemicals, including peroxides, hydroxide radicals and hydrogen
peroxides. In addition, DOX is hypothesized to combine with lipids
on the cell membrane, interfering with a number of cell functions.
DOX yields antitumor effects through one or more of the
aforementioned mechanisms and cells in all stages of the cell cycle
are sensitive to DOX, particularly those in hyperplastic tissues,
including tumors. Despite this, the application of DOX is limited
due to the side effects, including gastrointestinal adverse
reactions, alopecia, allergy, myelosuppression and heart toxicity,
among which dose-dependent heart failure is the most significant
(8). In addition, specific
resistance may occur more easily in single drug therapy. These
factors hinder the clinical utility of DOX. However, one method of
reducing the side effects and resistance is to use the drug in
conjunction with other agents.
Telomeres are nucleoprotein complexes composed of
(TTAGGG)n repeats and a specialized protein complex that
protect the chromosome-ends from being recognized as deleterious
DNA double-stranded breaks, which activates DNA damage responses
(9). Telomere length is maintained
by telomerase, a specialized reverse transcriptase, that adds
TTAGGG repeats to the ends of the chromosomes. In humans and other
long-living mammals, telomerase expression is repressed in the
majority of somatic cells. As a result, telomeres become
increasingly shorter with continuous cell divisions due to the
‘end-replication’ problem (10).
This progressive telomere shortening functions as a cell-autonomous
barrier against overproliferation, making a potent tumor-suppressor
mechanism. The re-expression of telomerase is a key event in
tumorigenesis, which is supported by the evidence that telomerase
is present in 90% of human cancers (11). The role of telomerase in glioma has
also been established, with the rat glioma C6 cell line included.
Therefore, telomerase is hypothesized to be a novel and effective
target for the therapy of gliomas (12,13).
With fast development of molecular biology and
genetics, numerous genes have been identified that are closely
associated with tumorigenesis, including a number of oncogenes and
tumor suppressor genes. PIN2-interacting protein 1 (PinX1), a
nucleolar protein associated with telomere/telomerase, is a
putative tumor suppressor. However, the role of PinX1 in
telomerase/telomere regulation and cancer remains unclear (14). The expression of the PinX1 mRNA
transcript is present in the majority of tested human tumors
(15–17). Certain studies consider PinX1 to be
an intrinsic telomerase/telomere inhibitor and a putative tumor
suppressor, as it binds to and suppresses telomerase enzymatic
activity (18). However, other
experiments have demonstrated that the depletion of PinX1
expression shortens telomere length and inhibits proliferation in
yeast cells (19). In addition,
Zhang et al demonstrated that silencing PinX1 induces
senescence in telomerase-positive cancer cells (20). Therefore, in the present study,
PinX1-short interfering (si)RNA was used to downregulate PinX1 mRNA
expression and subsequently study the effect on telomerase.
siRNA is rapidly becoming an important tool for gene
knockdown and the analysis of gene function (21). Knockdown of specific pathogenic
genes is a potent approach for treating diseases, including tumors.
In the present study, PinX1 was knocked-down in C6 cells and cell
viability was analyzed to confirm the potential of PinX1 as a
target gene for the treatment of gliomas. Furthermore, gliomas were
treated with a combination of PinX1-siRNA and DOX, with the aim of
increasing the efficiency of the treatment and decreasing the side
effects.
A major challenge in the study of siRNA is the
development of an appropriate delivery system with high efficiency
and low toxicity. In previous years, nonviral vectors have become
attractive options, among which nanoparticles are suitable
candidates for the siRNA delivery system (22). Nanoparticles are a series of
structures in the nanometer scale size range (≤100 nm) that retain
unique properties. Nanoparticles include polymeric micelles,
dendrimers, polymeric and ceramic nanoparticles, protein cage
architectures, viral-derived capsid nanoparticles, polyplexes and
liposomes (23). As siRNA
carriers, nanoparticles have overwhelming superiority since they
are biocompatible and biodegradable with strong penetrability and
good capacity, but have low toxicity and immunogenecity.
Furthermore, by functionalizing the surface with synthetic polymers
and appropriate ligands, nanoparticles can be targeted to specific
cells and locations within the body or be endowed with specific
properties. Superparamagnetic iron oxide nanoparticle (SPION), a
highly efficient T2 contrast agent for magnetic resonance imaging
(MRI), is widely used experimentally for agent delivery (24). The aforementioned attributes of the
SPION-based carrier system enable broad biomedical applications,
particularly in in vivo studies. In the present study, siRNA
molecules with SPION-labeled nanoparticles were delivered to C6
cells to confirm the suitability of this method as a transfection
tool. The results of the study may be the foundation for future
animal model experiments.
Materials and methods
Materials and reagents
Monomethoxy polyethylene glycol [mPEG; molecular
weight cut off (MWCO), 2 kDa] and N,N′-carbonyldiimidazole (CDI)
were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Hyperbranched polyethyleneimine (PEI; MWCO, 25 kDa) was purchased
from BASF (Ludwigshafen, Rheinland-Pfalz, Germany). Tetrahydrofuran
and chloroform (CHCl3) were dried over calcium hydride
and distilled prior to use. SPIONs, with an average diameter of 6
nm, were synthesized according to the method reported by Sun et
al (25). Glioma C6 cells were
obtained from the American Type Culture Collection (Rockville, MD,
USA) and cell culture media and fetal bovine serum (FBS) were
purchased from Invitrogen Life Technologies (Carlsbad, CA, USA).
Double-stranded oligonucleotides with homology to a desired target
region of PinX1 were synthesized by Guangzhou RiboBio Co., Ltd.
(Guangzhou, China) and the target sequence was GCTGTGGATCCCAGAAATA.
Cy3-control siRNA and negative control (NC) siRNA were also
supplied by Guangzhou RiboBio Co., Ltd. DOX was obtained from
Shenzhen Wanle Pharmaceutical Co., Ltd. (Shenzhen, China). Total
RNA was extracted with the TRIzol reagent (Invitrogen, Carlsbad,
CA, USA). A SYBR PrimeScript quantitative polymerase chain reaction
(qPCR) kit and primers were supplied by Takara Biotechnology, Co.,
Ltd. (Dalian, China). A bicinchoninic acid (BCA) kit was purchased
from Pierce Biotechnology, Inc. (Rockford, IL, USA) and a TeloTAGGG
Telomerase PCR ELISA kit was purchased from Roche Diagnostics
Corporation (Indianapolis, IN, USA). Cell Counting Kit-8 (CCK-8)
was obtained from Dojindo (Kumamoto, Japan) and the Hoechst 33243
stain was purchased from Sigma-Aldrich.
Synthesis of mPEG-PEI-SPION
The mPEG-PEI complex was prepared as previously
described (26). The hydroxyl
terminal groups of mPEG were activated by CDI in order to allow the
conjugation of mPEG to branched PEI. Next, CDI-activated mPEG was
conjugated to PEI. The mPEG-PEI-SPION complex was prepared by a
‘ligand exchange’ method as previously reported (27). In total, 200 mg mPEG-PEI and 10 mg
SPION were dissolved in 2 ml CHCl3. The solution was
stirred overnight at room temperature and then precipitated with
hexane. The precipitate was dispersed into double distilled water
under sonication. Large aggregates were removed by filtering
through a 220 nm membrane. The encapsulation efficiency of the
SPION was determined using a polarized Zeeman Atomic Absorption
Spectrophotometer (Z-2000 series).
Polyplex siRNA/mPEG-PEI-SPION
formation
siRNA and an appropriate amount of mPEG-PEI-SPION
(100 nmol siRNA:3.53 μg mPEG-PEI-SPION) (N/P=5) were dissolved
separately in double distilled water. The two solutions were mixed
by gentle pipetting and the mixture was maintained at room
temperature for 30 min to allow polyplex formation.
ζ-potential and size measurements
The ζ-potential measurements of mPEG-PEI-SPION and
siRNA/mPEG-PEI-SPION were conducted using a ZetaPlus instrument
(Brookhaven Instruments Corporation, Holtsville, NY, USA) at an
angle of 15° at 25°C. The average values plus the SDs were based on
the data of three runs. Nanoparticle size was determined using the
same instrument at 25°C. Scattering light was detected at a 90°
angle and the sizes determined were the mean values of three runs
plus the SD.
Cell culture
The rat glioma C6 cell line was maintained in high
glucose Dulbecco’s modified Eagle’s medium, supplemented with 10%
FBS and penicillin/streptomycin, at 37°C in a fully humidified
atmosphere of 5% CO2. When the cells reached confluence,
they were trypsinized and subcultured.
Transfection efficiency of
siRNA/mPEG-PEI-SPION
C6 glioma cells were plated in 24-well plates
(5×104 cells/well) and allowed to grow overnight.
Cy3-siRNA and mPEG-PEI-SPION (at N/P=5) were mixed by gentle
pipetting and then incubated for 30 min at room temperature. The
original cell culture medium was replaced with medium containing
the complexes (concentration of siRNA was 100 nM). The culture was
incubated for 4 h at 37°C. After 4 h, the medium was discarded and
the cells were washed twice with phosphate-buffered saline (PBS).
Next, cells were fixed with paraformaldehyde for 30 min. After
washing twice, the cells were stained with 10 μg/ml Hoechst 33243
for 15 min. The cells were observed under a fluorescence microscope
(Carl Zeiss AG, Jena, Germany) and fluorescent images were captured
and recorded.
qPCR
C6 cells were plated in 12-well plates
(1×105 cells/well) and allowed to grow overnight. Next,
the cells were transfected with 100 nM NC-siRNA/mPEG-PEI-SPION or
100 nM PinX1-siRNA/mPEG-PEI-SPION complexes (at N/P=5) as
aforementioned. Cells were washed with pre-chilled PBS and
collected in TRIzol reagent 48 h following transfection. Total RNA
was extracted with the TRIzol reagent according to the instructions
provided by the manufacturer. First strand cDNA was synthesized
from 1 μg total RNA. The reaction conditions were as follows: 37°C
for 15 min and 85°C for 5 sec. PCR was performed in a 20 μl volume
(1 μl cDNA) with SYBR green dye. Sequence-specific oligonucleotide
primers were as follows: PinX1, 5′-AACCACCTGGGACTTGGAGCTA-3′
(forward) and 5′-CCTGACCATGGCAAGTGTTGA-3′ (reverse); and β-actin,
5′-GGAGATTACTGCCCTGGCTCCTA-3′ (forward) and
5′-GACTCATCGTACTCCTGCTTGCTG-3′ (reverse). The reaction conditions
were as follows: 95°C for 30 sec, followed by 40 cycles of 95°C for
5 sec and 60°C for 34 sec. β-actin was amplified as the
housekeeping gene and qPCR assays of all the samples were performed
in triplicate.
Telomerase activity assay
Telomerase activity was measured with a TeloTAGGG
Telomerase PCR ELISA kit. C6 cells were transfected with
NC-siRNA/mPEG-PEI-SPION or PinX1-siRNA/mPEG-PEI-SPION as
aforementioned. After 48 h, cellular extracts were prepared with 1X
CHAPS lysis buffer. The protein concentration was determined using
a BCA kit. An extract equal to 1 μg protein was amplified with the
reaction mixture included in the kit. The extract and reaction
mixture were maintained at 25°C for 30 min, then heated at 90°C for
3 min and subjected to 30 cycles of PCR with specific programing
(94°C for 30 sec, 50°C for 30 sec and 72°C for 90 sec). Following
an additional 10 min at 72°C, the amplification products were
subjected to hybridization and ELISA, according to the instructions
provided by the manufacturer. The absorbance value of each sample
was calculated using a microplate reader at 450 nm, with a
reference of 690 nm. Each sample was tested in triplicate.
Cell viability
For the CCK-8 test, C6 cells were plated in 96-well
plates at an initial density of 1×104 cells/well.
PinX1-siRNA/mPEG-PEI-SPION and NC-siRNA/mPEG-PEI-SPION (at N/P=5)
complexes were constructed as aforementioned. The original cell
culture medium was replaced with medium containing one of the
complexes (100 nM siRNA) and the cells were incubated for 4 h at
37°C. After 4 h, the medium was replaced with fresh complete
medium, with or without 10 μg/ml DOX, and cells were allowed to
grow for 20 h. After 24 h of DOX incubation, the CCK-8 reagent was
added to the wells and incubated at 37°C for 1 h. Absorbance at 570
nm of each well was recorded with the microplate reader. Each
treatment group was replicated in three wells.
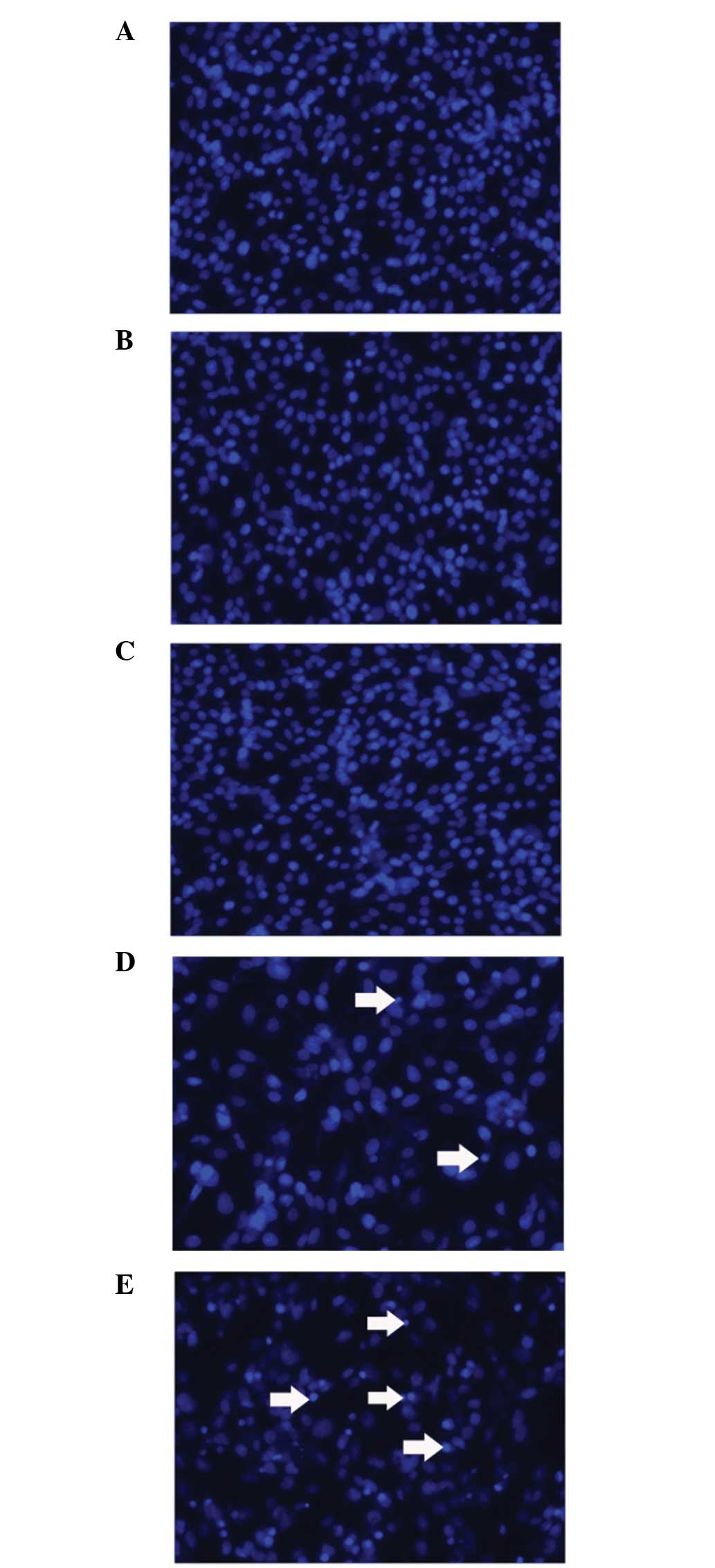
Cell apoptosis
C6 cells were plated in 24-well plates
(5×104 cells/well) and allowed to grow overnight.
Transfection and DOX incubation were performed as aforementioned.
Following DOX incubation for 24 h, the medium was discarded and the
cells were washed with PBS three times. Next, cells were fixed with
paraformaldehyde for 30 min and washed twice. The cells were
stained with 10 μg/ml Hoechst 33243 for 15 min and a fluorescence
microscope was used for observation.
Statistical analysis
Results were analyzed using SPSS version 12.0 (SPSS,
Inc., Chicago, IL, USA) and compared using one way analysis of
variance with Fisher’s least significant difference post hoc test.
Data are presented as the mean ± SD and P<0.05 was considered to
indicate a statistically significant difference.
Results
Size and ζ-potential of the
nanoparticles
The average size and ζ-potential of the
mPEG-PEI-SPION and siRNA/mPEG-PEI-SPION complexes are shown in
Table I. Sizes of
PinX1-siRNA/mPEG-PEI-SPIONs ranged between 100 and 150 nm, which
was a suitable size for higher specific surface area and better
penetrability. The average ζ-potential of
PinX1-siRNA/mPEG-PEI-SPIONs was 25.27±1.75 mV, which enabled the
absorption of PinX1-siRNA/mPEG-PEI-SPIONs through the cell membrane
which has a negative potential.
 | Table Iζ-potential and size of mPEG-PEI-SPION
and PinX1-siRNA/mPEG-PEI-SPION (N/P=5). |
Table I
ζ-potential and size of mPEG-PEI-SPION
and PinX1-siRNA/mPEG-PEI-SPION (N/P=5).
| Nanoparticle | ζ-potential, mV | Size, nm |
|---|
| mPEG-PEI-SPION | 34.42±0.78 | 39.6±1.2 |
|
PinX1-siRNA/mPEG-PEI-SPION (N/P=5) | 25.27±1.75 | 126.3±2.3 |
Transfection and knockdown
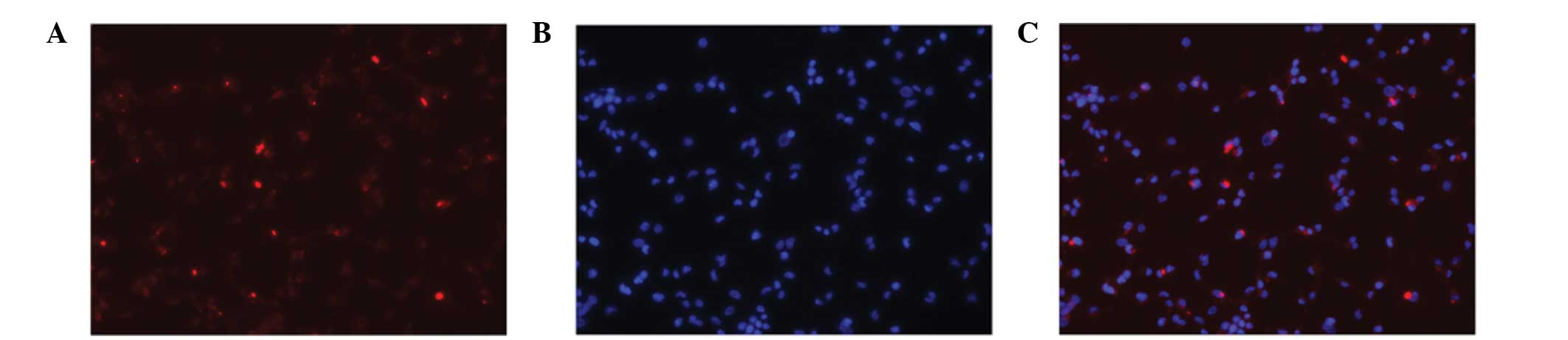
In the transfection of Cy3-siRNA/mPEG-PEI-SPION into
C6 cells, Cy3-siRNA/mPEG-PEI-SPION was shown to be efficiently
delivered into C6 cells. As shown in Fig. 1, almost all the C6 cells were
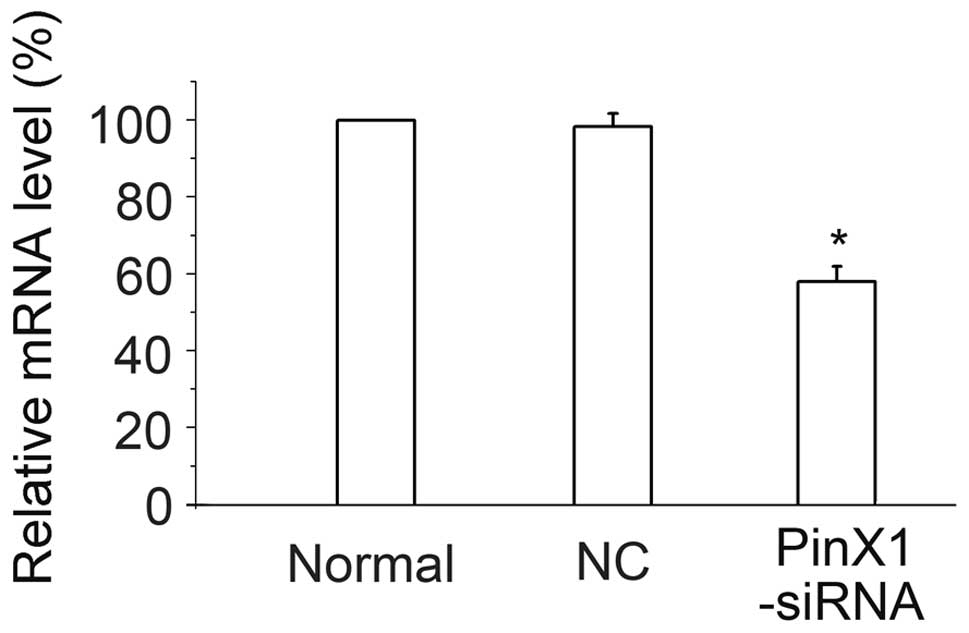
transfected with Cy3-siRNA/mPEG-PEI-SPION. The following qPCR
experiment demonstrated that PinX1-siRNA/mPEG-PEI-SPION
transfection resulted in a knockdown of PinX1 mRNA expression
(Fig. 2), while
NC-siRNA/mPEG-PEI-SPION did not have this effect. Therefore,
mPEG-PEI-SPION is an ideal tool for delivering siRNA into C6 cells.
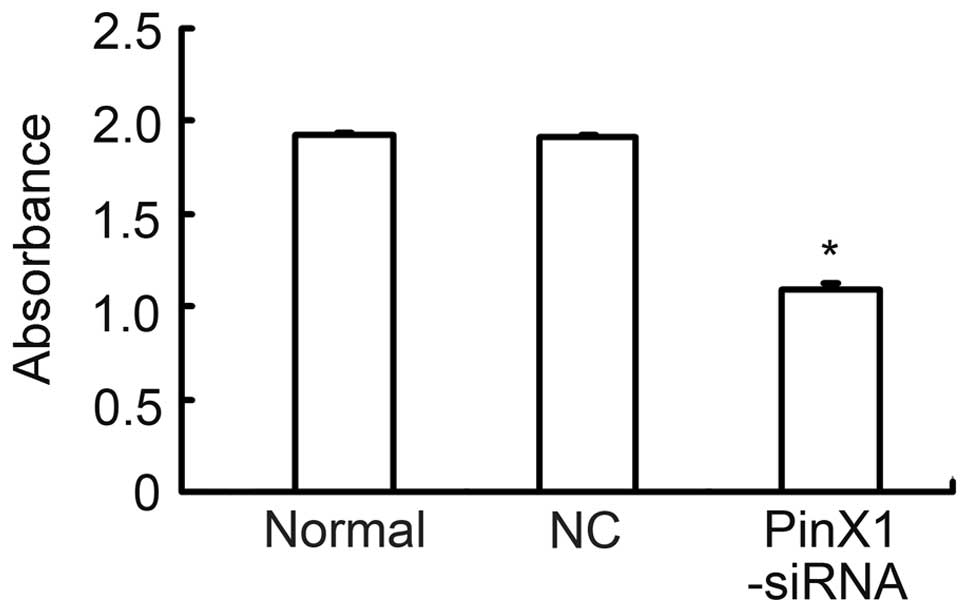
Furthermore, transfection of 100 nM PinX1-siRNA/mPEG-PEI-SPION
significantly decreased telomerase activity in C6 cells (Fig. 3).
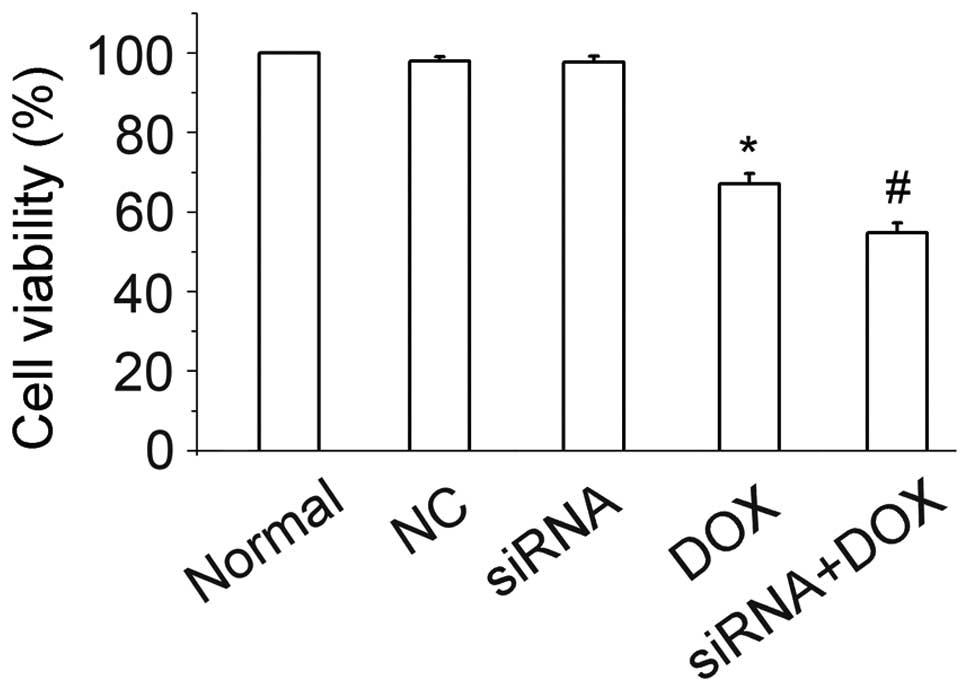
Cell viability
Administration of 10 μg/ml DOX resulted in a
prominent decrease in cell viability when compared with the normal
controls. PinX1-siRNA alone was unable to cause cytotoxicity in the
observed time period, however, when used in combination with DOX,
PinX1-siRNA was able to sensitize the inhibition effect of DOX.
PinX1-siRNA decreased cell viability by ~12.3%, as measured by the
CCK-8 method (Fig. 4). In
addition, PinX1-siRNA enhanced the cell toxicity of DOX by
promoting cell death and apoptosis, as shown by Hoechst 33243
staining (Fig. 5).
Discussion
The anthracycline chemical DOX is a common antitumor
agent, however, the drug is limited by its toxicity to healthy
organisms, of which acute or chronic heart failure is the most
severe (28).
Although siRNA has been shown to be a promising
therapy, the delivery system is one of the obstacles for siRNA
application, particularly in the nervous system which is difficult
to transfect. Viruses mediate transfection at a high efficiency,
but are limited by their immunogenicity. Nanomedicine is a rapidly
developing field, and due to the increasing interest, marked
progress has been made in the medical applications of nanoscale
devices. Nanoparticles have been applied in disease diagnosis,
treatment and prevention. Among them, cationic polymers are the
most common. PEG, a type of cationic polymer, has been increasingly
studied and identified to have a high transfection efficiency,
thus, currently PEG is considered to be the gold standard of
transfection efficiency (29).
Furthermore, PEG has been modified with PEI to obtain mPEG-PEI, an
improved nanoparticle with less toxicity, more target activity and
improved stability. In addition, mPEG-PEI can be labeled with other
molecules to obtain various characteristics, including SPION, a
magnetic nanoparticle which is detectable by MRI. Medarova et
al (30) successfully used
SPION-labeled nanoparticles to deliver siRNA to tumor cells. In the
present study, mPEG-PEI-SPION was competent in forming complexes
with siRNA and entering C6 cells, which facilitated further in
vivo study with MRI. To the best of our knowledge, the current
study is the first to apply this system in C6 cells.
Telomeres function as protective structures that cap
the ends of chromosomes. They consist of terminal TTAGGG repeats
and telomere-specific DNA binding proteins. With each cell
division, the 5′ end of the telomere is shortened by 50–200
nucleotides. Therefore, following several replications, the
telomeres reach a threshold and cell proliferation arrest or cell
death occurs. Telomerase is an RNA-dependent DNA polymerase that
functions as a reverse transcriptase which is responsible for the
synthesis of telomeres. Telomerase prevents the shortening of
telomeres and is essential for the maintenance of telomere length
and activity. The activation of telomerase is considered to be
critical in cell immortalization (11) and increasing evidence indicates
that telomere dysfunction is a common driver for the genomic
instability that is present in cancer (31). Ding et al reactivated
telomerase expression with the inducible mouse telomerase reverse
transcriptase transgene in a prostate cancer mouse model. The
authors found that re-expression of telomerase in the prostate
epithelium generated more aggressive tumors (32). These results further indicate that
telomerase may be a potent target for combating tumors.
PinX1 is a conserved nucleolar protein that has
complex roles in telomerase/telomere regulation and cancer. In the
study by Zhou and Lu, downregulation of PinX1 expression via
antisense cDNA transfection resulted in increased telomerase
activity, increased telomere length and enhanced tumor malignancy
in the HT1080 (telomerase-positive) cancer cell line. These results
indicated that PinX1 is a telomerase/telomere inhibitor and a
putative tumor suppressor in humans (18). However, whether PinX1 is
tumor-suppresive or promotive remains elusive. In the present
study, transfection of PinX1-siRNA/mPEG-PEI-SPION into C6 cells not
only resulted in the significant downregulation of PinX1 mRNA
expression, but transfection also weakened telomerase activity.
These observations support the hypothesis that PinX1 may be a
target for suppressing tumor growth, in accordance with the study
by Zhang et al (20).
The aims of the present study were to determine
whether mPEG-PEI-SPION may be a device for siRNA delivery into C6
cells and whether the specific silencing of PinX1 by siRNA may
improve the cytotoxic effect of DOX in C6 glioma cells. As
demonstrated, a combination of PinX1-siRNA/mPEG-PEI-SPION and DOX
resulted in increased cell loss when compared with DOX
administration alone. Therefore, PinX1-siRNA/DOX is more efficient
for inhibiting C6 growth. The results of the present study support
the hypothesis that combined therapy with PinX1-siRNA/DOX can
successfully maintain the tumor inhibition effect with reduced side
effects.
In conclusion, the current study has demonstrated
that mPEG-PEI-SPION is viable in delivering siRNA to C6 cells for
the purpose of treatment. Furthermore, PinX1 may regulate
telomerase activity and is therefore a potential target for
inhibiting tumors. To the best of our knowledge, this is the first
study to treat gliomas with a combination of DOX and PinX1-siRNA.
In addition, since SPIONs can be monitored by MRI, the present
study may be used as a pilot study for further investigation into
the application of siRNA/mPEG-PEI-SPION for brain tumors in
vivo.
Acknowledgements
The study was supported by grants from the National
Natural Science Foundation of China (nos. 30672411, 30973479 and
81301088), the ‘863’ Programs of China (no. 2007AA021101) and the
Science and Technology Planning Project of Guangdong Province,
China (nos. 2009B060700040 and 2011B031800141).
References
|
1
|
Sengupta S, Marrinan J, Frishman C and
Sampath P: Impact of temozolomide on immune response during
malignant glioma chemotherapy. Clin Dev Immunol. 2012:8310902012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shinojima N, Tada K, Shiraishi S, Kamiryo
T, Kochi M, Nakamura H, Makino K, Saya H, Hirano H, Kuratsu J, Oka
K, Ishimaru Y and Ushio Y: Prognostic value of epidermal growth
factor receptor in patients with glioblastoma multiforme. Cancer
Res. 63:6962–6970. 2003.PubMed/NCBI
|
|
3
|
Jagannathan J, Prevedello DM, Dumont AS
and Laws ER: Cellular signaling molecules as therapeutic targets in
glioblastoma multiforme. Neurosurg Focus. 20:E82006.PubMed/NCBI
|
|
4
|
De Ridder L: Behaviour of gliomas in vitro
vs histopathological grading. Int J Dev Neurosci. 17:541–546.
1999.PubMed/NCBI
|
|
5
|
Doroshow JH: Anthracycline
antibiotic-stimulated superoxide, hydrogen peroxide, and hydroxyl
radical production by NADH dehydrogenase. Cancer Res. 43:4543–4551.
1983.
|
|
6
|
Tewey KM, Rowe TC, Yang L, Halligan BD and
Liu LF: Adriamycin-induced DNA damage mediated by mammalian DNA
topoisomerase II. Science. 226:466–468. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mukhopadhyay P, Rajesh M, Bátkai S,
Kashiwaya Y, Haskó G, Liaudet L, Szabó C and Pacher P: Role of
superoxide, nitric oxide, and peroxynitrite in doxorubicin-induced
cell death in vivo and in vitro. Am J Physiol Heart Circ Physiol.
296:H1466–H1483. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rafiyath SM, Rasul M, Lee B, Wei G, Lamba
G and Liu D: Comparison of safety and toxicity of liposomal
doxorubicin vs. conventional anthracyclines: a meta-analysis. Exp
Hematol Oncol. 1:102012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Carneiro T, Khair L, Reis CC, Borges V,
Moser BA, Nakamura TM and Ferreira MG: Telomeres avoid end
detection by severing the checkpoint signal transduction pathway.
Nature. 467:228–232. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
de Lange T: How telomeres solve the
end-protection problem. Science. 326:948–952. 2009.PubMed/NCBI
|
|
11
|
Shay JW and Bacchetti S: A survey of
telomerase activity in human cancer. Eur J Cancer. 33:787–791.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Falchetti ML, Fiorenzo P, Mongiardi MP,
Petrucci G, Montano N, Maira G, Pierconti F, Larocca LM, Levi A and
Pallini R: Telomerase inhibition impairs tumor growth in
glioblastoma xenografts. Neurol Res. 28:532–537. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Harada K, Kurisu K, Tahara H and Tahara E,
Ide T and Tahara E: Telomerase activity in primary and secondary
glioblastomas multiforme as a novel molecular tumor marker. J
Neurosurg. 93:618–625. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lai XF, Shen CX, Wen Z, Qian YH, Yu CS,
Wang JQ, Zhong PN and Wang HL: PinX1 regulation of telomerase
activity and apoptosis in nasopharyngeal carcinoma cells. J Exp
Clin Cancer Res. 31:122012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chang Q, Pang JC, Li J, Hu L, Kong X and
Ng HK: Molecular analysis of PinX1 in medulloblastomas. Int J
Cancer. 109:309–314. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hawkins GA, Chang BL, Zheng SL, Isaacs SD,
Wiley KE, Bleecker ER, Walsh PC, Meyers DA, Xu J and Isaacs WB:
Mutational analysis of PINX1 in hereditary prostate cancer.
Prostate. 60:298–302. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Oh BK, Chae KJ, Park C and Park YN:
Molecular analysis of PinX1 in human hepatocellular carcinoma.
Oncol Rep. 12:861–866. 2004.PubMed/NCBI
|
|
18
|
Zhou XZ and Lu KP: The
PIN2/TRF1-interacting protein PINX1 is a potent telomerase
inhibitor. Cell. 107:347–359. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guglielmi B and Werner M: The yeast
homolog of human PinX1 is involved in rRNA and small nucleolar RNA
maturation, not in telomere elongation inhibition. J Biol Chem.
277:35712–35719. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang B, Bai YX, Ma HH, Feng F, Jin R,
Wang ZL, Lin J, Sun SP, Yang P, Wang XX, Huang PT, Huang CF, Peng
Y, Chen YC, Kung HF and Huang JJ: Silencing PinX1 compromises
telomere length maintenance as well as tumorigenicity in
telomerase-positive human cancer cells. Cancer Res. 69:75–83. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jackson AL, Bartz SR, Schelter J,
Kobayashi SV, Burchard J, Mao M, Li B, Cavet G and Linsley PS:
Expression profiling reveals off-target gene regulation by RNAi.
Nat Biotechnol. 21:635–637. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Malek A, Merkel O, Fink L, Czubayko F,
Kissel T and Aigner A: In vivo pharmacokinetics, tissue
distribution and underlying mechanisms of various PEI(−PEG)/siRNA
complexes. Toxicol Appl Pharmacol. 236:97–108. 2009.
|
|
23
|
Moghimi SM, Hunter AC and Murray JC:
Nanomedicine: current status and future prospects. FASEB J.
19:311–330. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Alwi R, Telenkov S, Mandelis A, Leshuk T,
Gu F, Oladepo S and Michaelian K: Silica-coated super paramagnetic
iron oxide nanoparticles (SPION) as biocompatible contrast agent in
biomedical photoacoustics. Biomed Opt Express. 3:2500–2509. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun S, Zeng H, Robinson DB, Raoux S, Rice
PM, Wang SX and Li G: Monodisperse MFe2O4 (M
= Fe, Co, Mn) nanoparticles. J Am Chem Soc. 126:273–279. 2004.
|
|
26
|
Chen G, Chen W, Wu Z, Yuan R, Li H, Gao J
and Shuai X: MRI-visible polymeric vector bearing CD3 single chain
antibody for gene delivery to T cells for immunosuppression.
Biomaterials. 30:1962–1970. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tromsdorf UI, Bigall NC, Kaul MG, Bruns
OT, Nikolic MS, Mollwitz B, Sperling RA, Reimer R, Hohenberg H,
Parak WJ, Förster S, Beisiegel U, Adam G and Weller H: Size and
surface effects on the MRI relaxivity of manganese ferrite
nanoparticle contrast agents. Nano Lett. 7:2422–2427. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ito H, Miller SC, Billingham ME, Akimoto
H, Torti SV, Wade R, Gahlmann R, Lyons G, Kedes L and Torti FM:
Doxorubicin selectively inhibits muscle gene expression in cardiac
muscle cells in vivo and in vitro. Proc Natl Acad Sci USA.
87:4275–4279. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zwiorek K, Kloeckner J, Wagner E and
Coester C: Gelatin nanoparticles as a new and simple gene delivery
system. J Pharm Pharm Sci. 7:22–28. 2005.PubMed/NCBI
|
|
30
|
Medarova Z, Pham W, Farrar C, Petkova V
and Moore A: In vivo imaging of siRNA delivery and silencing in
tumors. Nat Med. 13:372–377. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Murnane JP: Telomere dysfunction and
chromosome instability. Mutat Res. 730:28–36. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ding Z, Wu CJ, Jaskelioff M, Ivanova E,
Kost-Alimova M, Protopopov A, Chu GC, Wang G, Lu X, Labrot ES, Hu
J, Wang W, Xiao Y, Zhang H, Zhang J, Zhang J, Gan B, Perry SR,
Jiang S, Li L, Horner JW, Wang YA, Chin L and DePinho RA:
Telomerase reactivation following telomere dysfunction yields
murine prostate tumors with bone metastases. Cell. 148:896–907.
2012. View Article : Google Scholar : PubMed/NCBI
|