Introduction
Crohn’s disease (CD), also identical to Crohn
syndrome or regional enteritis, refers to one form of inflammatory
bowel disease that may affect any part of the gastrointestinal
tract between the mouth and the anus, resulting in various symptoms
(1). In recent years the incidence
and prevalence rates of CD have rapidly increased, contributing
significantly to the burden on the health care system and
exhibiting high morbidity and mortality rates (2). The underlying pathogenesis of CD
remains unclear, but may result from interactions between
environmental, immunological and bacterial factors (3). Various countries and nationalities
have demonstrated different incidence rates of CD, which cannot be
explained by living habits or other risk factors, suggesting that
genetic polymorphisms may be crucial in the development of CD
(4,5). Recently, studies have indicated that
polymorphism of the immunity-related GTPase M (IRGM) gene is
associated with an increased CD risk (6,7).
The IRGM protein is an atypical member of the
interferon-inducible GTPase family, which is characteristically
induced by interferons and provides resistance to intracellular
pathogens (8). The human
IRGM gene is located on chromosome 5q33.1 and contains five
exons (9,10). Previous studies have shown that
IRGM may have a key function in the innate immune response by
regulating autophagy formation in response to intracellular
pathogens (11,12). Furthermore, certain studies have
demonstrated that autophagy is a potential pathogenic mechanism in
CD (13,14). Therefore, it was hypothesized that
single nucleotide polymorphisms (SNPs) in the IRGM gene may
be important in the development of CD (15,16).
Certain studies have indicated that a common polymorphism,
rs13361189 C>T, in the IRGM gene may increase the risk of
CD (17,18); however, individually published
studies provided inconclusive results (19,20).
Therefore, in the present study a meta-analysis of all eligible
case-control studies was conducted to evaluate the correlation
between the IRGM rs13361189 polymorphism and susceptibility
to CD.
Methods
Literature search
The PubMed, CISCOM, CINAHL, Web of Science, Google
Scholar, EBSCO, Cochrane Library and CBM databases were searched
from inception through to October 1, 2013 without the application
of any language restrictions. The following keywords and medical
subject headings were used: (‘SNP’ or ‘mutation’ or ‘genetic
polymorphism’ or ‘variation’ or ‘polymorphism’ or ‘single
nucleotide polymorphism’ or ‘variant’) and (‘Crohn’s disease’ or
‘CD’) and (‘human immunity-related GTPase M’ or ‘IRGM’). In
addition, a manual search was performed to obtain other potential
articles.
Selection criteria
In the present meta-analysis, studies were included
when the following criteria were met: i) The study design was a
clinical cohort or case-control study; ii) the study concerned the
correlation between the IRGM rs13361189 polymorphism and
susceptibility to CD; iii) the patients conformed to the diagnostic
criteria of CD; and iv) the study provided sufficient information
regarding the frequency of the IRGM rs13361189 polymorphism.
Studies that did not meet the inclusion criteria were excluded. The
most recent or the largest sample-size study was included when the
authors published several studies regarding the same subject
matter.
Data extraction
Using a standardized form, the relevant data were
systematically extracted from all the included studies by two
researchers. The standardized form included the following items:
Language of publication, publication year of article, first
author’s surname, geographical location, design of the study,
sample size, country of origin of the subjects, allele frequencies,
source of the samples, genotyping method of the SNPs and evidence
of Hardy-Weinberg equilibrium (HWE) in the healthy control
subjects.
Quality assessment
The methodological quality of the included studies
was evaluated according to the Newcastle-Ottawa Scale (NOS)
(21). The NOS criteria comprised:
i) Subject selection (scores, 0–4); ii) comparability of subjects
(scores, 0–2); and iii) clinical outcomes (scores, 0–3). The NOS
scores ranged between 0 and 9 and a score ≥7 indicated that a study
was of good quality.
Statistical analysis
The meta-analysis was performed using STATA 12.0
software (StataCorp, College Station, TX, USA). The relative risk
(RR) and the 95% confidence intervals (CI) were estimated. The Z
test was used to estimate the statistical significance of the RRs,
and the power calculations were conducted using power and sample
size calculations (22). Cochran’s
Q test and the I2 test were used to evaluate potential
heterogeneity between the studies (23). When the Q-test result was P<0.05
or the I2 test result was >50% this indicated
significant heterogeneity and the random-effect model was
conducted; otherwise, the fixed-effects model was used. Subgroup
and meta-regression analyses were conducted to investigate the
potential sources of heterogeneity. Sensitivity analysis was
performed by omitting each study in turn to evaluate the influence
of single studies on the overall estimation. Begg’s funnel plots
and Egger’s linear regression test were conducted to identify any
publication bias (24).
Results
Characteristics of the included
studies
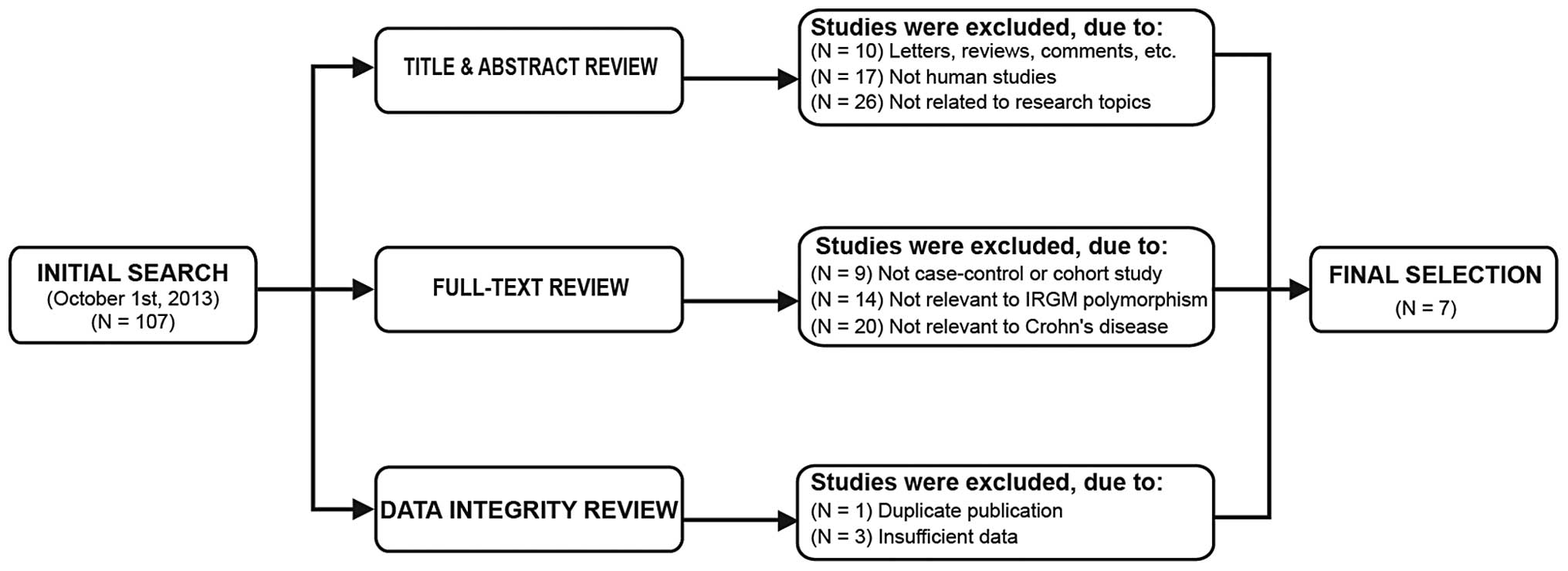
A total of 107 articles were initially identified
using the aforementioned keywords. The titles and abstracts of the
articles were reviewed and 53 articles were subsequently excluded;
the full texts and data integrity for the remaining articles were
reviewed and a further 47 studies were excluded. Finally, seven
case-control studies were included in the present meta-analysis
(17–20,25–27),
with publication years that ranged from 2008 to 2013. The selection
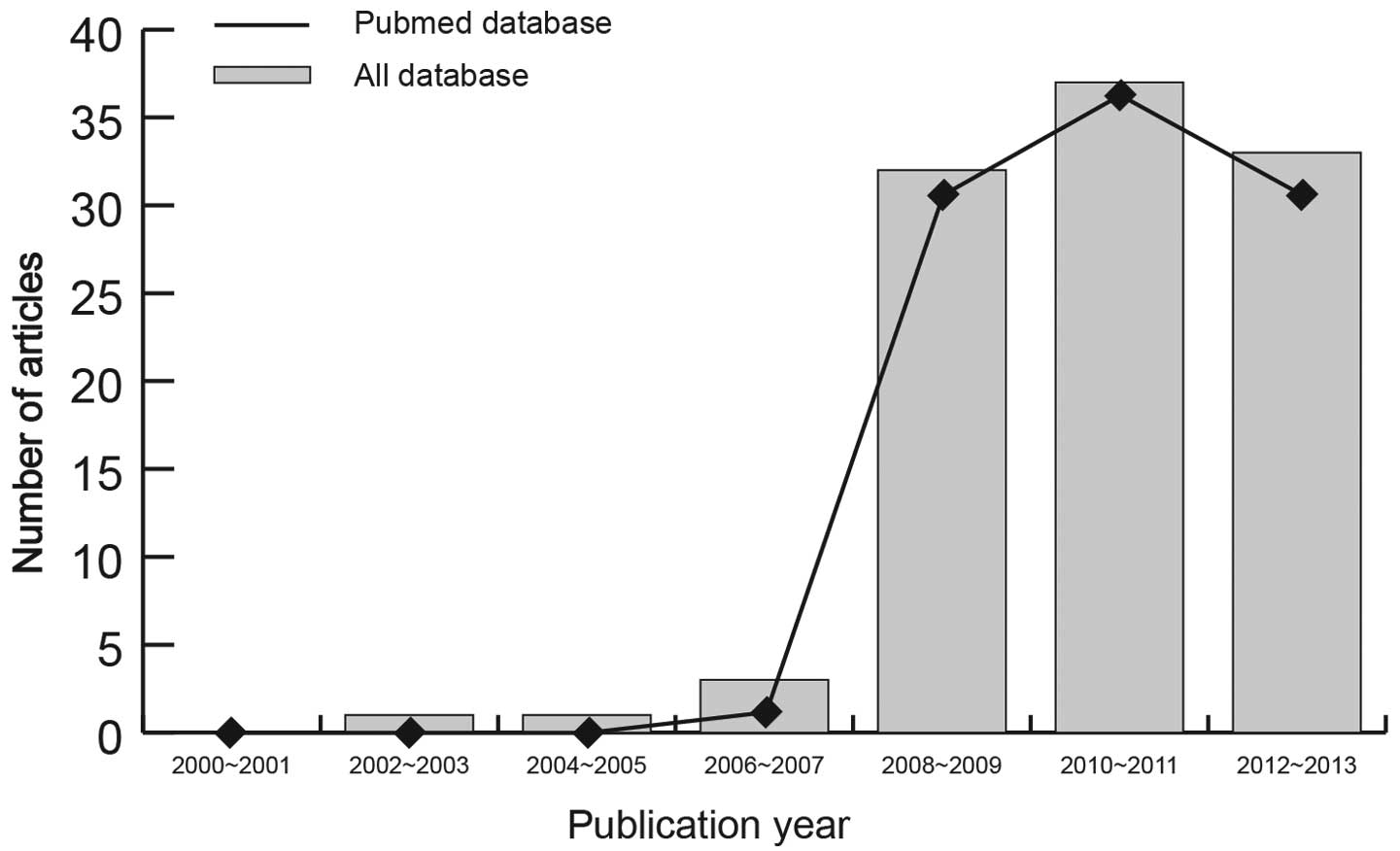
process of the eligible studies is shown in Fig. 1. The distribution of the number of
topic-related studies in the electronic databases during the last
decade is demonstrated in Fig. 2.
A total of 6,320 subjects were included in the meta-analysis, which
included 3,093 CD patients and 3,227 healthy control subjects. The
power values that were calculated for the sample size of the
included studies were >0.70. Six studies were conducted with
Caucasian populations, whereas only one study was performed with an
Asian population. The TaqMan® method was conducted in
five studies and the other two studies used polymerase chain
reaction-restriction fragment length polymorphism (PCR-RFLP) and
direct sequencing methods, respectively. The genotype frequencies
of the controls were all in HWE (P>0.05) and the NOS scores of
the included studies were >5 (moderate-to-high quality). The
study characteristics and methodological quality are summarized in
Table I.
 | Table IBaseline characteristics and
methodological quality of the included studies. |
Table I
Baseline characteristics and
methodological quality of the included studies.
First
author
(Ref.) | Year | Country | Ethnicity | Sample size | Gender (M/F) | Age (years) | Genotyping
method | HWE test
(P-value) | NOS score |
|---|
|
|
|
|---|
| Case | Control | Power | Case | Control | Case | Control |
|---|
Durães
(25) | 2013 | Portugal | Caucasian | 511 | 626 | 0.814 | 236/275 | 241/385 | 28.6±11.2 | 30.5 (9–83) | TaqMan | 0.356 | 8 |
Zheng
(19) | 2012 | China | Asian | 318 | 318 | 0.764 | 154/164 | 156/162 | 37.2±11.4 | 36.7±12.3 | Direct
sequencing | 0.142 | 8 |
Wang
(27) | 2012 | USA | Caucasian | 227 | 201 | 0.743 | 78/149 | 86/115 | 26.7±12.9 | - | TaqMan | 0.373 | 7 |
Prager
(26) | 2012 | Germany | Caucasian | 464 | 508 | 0.797 | 174/290 | 295/213 | 29.5±11.6 | 60.0±16.2 | TaqMan | 0.503 | 7 |
Wolfkamp
(18) | 2010 | Netherlands | Caucasian | 256 | 529 | 0.779 | - | - | - | - | PCR-RFLP | 0.239 | 6 |
Meggyesi
(20) | 2010 | Hungary | Caucasian | 810 | 469 | 0.828 | 434/376 | 251/218 | 37.1±12.6 | 40.5±11.5 | TaqMan | 0.743 | 8 |
Roberts
(17) | 2008 | New Zealand | Caucasian | 507 | 576 | 0.808 | - | 236/340 | - | - | TaqMan | 0.121 | 6 |
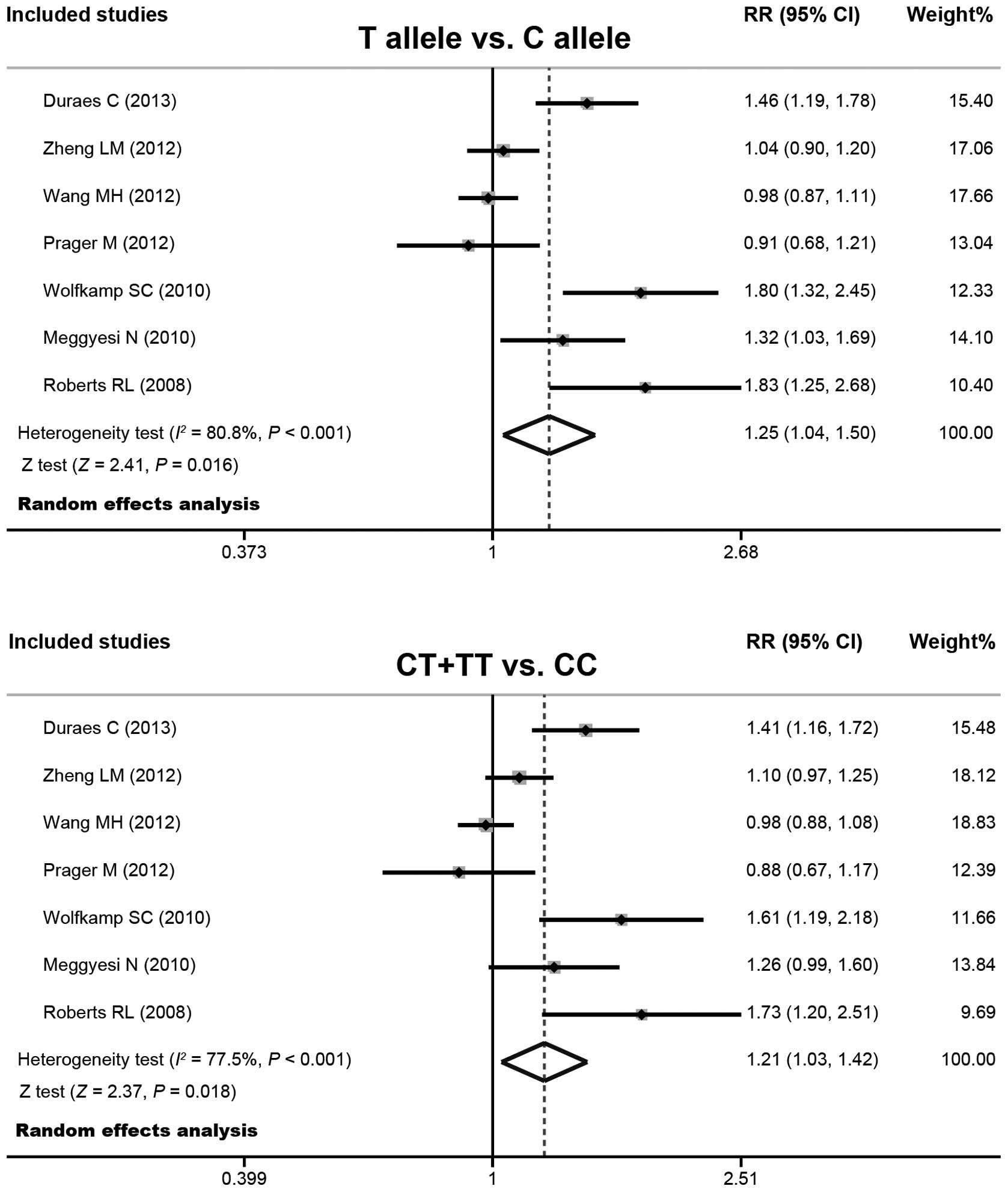
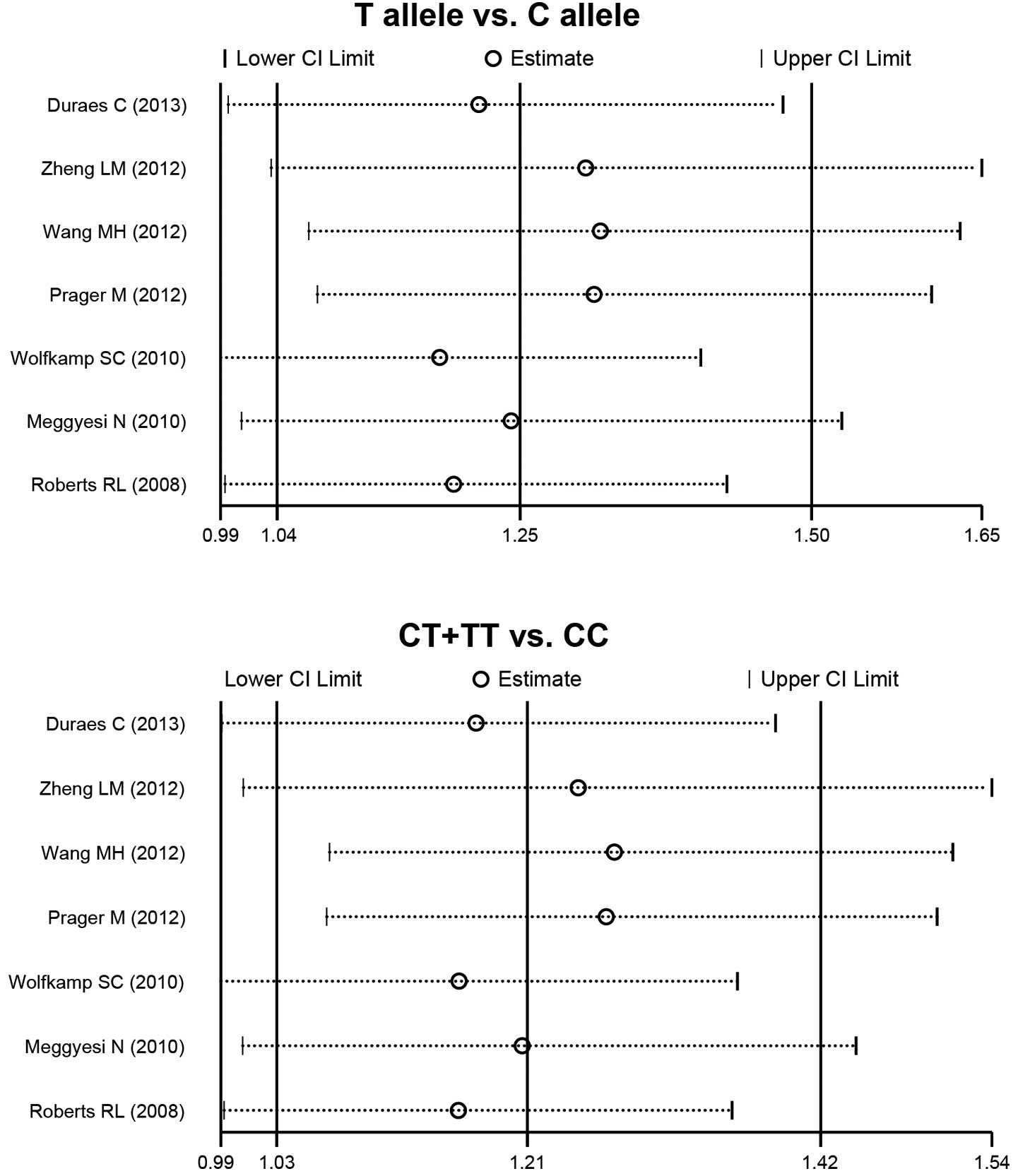
Quantitative data synthesis
The random effects model was conducted due to the
significant heterogeneity that existed between the studies. The
present meta-analysis results revealed that the IRGM
rs13361189 polymorphism correlates with an increased risk of CD (T
allele versus C allele: RR=1.25 with 95% CI, 1.04–1.50; P=0.016 and
CT + TT versus CC: RR=1.21 with 95% CI, 1.03–1.42; P=0.018;
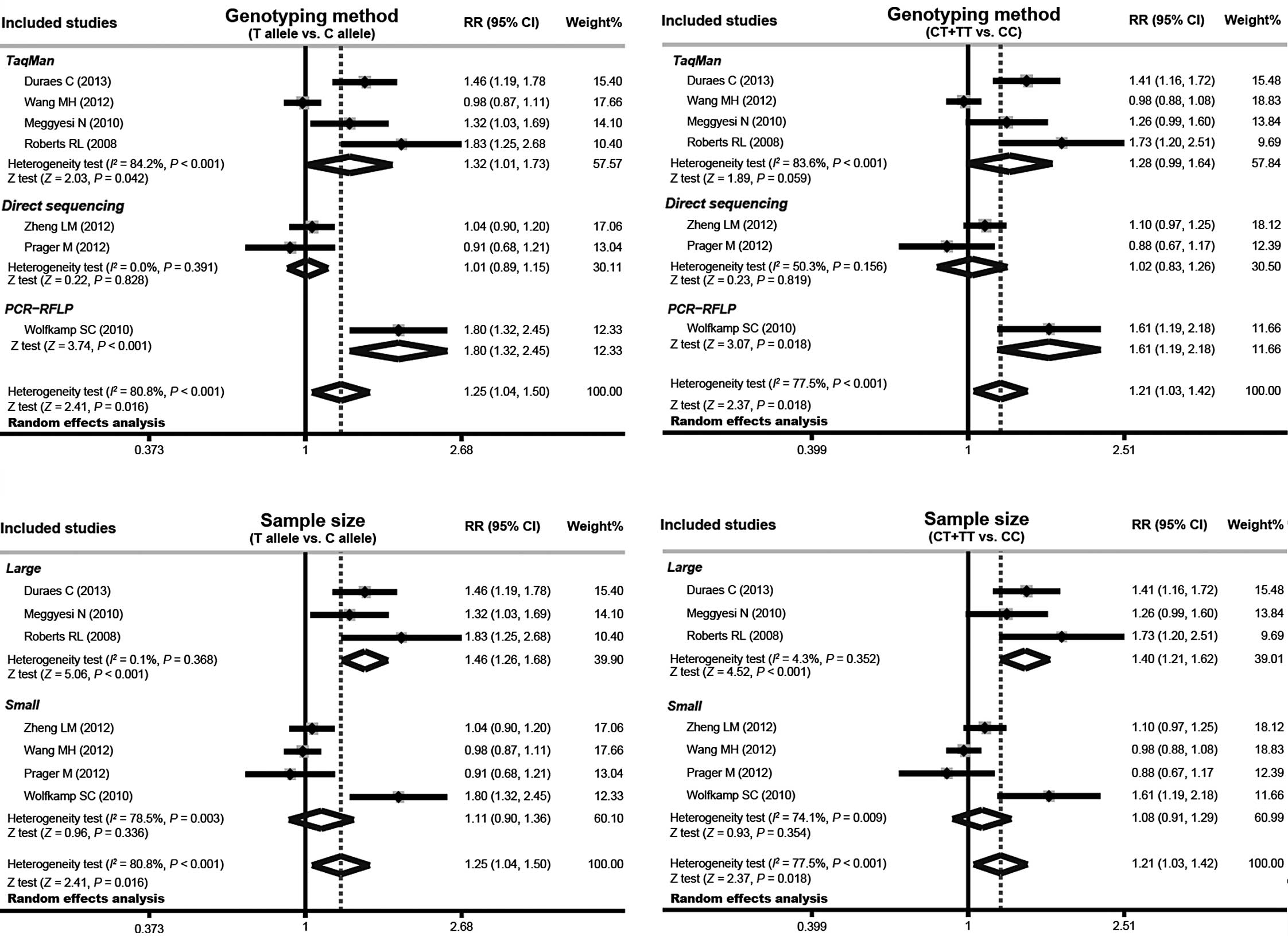
Fig. 3). The subgroup analysis
that was conducted using the genotyping method indicated that the
IRGM rs13361189 polymorphism was correlated with an
increased risk of CD in the TaqMan (T allele versus C allele:
RR=1.32 with 95% CI, 1.01–1.73; P=0.042) and PCR-RFLP subgroups (T
allele versus C allele: RR=1.80 with 95% CI, 1.32–2.45; P<0.001
and CT + TT versus CC: RR=1.61 with 95% CI, 1.19–2.18; P=0.018),
but not in the direct sequencing subgroup (P>0.05; Fig. 4). Further subgroup analysis by
sample size demonstrated significant correlations between the
IRGM rs13361189 polymorphism and an increased risk of CD in
the large sample-size subgroup (T allele versus C allele: RR=1.46
with 95% CI, 1.26–1.68; P<0.001 and CT + TT versus CC: RR=1.40
with 95% CI, 1.21–1.62; P<0.001). However, no correlation was
identified between the IRGM rs13361189 polymorphism and CD
risk in the small sample-size subgroup (P>0.05).
The results of the sensitivity analysis indicated
that no single study influenced the overall pooled odds ratio
(Fig. 5). Univariate and
multivariate meta-regression analyses showed that sample size may
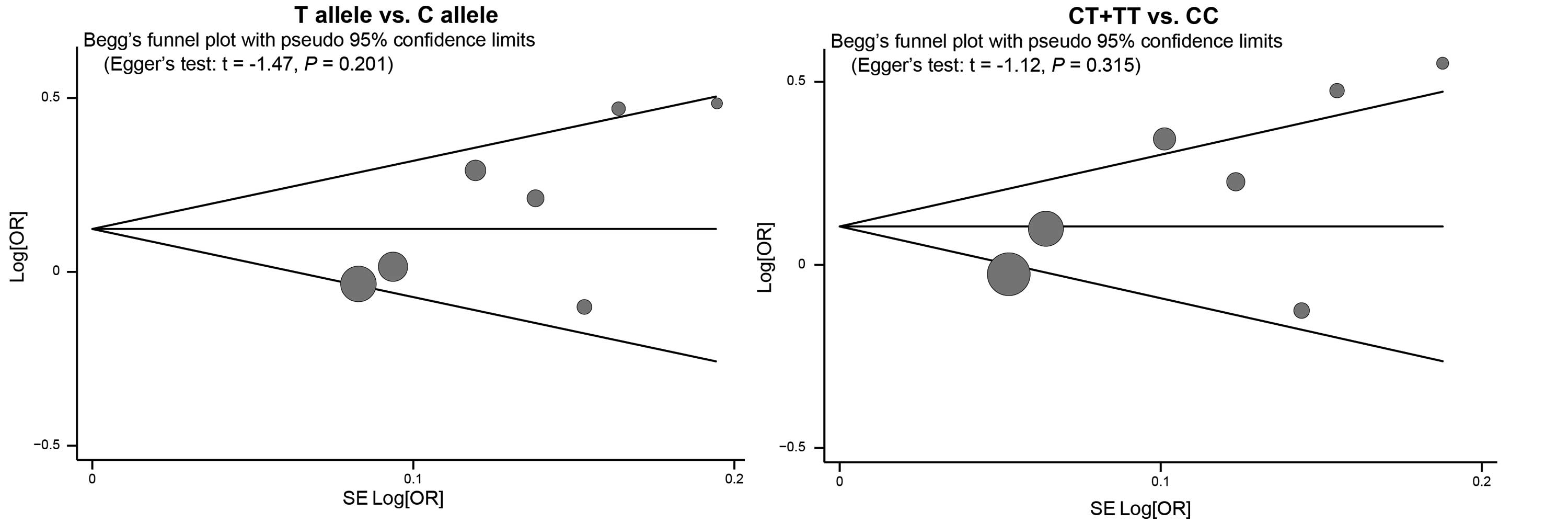
be a predominant source of heterogeneity (P=0.003; Table II). There was no evidence of
obvious asymmetry in the Begg’s funnel plots (Fig. 6) and the Egger’s test did not
display strong statistical evidence for publication bias (T allele
versus C allele: t=−1.47, P=0.201 and CT + TT versus CC: t=−1.12,
P=0.315).
 | Table IIUnivariate and multivariate
meta-regression analysis of potential source of heterogeneity. |
Table II
Univariate and multivariate
meta-regression analysis of potential source of heterogeneity.
| | | | | 95% CI |
|---|
| | | | |
|
|---|
| Heterogeneity
factor | Coefficient | SE | Z-value | P-value | LL | UL |
|---|
| Publication
year |
| Univariate | −0.589 | 0.039 | −1.49 | 0.137 | −0.136 | 0.019 |
| Multivariate | −0.062 | 0.066 | −0.94 | 0.348 | −0.192 | 0.068 |
| Genotyping
method |
| Univariate | 0.053 | 0.150 | 0.35 | 0.724 | −0.240 | 0.346 |
| Multivariate | 0.130 | 0.227 | 0.57 | 0.567 | −0.315 | 0.575 |
| Sample size |
| Univariate | −0.258 | 0.140 | −1.84 | 0.065 | −0.532 | 0.016 |
| Multivariate | −0.534 | 0.182 | −2.94 | 0.003 | −0.890 | −0.177 |
Discussion
IRGM is responsible for the innate immune response
via regulation of autophagy formation in response to intracellular
pathogens (28). Cell homeostasis
is dependent on the biosynthesis of macromolecules and the balance
between catabolism and autophagy (29). In addition, autophagy is considered
to be a key regulatory mechanism in the update, development and
differentiation of cellular components and tissue remodeling
(30). However, the interfered
autophagy process may result in failure of the timely removal of
injured cell structures, aging organelles, abandoned biological
macromolecules and intracellular bacteria, which may trigger
inappropriate immune responses, thus leading to the pathogenesis of
chronic intestinal inflammation (31,32).
Therefore, IRGM genetic polymorphisms that influence the
normal expression of IRGM have been indicated to be associated with
an increased risk of CD (7,16).
In the present meta-analysis, the correlation
between the IRGM rs13361189 polymorphism and susceptibility
to CD was evaluated. Seven independent case-control studies were
included with a total of 3,093 CD patients and 3,227 healthy
control subjects. The meta-analysis results demonstrated that the
IRGM rs13361189 polymorphism correlates with an increased
risk of CD, indicating that rs13361189 may be a causative factor
for the incidence of CD. Although the exact function of the
IRGM genetic polymorphism in CD is not fully understood, a
potential explanation is that the IRGM genetic polymorphism
may alter the function of IRGM, leading to the invasion of bacteria
in vivo, resulting in tissue damage and chronic intestinal
inflammation (33). As
heterogeneity was clearly identified, stratified analyses based on
the genotyping method and sample size were performed. The subgroup
analysis using the genotyping method showed significant
correlations between the IRGM rs13361189 polymorphism and an
increased risk of CD in the TaqMan® and the PCR-RFLP
subgroups. However, no correlation was identified in the direct
sequencing subgroup; this result was unreliable due to the small
sample size. Further subgroup analysis by sample size showed a
significant correlation between the IRGM rs13361189
polymorphism and an increased risk of CD in the subgroup with a
large sample size. However, no correlation between the IRGM
rs13361189 polymorphism and CD risk was identified in the subgroup
with a small sample size. These results indicated that sample size
may be a potential source of heterogeneity. The findings were
predominantly consistent with previous studies, which demonstrated
that the IRGM rs13361189 polymorphism may be strongly
associated with the development and progression of CD, indicating
that the IRGM rs13361189 polymorphism may be utilized as a
biomarker for the early diagnosis of CD.
However, the present meta-analysis had certain
limitations. Firstly, the results may not provide sufficient
statistical power to estimate the correlations between the
IRGM rs13361189 polymorphism and CD risk due to the
relatively small sample size. Secondly, meta-analysis is a type of
retrospective study that may lead to subject selection bias,
thereby affecting the reliability of the results. Thirdly, the
present meta-analysis failed to obtain the original data from the
included studies, which may have further limited the evaluation of
the potential roles of the IRGM genetic polymorphism in the
development of CD. Furthermore, the inclusion criteria of the cases
and controls were not well defined in all the included studies,
which may have influenced the results.
In conclusion, the present meta-analysis indicated
that the IRGM rs13361189 polymorphism may contribute to
susceptibility to CD. Thus, the IRGM rs13361189 polymorphism
may be a potential biomarker for the early diagnosis of CD.
However, due to the abovementioned limitations, additional detailed
studies are required to confirm these findings.
Acknowledgements
The authors would like to acknowledge the reviewers
for their helpful comments regarding this meta-analysis and would
also like to thank their colleagues at the Department of Anus and
Intestines of The Fourth Affiliated Hospital of China Medical
University.
References
|
1
|
Frank DN, Robertson CE, Hamm CM, et al:
Disease phenotype and genotype are associated with shifts in
intestinal-associated microbiota in inflammatory bowel diseases.
Inflamm Bowel Dis. 17:179–184. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zheng JJ, Shi XH, Zhu XS, et al: A
comparative study of incidence and prevalence of Crohn’s disease in
mainland China in different periods. Zhonghua Nei Ke Za Zhi.
50:597–600. 2011.(In Chinese).
|
|
3
|
Prescott NJ, Dominy KM, Kubo M, et al:
Independent and population-specific association of risk variants at
the IRGM locus with Crohn’s disease. Hum Mol Genet. 19:1828–1839.
2010.PubMed/NCBI
|
|
4
|
Sivaram G, Tiwari SK, Bardia A, et al:
Macrophage migration inhibitory factor, Toll-like receptor 4, and
CD14 polymorphisms with altered expression levels in patients with
ulcerative colitis. Hum Immunol. 73:201–205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Joossens M, Van Steen K, Branche J, et al:
Familial aggregation and antimicrobial response dose-dependently
affect the risk for Crohn’s disease. Inflamm Bowel Dis. 16:58–67.
2010.PubMed/NCBI
|
|
6
|
Brest P, Lapaquette P, Mograbi B, et al:
Risk predisposition for Crohn disease: a ‘ménage à trois’ combining
IRGM allele, miRNA and xenophagy. Autophagy. 7:786–787. 2011.
|
|
7
|
Glas J, Seiderer J, Bues S, et al: IRGM
variants and susceptibility to inflammatory bowel disease in the
German population. PLoS One. 8:e543382013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Taylor GA: IRG proteins: key mediators of
interferon-regulated host resistance to intracellular pathogens.
Cell Microbiol. 9:1099–1107. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Moon CM, Shin DJ, Kim SW, et al:
Associations between genetic variants in the IRGM gene and
inflammatory bowel diseases in the Korean population. Inflamm Bowel
Dis. 19:106–114. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nord H: Application of genomic and
expression arrays for identification of new cancer genes.
Dissertation. Uppsala University. Acta Universitatis Upsaliensis;
ISSN: 1651-6206Uppsala, Sweden: 2010
|
|
11
|
Mizushima N, Levine B, Cuervo AM and
Klionsky DJ: Autophagy fights disease through cellular
self-digestion. Nature. 451:1069–1075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Strausberg RL, Feingold EA, Grouse LH, et
al; Mammalian Gene Collection Program Team. Generation and initial
analysis of more than 15,000 full-length human and mouse cDNA
sequences. Proc Natl Acad Sci U S A. 99:16899–16903. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Deretic V: Autophagy, an immunologic magic
bullet: Mycobacterium tuberculosis phagosome maturation block and
how to bypass it. Future Microbiol. 3:517–524. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Eskelinen EL and Saftig P: Autophagy: a
lysosomal degradation pathway with a central role in health and
disease. Biochim Biophys Acta. 1793:664–673. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xavier RJ, Huett A and Rioux JD: Autophagy
as an important process in gut homeostasis and Crohn’s disease
pathogenesis. Gut. 57:717–720. 2008.PubMed/NCBI
|
|
16
|
Palomino-Morales RJ, Oliver J,
Gómez-García M, et al: Association of ATG16L1 and IRGM genes
polymorphisms with inflammatory bowel disease: a meta-analysis
approach. Genes Immun. 10:356–364. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Roberts RL, Hollis-Moffatt JE, Gearry RB,
et al: Confirmation of association of IRGM and NCF4 with ileal
Crohn’s disease in a population-based cohort. Genes Immun.
9:561–565. 2008.PubMed/NCBI
|
|
18
|
Wolfkamp SC, Te Velde AA, Weersma RK, et
al: Is there a role for Crohn’s disease-associated autophagy genes
ATG16L1 and IRGM in formation of granulomas? Eur J Gastroenterol
Hepatol. 22:933–937. 2010.
|
|
19
|
Zheng LM and Pang Z: Association of IRGM
and ATG16L1 gene polymorphisms with Crohn’s disease in the Chinese
Han population. Chin J Gastroenterol Hepatol. 21:437–440. 2012.(In
Chinese).
|
|
20
|
Meggyesi N, Kiss LS, Koszarska M, et al:
NKX2–3 and IRGM variants are associated with disease susceptibility
to IBD in Eastern European patients. World J Gastroenterol.
16:5233–5240. 2010.
|
|
21
|
Stang A: Critical evaluation of the
Newcastle-Ottawa scale for the assessment of the quality of
nonrandomized studies in meta-analyses. Eur J Epidemiol.
25:603–605. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dupont WD and Plummer WD Jr: Power and
sample size calculations. A review and computer program. Control
Clin Trials. 11:116–128. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zintzaras E and Ioannidis JP: HEGESMA:
genome search meta-analysis and heterogeneity testing.
Bioinformatics. 21:3672–3673. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Peters JL, Sutton AJ, Jones DR, et al:
Comparison of two methods to detect publication bias in
meta-analysis. JAMA. 295:676–680. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Durães C, Machado JC, Portela F, et al:
Phenotype-genotype profiles in Crohn’s disease predicted by genetic
markers in autophagy-related genes (GOIA study II). Inflamm Bowel
Dis. 19:230–239. 2013.PubMed/NCBI
|
|
26
|
Prager M, Büttner J, Haas V, et al: The
JAK2 variant rs10758669 in Crohn’s disease: altering the intestinal
barrier as one mechanism of action. Int J Colorectal Dis.
27:565–573. 2012.
|
|
27
|
Wang MH, Okazaki T, Kugathasan S, et al:
Contribution of higher risk genes and European admixture to Crohn’s
disease in African Americans. Inflamm Bowel Dis. 18:2277–2287.
2012.PubMed/NCBI
|
|
28
|
Deretic V: Autophagy as an innate immunity
paradigm: expanding the scope and repertoire of pattern recognition
receptors. Curr Opin Immunol. 24:21–31. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Folmes CD, Dzeja PP, Nelson TJ and Terzic
A: Metabolic plasticity in stem cell homeostasis and
differentiation. Cell Stem Cell. 11:596–606. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
He C and Klionsky DJ: Regulation
mechanisms and signaling pathways of autophagy. Annu Rev Genet.
43:67–93. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Caramés B, Taniguchi N, Otsuki S, et al:
Autophagy is a protective mechanism in normal cartilage, and its
aging-related loss is linked with cell death and osteoarthritis.
Arthritis Rheum. 62:791–801. 2010.PubMed/NCBI
|
|
32
|
Deretic V, Saitoh T and Akira S: Autophagy
in infection, inflammation and immunity. Nat Rev Immunol.
13:722–737. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
McCarroll SA, Huett A, Kuballa P, et al:
Deletion polymorphism upstream of IRGM associated with altered IRGM
expression and Crohn’s disease. Nat Genet. 40:1107–1112. 2008.
|