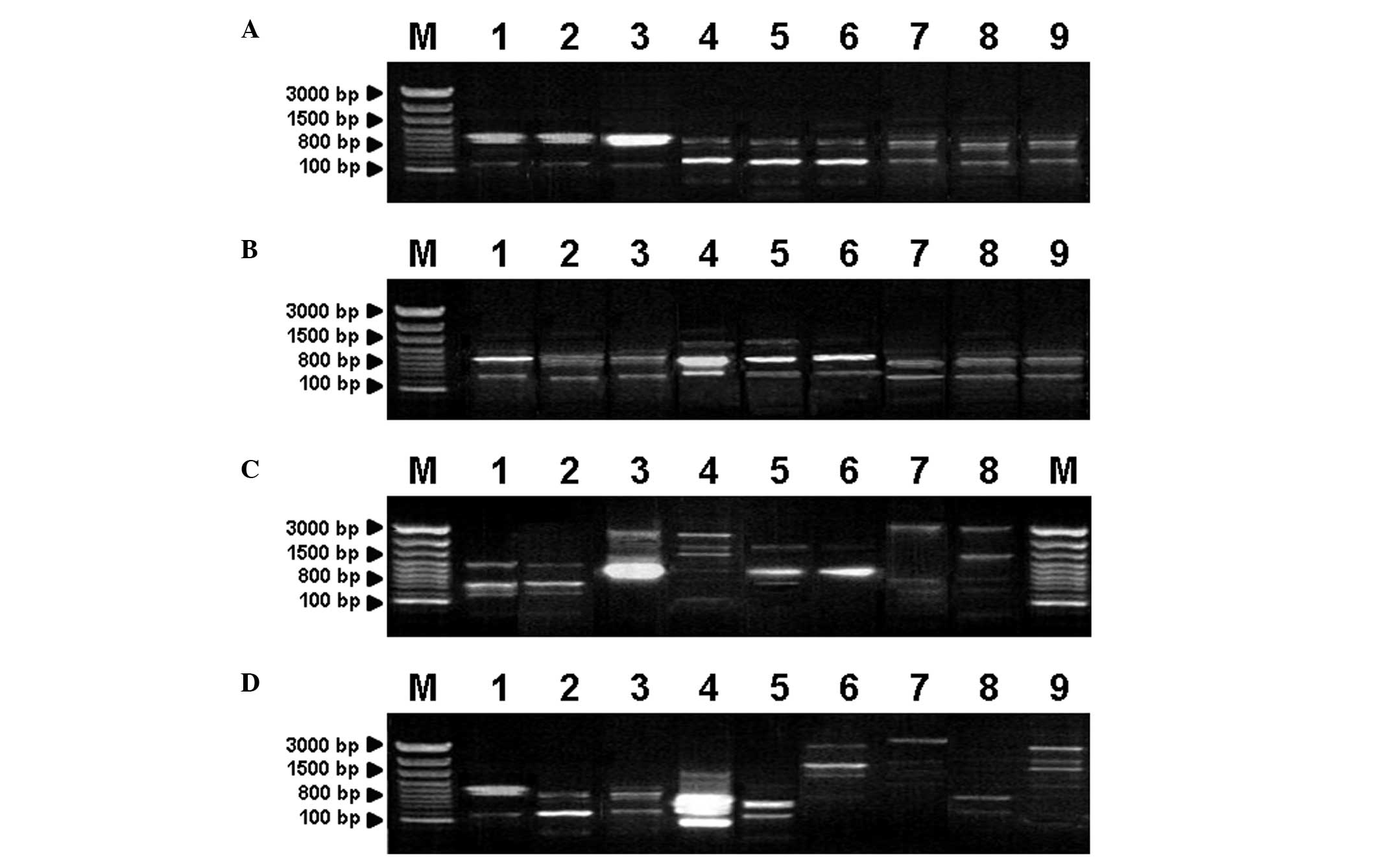
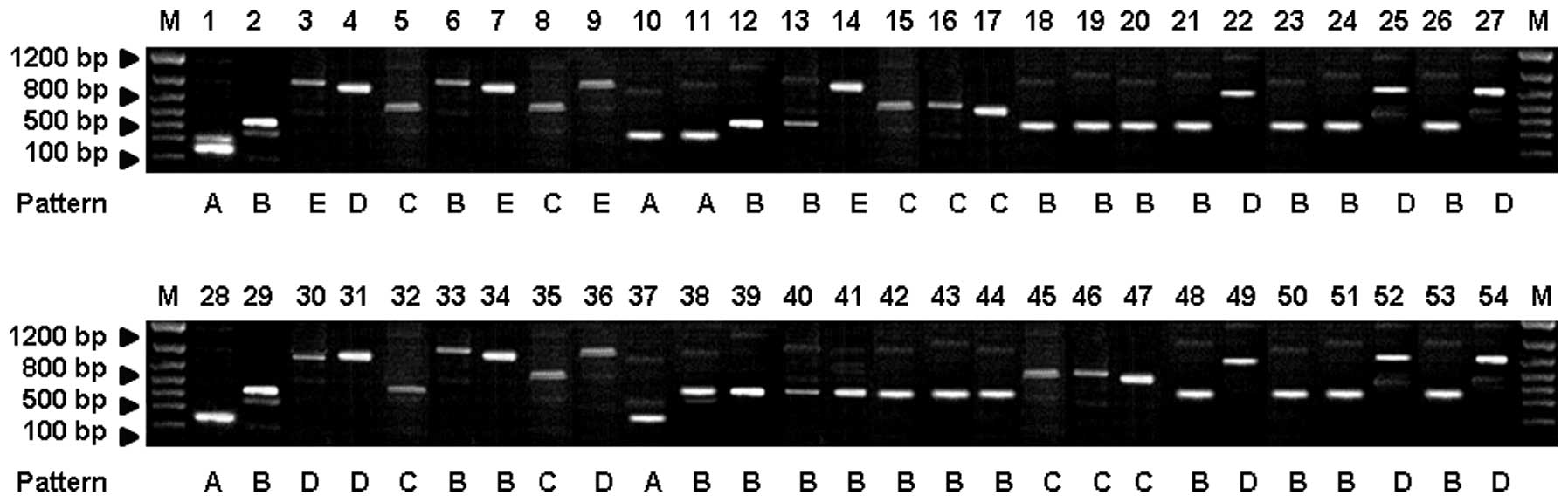
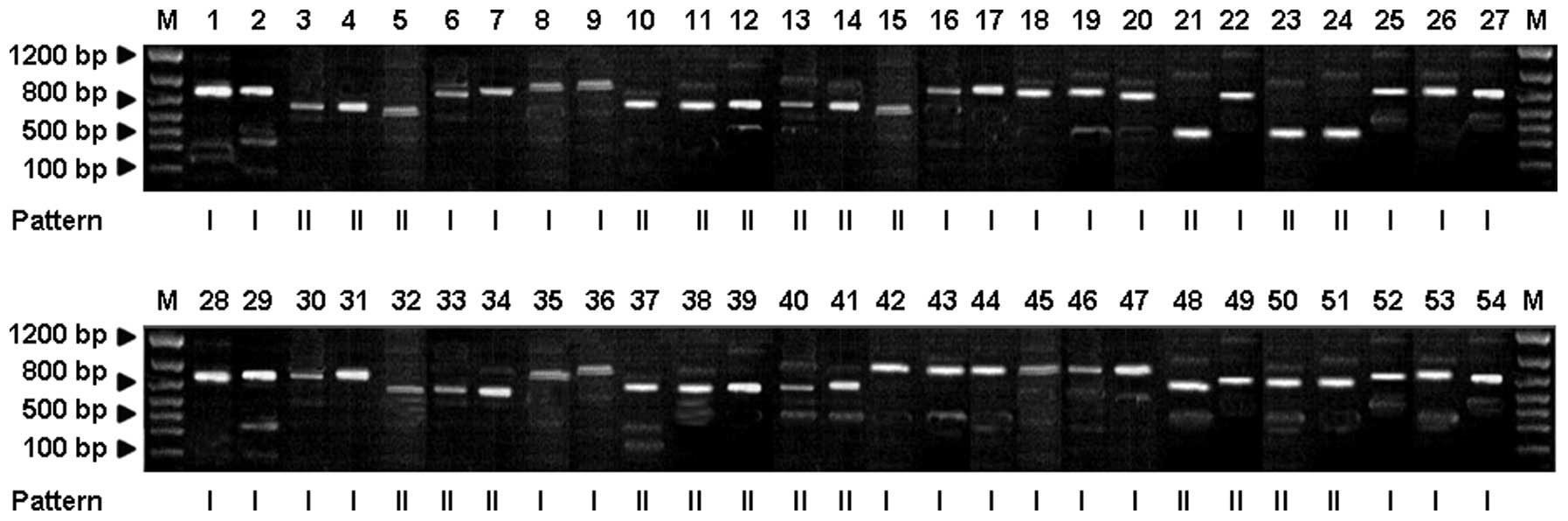
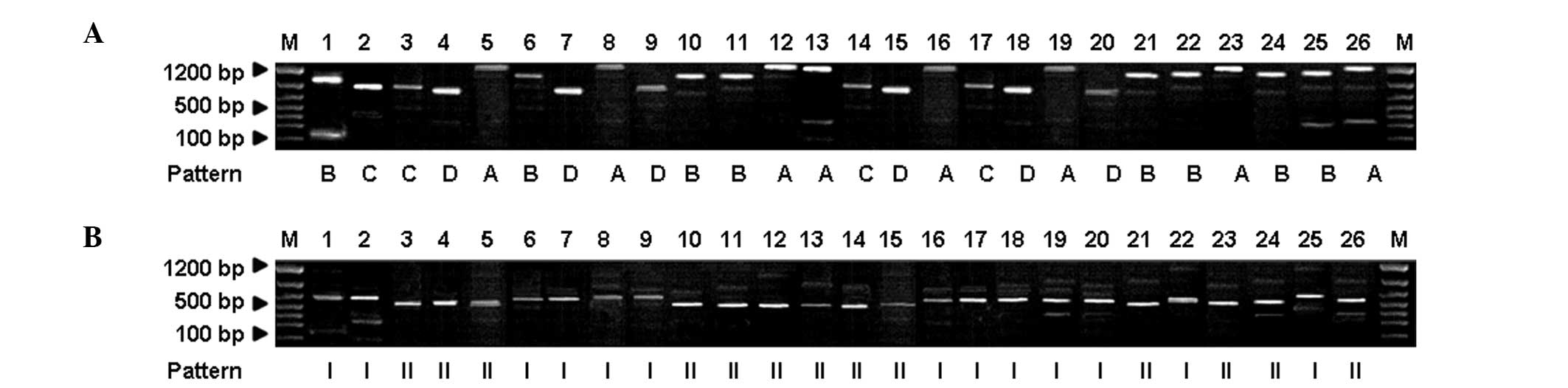
|
1
|
Moretti A, Agnetti F, Mancianti F, et al:
Dermatophytosis in animals: epidemiological, clinical and zoonotic
aspects. G Ital Dermatol Venereol. 148:563–572. 2013.PubMed/NCBI
|
|
2
|
Gaedigk A, Gaedigk R and Abdel-Rahman SM:
Genetic heterogeneity in the rRNA gene locus of Trichophyton
tonsurans. J Clin Microbiol. 41:5478–5487. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kac G: Molecular approaches to the study
of dermatophytes. Med Mycol. 38:329–336. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shehata AS, Mukherjee PK, Aboulatta HN, et
al: Single-step PCR using (GACA)4 primer: utility for
rapid identification of dermatophyte species and strains. J Clin
Microbiol. 46:2641–2645. 2008.PubMed/NCBI
|
|
5
|
Zhu H, Wen H and Liao W: Identification of
Trichophyton rubrum by PCR fingerprinting. Chin Med J
(Engl). 115:1218–1220. 2002.
|
|
6
|
Orsini M and Romano-Spica V: A
microwave-based method for nucleic acid isolation from
environmental samples. Lett Appl Microbiol. 33:17–20. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cassago A, Panepucci R, Baião A and
Henrique-Silva F: Cellophane based mini-prep method for DNA
extraction from the filamentous fungus Trichoderma reesei.
BMC Microbiol. 2:142002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kac G, Bougnoux ME, Feuilhade De Chauvin
M, Sene S and Derouin F: Genetic diversity among Trichophyton
mentagrophytes isolates using random amplified polymorphic DNA
method. Br J Dermatol. 140:839–844. 1999.
|
|
9
|
Summerbell RC, Haugland RA, Li A and Gupta
AK: rRNA gene internal transcribed spacer 1 and 2 sequences of
asexual, anthropophilic dermatophytes related to Trichophyton
rubrum. J Clin Microbiol. 37:4005–4011. 1999.PubMed/NCBI
|
|
10
|
Makimura K, Mochizuki T, Hasegawa A, et
al: Phylogenetic classification of Trichophyton
mentagrophytes complex strains based on DNA sequences of
nuclear ribosomal internal transcribed spacer 1 regions. J Clin
Microbiol. 36:2629–2633. 1998.
|
|
11
|
Apodaca G and McKerrow JH: Regulation of
Trichophyton rubrum proteolytic activity. Infect Immun.
57:3081–3090. 1989.PubMed/NCBI
|
|
12
|
Jackson CJ, Barton RC, Kelly SL and Evans
EG: Strain identification of Trichophyton rubrum by specific
amplification of subrepeat elements in the ribosomal DNA
nontranscribed spacer. J Clin Microbiol. 38:4527–4534. 2000.
|
|
13
|
Mochizuki T, Kawasaki M, Ishizaki H, et
al: Molecular epidemiology of Arthroderma benhamiae, an
emerging pathogen of dermatophytoses in Japan, by polymorphisms of
the non-transcribed spacer region of the ribosomal DNA. J Dermatol
Sci. 27:14–20. 2001.
|
|
14
|
Zhu HM, Liao WQ and Dai JX: PCR
fingerprint analysis of Trichophyton rubrum and
Trichophyton mentagrophytes. Chin J Dermatol. 33:3492000.(In
Chinese).
|
|
15
|
Shin JH, Sung JH, Park SJ, et al: Species
identification and strain differentiation of dermatophyte fungi
using polymerase chain reaction amplification and restriction
enzyme analysis. J Am Acad Dermatol. 48:857–865. 2003. View Article : Google Scholar
|
|
16
|
Li Q, Liu WD, Yang GL and Li CJ: DNA
typing of Trichophyton rubrum. Chin J Dermatol. 35:352–354.
2002.(In Chinese).
|
|
17
|
Jackson CJ, Mochizuki T and Barton RC: PCR
fingerprinting of Trichophyton mentagrophytes var.
interdigitale using polymorphic subrepeat loci in the rDNA
nontranscribed spacer. J Med Microbiol. 55:1349–1355. 2006.
|
|
18
|
Takeda K, Nishibu A, Anzawa K and
Mochizuki T: Molecular epidemiology of a major subgroup of
Arthroderma benhamiae isolated in Japan by restriction
fragment length polymorphism analysis of the non-transcribed spacer
region of ribosomal RNA gene. Jpn J Infect Dis. 65:233–239.
2012.PubMed/NCBI
|
|
19
|
Fan JF, Li HJ, Suo JJ, Li RY and Wan Z:
Strain typing in Trichophyton rubrum. Academic Journal of Chinese
Pla Medical School. 25:225–226. 2004.(In Chinese).
|
|
20
|
Nishio K, Kawasaki M and Ishizaki H:
Phylogeny of the genera Trichophyton using mitochondrial DNA
analysis. Mycopathologia. 117:127–132. 1992.
|
|
21
|
Jackson CJ, Barton RC and Evans EG:
Species identification and strain differentiation of dermatophyte
fungi by analysis of ribosomal-DNA intergenic spacer regions. J
Clin Microbiol. 37:931–936. 1999.PubMed/NCBI
|