1. Introduction
Schizophrenia is a common mental
disorder
Schizophrenia is a severe psychiatric disorder that
has been found to have a lifetime prevalence of ~1% in a number of
population studies (1–3). The condition is characterized by
impaired cognition, positive psychotic symptoms, including
hallucinations, delusions and disorganized behavior, and negative
symptoms, such as social withdrawal and apathy (2). Schizophrenia is a ‘complex genetic
disease’, with an etiology involving multiple genetic and
environmental factors. The genetic contribution is significant,
since schizophrenia has been shown to have a heritability risk of
~80%, with the risk decreasing by ~50% for each degree of family
relation (3).
microRNAs (miRNA) are the unseen
regulators of gene expression
miRNAs are a class of small RNAs found only in
eukaryotes, which were first identified less than two decades ago.
Modulation of gene expression does not rely solely on regulatory
proteins. miRNAs are single-stranded RNA molecules, consisting of
21–23 nucleotides, that are not translated into proteins, but are
key to the complex cell machinery responsible for gene expression
(4). A previous study estimated
that one third of human genes are directly targeted by miRNAs
(5). In an attempt to clone the
gene responsible for the lin-4 phenotype in roundworms, Lee et
al identified miRNAs for the first time as small non-coding RNA
molecules (6). To date, 2,652
mature human miRNAs have been identified (www.mirbase.org; accessed 20, June 2013).
The biogenesis of miRNAs is initiated by
transcription from intergenic or intron genomic regions into
primary-miRNA molecules (pri-miRNAs). Pri-miRNAs are cleaved inside
the nucleus by the components of the microprocessor complex,
consisting of Drosha and DGCR8, to generate RNA hairpins, known as
precursor-miRNA molecules (pre-miRNAs). Pre-miRNA is exported into
the cytoplasm and is further cleaved by the RNaseIII enzyme, Dicer;
therefore, a duplex of two miRNA strands is formed. Next, the miRNA
duplex is unwound and one of the strands is incorporated into a
large miRNA-induced silencing complex, which participates in the
detection and binding to the 3′-untranslated region of messenger
RNAs (mRNAs). As a result, translation is inhibited. However, the
underlying mechanisms are yet to be fully understood (Fig. 1) (7).
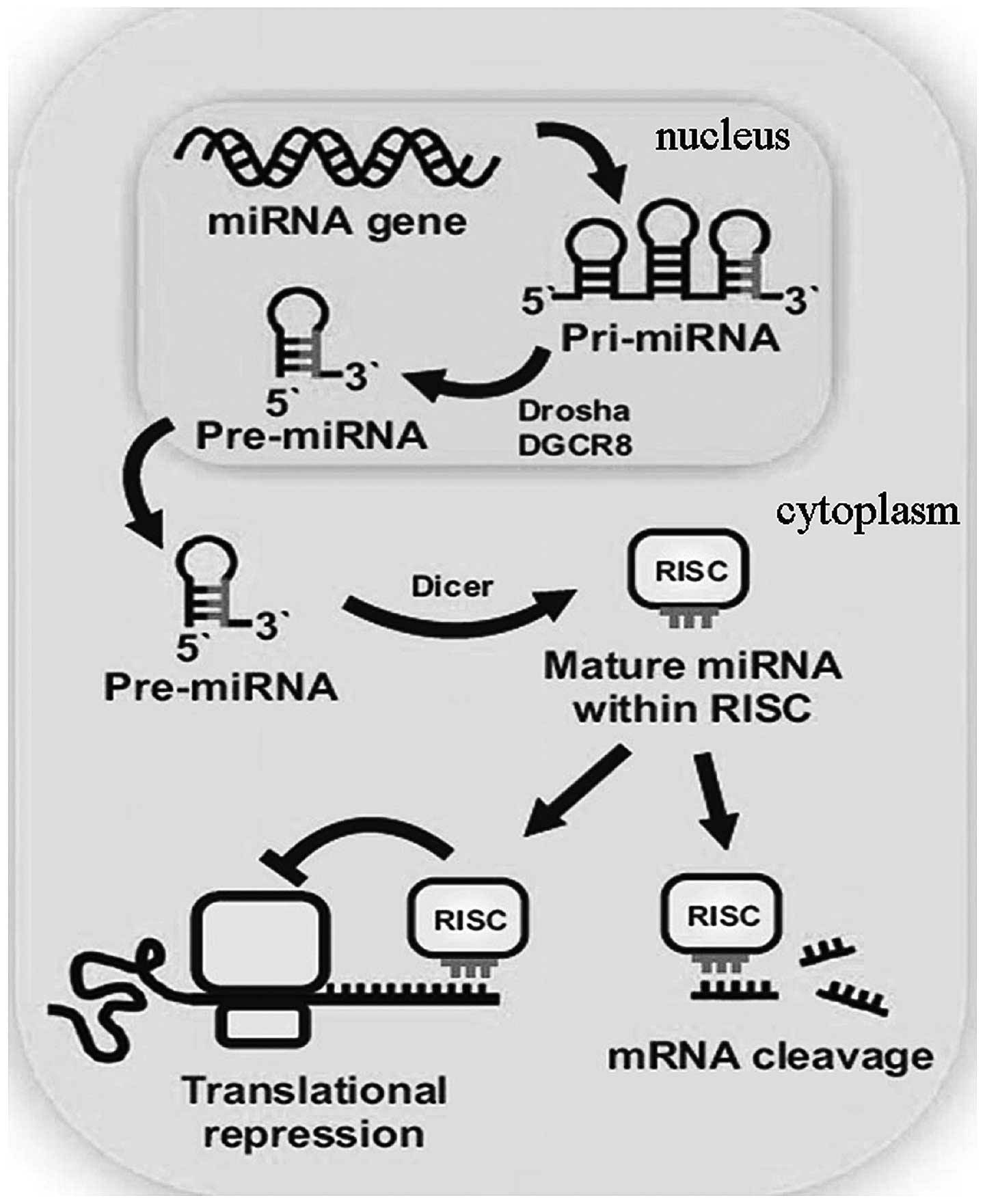 | Figure 1Biogenesis of miRNA [adapted from
Bravo and Dinan, 2011 (3)]. Mature
miRNA is much smaller compared with the gene involved in encoding
the molecule. Pri-miRNA has a hairpin structure with a poly(A) tail
and cap. The hairpin molecule is processed by the nuclease, Drosha,
and the RNA-binding protein, DGCR8 (known as Pasha in
invertebrates), and the resulting molecule is known as pre-miRNA.
The small pre-miRNA passes from the nucleus into the cytosol and is
processed by the endonuclease, Dicer. Mature miRNA forms part of
the RISC and the complex provides the gene silencing capacity of
miRNA. miRNA, microRNA; pri-miRNA, primary-miRNA; pre-miRNA,
precursor-miRNA; RISC, RNA-induced silencing complex. |
Genetic basis of miRNA abnormalities in
schizophrenia
A hemizygous deletion of a 1.5–3-Mb region of
chromosome 22 can lead to the 22q11 deletion syndrome (22q11DS),
which is characterized by multiple physical and psychiatric
abnormalities. A previous study determined that ~30% of 22q11DS
patients may develop schizophrenia (8). miR-25 and miR-185 are regulators of
the sarco/endoplasmic reticulum Ca2+ ATPase (SERCA2),
which is responsible for loading Ca2+ into the
endoplasmic reticulum. Earls et al found that miR-25 and
miR-185 were depleted in mouse models of 22q11DS and restoration of
these miRNAs to presynaptic neurons rescued the long-term
potentiation of DGCR8+/− mice (1). The authors concluded that
miRNA-dependent SERCA2 dysregulation is a pathogenic event in
22q11DS and schizophrenia.
2. miRNAs and schizophrenia
miRNA function in the nervous system
The intricate architecture of the nervous system and
the ability of the neurons for postsynaptic remodeling requires the
coordination of complex intracellular networks consisting of
molecular signal transduction systems. Due to the abundance of
neural networks, gene variants are able to cause system
dysfunctions, manifesting as associated neurobehavioral syndromes.
Previous studies revealed that post-transcriptional gene regulation
by miRNA is an important factor shaping the topography of the
neural networks. Over half of miRNAs identified have been shown to
be highly or exclusively expressed in the brain, a number of which
have been implicated in important aspects of neuronal function
(9). miR-124 and miR-9 play a
crucial role in neurogenesis; overexpression of these miRNAs
decreases the number of astrocytes, whereas inhibition of these
miRNAs reduces the number of neurons (10). miR-134 was found to regulate the
size of dendritic spines and enrich the synapse dendritic region of
rat hippocampal neurons (11). In
addition, miR-134 regulates the translation of Limk1, which is a
protein kinase that affects dendritic spine morphology via the
regulation of actin filaments (5).
Dysregulation of miRNA in
schizophrenia
miRNA has received increasing attention in genetic
studies of schizophrenia. A number of studies support the
hypothesis that miRNA plays an important role not only in human
brain development, but also in brain diseases (12–16).
Hunsberger et al hypothesized that miRNA may serve as a
unifying link among the structural developmental anomalies,
neurotransmitter alterations and response to treatment in
schizophrenia (17). The
schizophrenia-associated miRNAs are summarized in Table I.
 | Table ISchizophrenia-associated miRNA.
Adapted from the study by Beveridge and Cairns (36). |
Table I
Schizophrenia-associated miRNA.
Adapted from the study by Beveridge and Cairns (36).
| Study | Type | Brain region |
Schizophrenia-associated miRNA |
|---|
| Lett et al,
2013 (20) | GWAS | Hippocampi, white
matter | miR-137 |
| Gardiner et
al, 2011 (21) | Peripheral tissue
(PBMC) | N/A |
[Downregulated]miR-107,miR-1275,miR-128,miR-130b*,miR-134,miR-148b,
miR-150*, miR-151-3p, miR-16-2*, miR-181a,
miR-200c, miR-224, miR-28-3p, miR-28-5p, miR-29b-1*,
miR-30e*, miR-31, miR-329, miR-335*,
miR-342-5p,miR-409-3p,miR-431,miR-432,miR-486-3p,miR-487b,miR-544,
miR-574-3p, miR-576-5p, miR-584, miR-625*, miR-664,
miR-877, miR-99b |
| Lai et al,
2011 (22) | Peripheral tissue
(PBMC) | N/A | [Upregulated]
miR-34a, miR-449a, miR-548d, miR-564, miR-572, miR-652
[Downregulated] miR-432 |
| Moreau et
al, 2011 (23) | Postmortem
brain | DLPFC (BA9) | [Upregulated]
miR-148b, miR-151 miR-27b, miR-301, miR-545, miR-639
[Downregulated] miR-106b, miR-138, miR-193b, miR-210, miR-22,
miR-324-3p, miR-338, miR-339, miR-425 |
| Santarelli et
al, 2011 (24) | Postmortem
brain | DLPFC (BA9) | [Upregulated]
miR-105, miR-134, miR-148b, miR-150, miR-152, miR-154, miR-17-5p,
miR-187, miR-193a, miR-199a*, miR-199b, miR-222, miR-25,
miR-328, miR-382, miR-409-3p, miR-423, miR-425-5p, miR-433,
miR-452*, miR-487a, miR-495, miR-502, miR-512-3p,
miR-519c, miR-532, miR-542-3p, miR-548b, miR-590, miR-592, miR-652,
miR-767-5p, miR-92b |
| The Schizophrenia
Psychiatric GWAS Consortium, 2011 | GWAS | N/A | miR-137 |
| Beveridge et
al, 2010 (25) | Postmortem
brain | STG (BA22) | [Upregulated]
let-7e, miR-107, miR-125b, miR-128a, miR-128b, miR-129, miR-130a,
miR-133b, miR-138, miR-146b, miR-148a, miR-150, miR-152, miR-155,
miR-15a, miR-15b, miR-16, miR-17-3p, miR-17-5p, miR-181b, miR-195,
miR-197, miR-199a*, miR-19a, miR-20a, miR-222, miR-23a,
miR-24, miR-26b, miR-26b, miR-27b, miR-28, miR-296, miR-328,
miR-330, miR-335, miR-338, miR-339, miR-340, miR-373*,
miR-381, miR-409-5p, miR-432*, miR-452*,
miR-455, miR-484, miR-485-5p, miR-486, miR-487a, miR-489, miR-494,
miR-499, miR-502, miR-517a, miR-517c, miR-518b, miR-519d,
miR-520a*, miR-520g, miR-9*, miR-99a |
| Beveridge et
al, 2010 (25) | Postmortem
brain | DLPFC (BA9) | [Upregulated]
let-7d, miR-101, miR-105, miR-107, miR-126*, miR-128a,
miR-153, miR-15a, miR-15b, miR-16, miR-16, miR-181a, miR-181b,
miR-181b, miR-181d, miR-184, miR-195, miR-199a, miR-20a, miR-219,
miR-223, miR-26b, miR-27a, miR-29c, miR-302a*,
miR-302b*, miR-31, miR-33, miR-338, miR-409-3p,
miR-512-3p, miR-519b, miR-7 |
| Kim et al,
2010 (26) | Postmortem
brain | DLPFC (BA46) | [Upregulated]
miR-132, miR-132*, miR-154*, miR-212,
miR-34a, miR-544, miR-7 |
| Mellios et
al, 2010 (27) | Postmortem
brain | Frontal cortex
(BA10) | [Downregulated]
miR-30b |
| Xu et al,
2010 (28) | miR-SNP | N/A | miR-24,
pre-miR-30e, miR-30e |
| Feng et al,
2009 (29) | miR-SNP | N/A | let-7f-2,
miR-188-3p, pre-miR-18b, miR-325-3p, pre-miR-502, pre-miR-505,
miR-509-3p, miR-510-3p, miR-660 |
| Mellios et
al, 2009 (30) | Postmortem
brain | Frontal cortex
(BA10) | [Downregulated]
miR-30e, miR-195 |
| Sun et al,
2009 (31) | miR-SNP | N/A | miR-502,
miR-510 |
| Tabares-Seisdedos
et al, 2009 (32) | CNV | N/A | miR-124-1, miR-320,
miR-383, miR-486, miR-596, miR-597, miR-598 |
| Zhu et al,
2009 (3) | Postmortem
brain | | DLPFC (BA46)
miR-346 |
| Beveridge et
al, 2008 (33) | Postmortem
brain | | STG (BA22)
miR-181b |
| Hansen et
al, 2007 (34) | miR-SNP | N/A | miR-198,
miR-206 |
| Perkins et
al, 2007 (18) | Postmortem
brain | DLPFC (BA9) | [Upregulated]
miR-106b, miR-7
[Downregulated] miR-26b, miR-30b, miR-29b, miR-195, miR-92,
miR-30a, miR-30d, miR-20b, miR-29c, miR-29a, miR-212, miR-24,
miR-30e, miR-9* |
Alterations in the sequence of certain miRNAs leads
to the alteration of gene regulation, which contributes to the
development of a psychiatric disorder. Perkins et al
compared the expression of 264 miRNAs from the prefrontal cortex of
patients diagnosed with schizophrenia and 21 individuals not
suffering from a psychiatric illness, used as controls (18). Using a custom-made miRNA
microarray, the authors identified that the expression of 15 miRNAs
decreased and the expression of one miRNA increased in the
prefrontal cortex of the schizophrenia patients, when compared with
the control individuals.
In addition, Guo et al demonstrated that
miR-195 is involved in a complex regulatory network, which affects
the signaling pathways considered to be significant in the
development of schizophrenia (19).
The gene encoding miR-346 is located in the intron
of the glutamate receptor ionotropic δ1 (GRID1) gene, which is
known to be involved in schizophrenia susceptibility (5). Using quantitative polymerase chain
reaction, Zhu et al detected the expression levels of
miR-346 and GRID1 in brain RNA samples of 35 patients with
schizophrenia and 34 controls, obtained from the Stanley Medical
Research Institute (Chevy Chase, MD, USA) (3). The expression levels of miR-346 and
GRID1 were found to be lower in the schizophrenia patients compared
with the controls.
In a study of the Chinese population, Xu et
al described a potentially functional variant that affected
pre-miR-30e and was closely associated with schizophrenia (28). The variant affected the predicted
structure and the release of pre-miRNA, as well as the accuracy of
the mature miRNA. Despite the samples being obtained from the
peripheral blood, the findings of Xu et al were comparable
to the observations of Perkins et al (18), who detected an increase in the
expression level of miR-30e in the prefrontal cortex of patients
with schizophrenia.
Smrt et al demonstrated that miR-137 serves
as a regulator of adult neural stem cell maturation and migration
to the subventricular zone, located around the lateral ventricles,
and the subgranular zone of the hippocampus (35). In addition, miR-137 was shown to be
highly associated (P=1.6×10−11) with schizophrenia in
one of the largest genome-wide association studies (36). Since miRNAs are crucial regulators
of gene expression, important genetic mechanisms may contribute to
the phenotypic heterogeneity. Lett et al made demographics
for age-at-onset samples, as well as healthy controls, and their
findings indicated that miR-137 plays a considerable role in the
variation in phenotypes that is believed to have an important role
in clinical outcome and treatment response (20). The authors concluded that the
effects of miR-137 on the phenotypic heterogeneity of schizophrenia
may occur via neurodevelopmental gene networks. These observations
may provide a model for the role of miRNAs in the phenotypic
heterogeneity of psychiatric disorders.
3. miRNAs may be potential biomarkers in the
diagnosis of schizophrenia
The diagnosis of schizophrenia is currently based
exclusively on signs and symptoms; therefore, a diagnosis requires
qualified psychiatric assessment. The majority of studies
investigating the genetics of schizophrenia have been limited to
protein-coding genes. However, the regulatory role of miRNAs has
received increasing attention, with results indicating that miRNAs
may contribute to the etiology of schizophrenia. miRNAs are known
to influence complex gene networks and pathways, which suggests
that they may be potential biomarkers when dysregulated. miRNAs as
biomarkers have been shown to be useful in the clinical
stratification of neoplasms, and even have a greater prognostic
significance compared with mRNAs, which may result from miRNAs
being discrete functional entities (36). The expression of peripheral miRNA
has also been examined in other neurological disorders. For
instance, in Alzheimer’s disease, miRNA was found to be altered in
peripheral blood mononuclear cells (PBMCs) (37), cerebrospinal fluid and brain tissue
(38).
Gardiner et al investigated the expression
profile of miRNA in PBMCs of 112 patients with schizophrenia and 76
non-psychiatric controls (21).
The authors identified 83 miRNAs that were significantly
downregulated in the schizoaffective group, including a large
subgroup of miRNAs (20%) transcribed from a single imprinted locus
at the maternally expressed DLK1–DIO3 region on chromosome 14q32.
Similarly, Lai et al identified a signature of seven miRNAs
in an initial cohort of 30 patients with schizophrenia and 30
controls, which included the upregulated miR-34a, miR-449a,
miR-564, miR-548d, miR-572 and miR-652, and downregulated miR-432
(22). The results of the study
were subsequently validated in an extended cohort of 60
schizophrenia patients and 30 controls. The expression levels of a
number of these miRNAs were found to be correlated with negative
symptoms, neurocognitive dysfunction and mismatched negativity
performances of the schizoaffective patients. Notably, miR-449a was
shown to be closely associated with the majority of features
examined in the Wisconsin Card Sorting Test (39), indicating the possible involvement
of miR-449a in the executive function of the brain (36).
The majority of previous blood-based gene expression
studies on schizophrenia have been limited to the expression of
protein-coding genes. A large number of mRNAs have been detected
using microarray platforms (40–43),
leading to a number of putative mRNAs being associated with
schizophrenia; however, the majority of associations have not been
possible to replicate in other studies (44). The regulatory role of miRNAs has
received increasing attention since miRNAs regulate the expression
levels of genes by inhibiting the translation of mRNAs and may be
potential biomarkers for schizophrenia (45–49).
Since each miRNA can regulate the expression levels of hundreds of
target genes, the number of discriminating miRNAs as biomarkers is
likely to be much less compared with mRNAs.
Therefore, the aforementioned studies have
demonstrated that the peripheral patterns of miRNAs may be used as
biomarkers for schizophrenia and are associated with
subphenotypes.
4. Summary and future prospects
Schizophrenia exhibits a complex neurobehavioral
phenotype that is considered to have developed through turbulences
in the neural circuitry and synaptic function. Due to the abundance
of neural networks, various combinations of gene variants can cause
system dysfunctions, manifesting as associated neurobehavioral
syndromes. miRNAs appear to be important in neural networks and are
highly expressed in the brain, emerging as key regulators of
numerous neurodevelopmental and neurological processes.
Dysregulation of miRNAs leads to pervasive changes and defects of
the nervous system, which may assist investigations into the
pathophysiology and neuropathology of schizophrenia. Due to their
tuning effect on numerous proteins, miRNA biomarkers for
schizophrenia and associated phenotypes may ultimately provide the
basis for early detection, disease stratification and prediction of
response to drugs and side-effects; however, mood swings can affect
the expression of certain miRNAs (50), and this may be a disadvantage of
using miRNAs to diagnose schizophrenia.
miRNAs have the following advantages: i) miRNA
remains stable despite changes in temperature, pH or physical
state; ii) the expression of miRNAs is specific with no difference
in gender or in individual; iii) miRNAs in serum and plasma can be
quantified in vitro (51);
thus, miRNAs are promising biomarkers instead of proteins. To date,
miRNAs extracted from brain tissue, cerebrospinal fluid and PBMCs
have been used as biomarkers in the diagnosis of schizophrenia;
furthermore, the decoding of the miRNA genome may contribute to
elucidating the etiology and improving the treatment of
schizophrenia. miRNAs extracted from other peripheral sources,
however, have not been investigated. Future studies should
investigate miRNAs from other peripheral fluids, including saliva
and urine, as these may also be potential biomarkers in the
diagnosis of schizophrenia.
References
|
1
|
Earls LR, Fricke RG, Yu J, Berry RB,
Baldwin LT and Zakharenko SS: Age-dependent microRNA control of
synaptic plasticity in 22q11 deletion syndrome and schizophrenia. J
Neurosci. 32:14132–14144. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Abdolmaleky HM and Thiagalingam S: Can the
schizophrenia epigenome provide clues for the molecular basis of
pathogenesis? Epigenomics. 3:679–683. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhu Y, Kalbfleisch T, Brennan MD and Li Y:
A microRNA gene is hosted in an intron of a
schizophrenia-susceptibility gene. Schizophr Res. 109:86–89. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Guarnieri DJ and DiLeone RJ: microRNAs: a
new class of gene regulators. Ann Med. 40:197–208. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bravo JA and Dinan TG: microRNAs: a novel
therapeutic target for schizophrenia. Curr Pharm Des. 17:176–188.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee RC, Feinbaum RL and Ambros V: The
C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854. 1993.
|
|
7
|
Bartel DP: microRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Williams NM: Molecular mechanisms in 22q11
deletion syndrome. Schizophr Bull. 37:882–889. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cao X, Yeo G, Muotri AR, Kuwabara T and
Gage FH: Noncoding RNAs in the mammalian central nervous system.
Annu Rev Neurosci. 29:77–103. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vo N, Klein ME, Varlamova O, et al: A
cAMP-response element binding protein-induced microRNA regulates
neuronal morphogenesis. Proc Natl Acad Sci USA. 102:16426–16431.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schratt GM, Tuebing F, Nigh EA, et al: A
brain-specific microRNA regulates dendritic spine development.
Nature. 439:283–289. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sepramaniam S, Tan JR, Tan KS, et al:
Circulating microRNAs as biomarkers of acute stroke. Int J Mol Sci.
15:1418–1432. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ma L, Wei L, Wu F, et al: Advances with
microRNAs in Parkinson’s disease research. Drug Des Devel Ther.
7:1103–1013. 2013.
|
|
14
|
Müller M, Kuiperij HB, Claassen JA, et al:
MicroRNAs in Alzheimer’s disease: differential expression in
hippocampus and cell-free cerebrospinal fluid. Neurobiol Aging.
35:152–158. 2014.
|
|
15
|
Kumar P, Dezso Z, MacKenzie C, et al:
Circulating miRNA biomarkers for Alzheimer’s disease. PLoS One.
8:e698072013.
|
|
16
|
Shi W, Du J, Qi Y, et al: Aberrant
expression of serum miRNAs in schizophrenia. J Psychiatr Res.
46:198–204. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hunsberger JG, Austin DR, Chen G and Manji
HK: microRNAs in mental health: from biological underpinnings to
potential therapies. Neuromolecular Med. 11:173–182. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Perkins DO, Jeffries CD, Jarskog LF, et
al: microRNA expression in the prefrontal cortex of individuals
with schizophrenia and schizoaffective disorder. Genome Biol.
8:R272007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guo AY, Sun J, Jia P and Zhao Z: A novel
microRNA and transcription factor mediated regulatory network in
schizophrenia. BMC Syst Biol. 4:102010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lett TA, Chakavarty MM, Felsky D, et al:
The genome-wide supported microRNA-137 variant predicts phenotypic
heterogeneity within schizophrenia. Mol Psychiatry. 18:443–450.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gardiner E, Beveridge NJ, Wu JQ, et al:
Imprinted DLK1–DIO3 region of 14q32 defines a
schizophrenia-associated miRNA signature in peripheral blood
mononuclear cells. Mol Psychiatry. 17:827–840. 2012.
|
|
22
|
Lai CY, Yu SL, Hsieh MH, et al: microRNA
expression aberration as potential peripheral blood biomarkers for
schizophrenia. PLoS One. 6:e216352011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Moreau MP, Bruce SE, David-Rus R, et al:
Altered microRNA expression profiles in postmortem brain samples
from individuals with schizophrenia and bipolar disorder. Biol
Psychiatry. 69:188–193. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Santarelli DM, Beveridge NJ, Tooney PA and
Cairns MJ: Upregulation of dicer and microRNA expression in the
dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia.
Biol Psychiatry. 69:180–187. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Beveridge NJ, Gardiner E, Carroll AP, et
al: Schizophrenia is associated with an increase in cortical
microRNA biogenesis. Mol Psychiatry. 15:1176–1189. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kim AH, Reimers M, Maher B, et al:
MicroRNA expression profiling in the prefrontal cortex of
individuals affected with schizophrenia and bipolar disorders.
Schizophr Res. 124:183–191. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mellios N, Galdzicka M, Ginns E, et al:
Gender-specific reduction of estrogen-sensitive small RNA, miR-30b,
in subjects with schizophrenia. Schizophr Bull. 38:433–443. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu Y, Li F, Zhang B, et al: microRNAs and
target site screening reveals a pre-microRNA-30e variant associated
with schizophrenia. Schizophr Res. 119:219–227. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Feng J, Sun G, Yan J, et al: Evidence for
X-chromosomal schizophrenia associated with microRNA alterations.
PLoS One. 4:e61212009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mellios N, Huang HS, Baker SP, et al:
Molecular determinants of dysregulated GABAergic gene expression in
the prefrontal cortex of subjects with schizophrenia. Biol
Psychiatry. 65:1006–1014. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sun G, Yan J, Noltner K, et al: SNPs in
human miRNA genes affect biogenesis and function. RNA.
15:1640–1651. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tabarés-Seisdedos R and Rubenstein JL:
Chromosome 8p as a potential hub for developmental neuropsychiatric
disorders: implications for schizophrenia, autism and cancer. Mol
Psychiatry. 14:563–589. 2009.PubMed/NCBI
|
|
33
|
Beveridge NJ, Tooney Pa, Carroll AP, et
al: Dysregulation of miRNA 181b in the temporal cortex in
schizophrenia. Hum Mol Genet. 17:1156–1168. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hansen T, Olsen L, Lindow M, et al: Brain
expressed microRNAs implicated in schizophrenia etiology. PLoS One.
2:e8732007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Smrt RD, Szulwach KE, Pfeiffer RL, et al:
microRNA miR-137 regulates neuronal maturation by targeting
ubiquitin ligase mind bomb-1. Stem Cells. 28:1060–1070. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Beveridge NJ and Cairns MJ: microRNA
dysregulation in schizophrenia. Neurobiol Dis. 46:263–271. 2012.
View Article : Google Scholar
|
|
37
|
Schipper HM, Maes OC, Chertkow HM and Wang
E: microRNA expression in Alzheimer blood mononuclear cells. Gene
Regul Syst Bio. 1:263–274. 2007.PubMed/NCBI
|
|
38
|
Cogswell JP, Ward J, Taylor IA, et al:
Identification of miRNA changes in Alzheimer’s disease brain and
CSF yields putative biomarkers and insights into disease pathways.
J Alzheimers Dis. 14:27–41. 2008.
|
|
39
|
Berg EA: A simple objective technique for
measuring flexibility in thinking. J Gen Psychol. 39:15–22. 1948.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Vawter MP, Ferran E, Galke B, Cooper K,
Bunney WE and Byerley W: microarray screening of lymphocyte gene
expression differences in a multiplex schizophrenia pedigree.
Schizophr Res. 67:41–52. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Glatt SJ, Everall IP, Kremen WS, et al:
Comparative gene expression analysis of blood and brain provides
concurrent validation of SELENBP1 up-regulation in schizophrenia.
Proc Natl Acad Sci USA. 102:15533–15538. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sullivan PF, Fan C and Perou CM:
Evaluating the comparability of gene expression in blood and brain.
Am J Med Genet B Neuropsychiatr Genet. 141B:261–268. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kuzman MR, Medved V, Terzic J and Krainc
D: Genome-wide expression analysis of peripheral blood identifies
candidate biomarkers for schizophrenia. J Psychiatr Res.
43:1073–1077. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yao Y, Schröder J and Karlsson H:
Verification of proposed peripheral biomarkers in mononuclear cells
of individuals with schizophrenia. J Psychiatr Res. 42:639–643.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Perkins DO, Jeffries C and Sullivan P:
Expanding the ‘central dogma’: the regulatory role of nonprotein
coding genes and implications for the genetic liability to
schizophrenia. Mol Psychiatry. 10:69–78. 2005.
|
|
46
|
Cheng HY, Papp JW, Varlamova O, et al:
microRNA modulation of circadian-clock period and entrainment.
Neuron. 54:813–829. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kocerha J, Faghihi MA, Lopez-Toledano MA,
et al: microRNA-219 modulates NMDA receptor-mediated
neurobehavioral dysfunction. Proc Natl Acad Sci USA. 106:3507–3512.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Coyle JT: microRNAs suggest a new
mechanism for altered brain gene expression in schizophrenia. Proc
Natl Acad Sci USA. 106:2975–2976. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Miller BH and Wahlestedt C: microRNA
dysregulation in psychiatric disease. Brain Res. 1338:89–99. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhou R, Yuan P, Wang Y, et al: Evidence
for selective microRNAs and their effectors as common long-term
targets for the actions of mood stabilizers.
Neuropsychopharmacology. 34:1395–1405. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, et al: Circulating microRNAs as
stable blood-based markers for cancer detection. Proc Natl Acad Sci
USA. 105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|















