Introduction
Mismatch repair (MMR) genes provide repair
mechanisms for gene replication (1,2).
These genes are not oncogenes or tumor suppressor genes, but an
additional type of cancer-related gene. Recently, MMR genes have
become increasingly studied to investigate the molecular mechanisms
underlying the onset of cancer. The main function of MMR is to
identify and repair the mismatched bases in base-specific DNA
replication, in order to ensure the high-fidelity of DNA
replication, control the occurrence of mutations and maintain the
genomic stability (3). Currently,
there are nine genes involved in MMR that encode a variety of MMR
proteins, which specifically recognize and repair the mismatched
bases (4,5). Mutations in any gene of this system
can cause the dysfunction of MMR and result in genetic instability,
presenting as replication errors or microsatellite instability,
consequently leading to cancer. hMSH2 was the first human MMR gene
to be isolated, whose mutations are the highest among the nine MMR
genes (6). hMSH2 has been shown to
recognize mismatched bases (7),
and play an important role in reducing the number of mutations and
maintaining the stability of genes.
Oral lichen planus (OLP) is a common non-infectious
oral disease, with an incidence rate of 1–2% in the adult Swedish
population (8). The disease is
more common among individuals aged >40 years, particularly in
females (9). The pathogenesis of
OLP is multifactorial, and may include autoimmunity, chronic local
irritation, mental factors, bacterial or viral infections and other
factors (10,11). However, the exact mechanism is
unknown. Although OLP is a benign disease, it does have the
possibility of cancerization (12). Thus, OLP has been classified as a
potentially malignant lesion by the World Health Organization
(13). The incidence rate of OLP
cancerization varies among different epidemiological surveys,
ranging between 0.4 and 5.6%, and commonly between 1 and 2%
(14,15). Thus, the cancer risk, influencing
factors and cancerization mechanisms of OLP have attracted
increasing attention.
In recent years, the single nucleotide database of
hMSH2 has been established (16,17),
which has resulted in increased research into the role of the hMSH2
gene in carcinogenesis. The expression of hMSH2 has been reported
to change in oral squamous cell carcinoma (18); however, the expression of hMSH2 in
OLP and its role in carcinogenesis has yet to be reported.
The aim of the present study was to investigate the
role of hMSH2 in the pathogenesis and carcinogenesis of OLP by
detecting the expression of hMSH2 in OLP tissues using
immunohistochemistry. The results may provide novel ideas for
accurately predicting the cancer risk of OLP and developing
personalized treatment.
Materials and methods
Tissue samples
Formalin-fixed, paraffin-embedded, human OLP
specimens were obtained from the Pathology Department at the Second
Affiliated Hospital of Hebei Medical University (Shijiazhuang,
China). The specimens included 24 cases of reticular type OLP and
27 cases of erosive/atrophy type OLP. The 51 patients comprising
the OLP group included 22 males and 29 females, aged between 36 and
72 years (mean age, 53.54 years). Tissue samples were obtained
during surgery, and none of the patients had received any treatment
prior to surgery.
In total, 40 cases of normal oral mucosa (NM),
obtained from the pericyst during cosmetics surgery of the alveolar
bone, were used as a control group (NM group). The group included
22 males and 18 females, aged between 11 and 68 years (mean age,
31.30 years). The study was conducted in accordance with the
Declaration of Helsinki and with approval from the Ethics Committee
of Hebei Medical University. Written informed consent was obtained
from all the participants.
Immunohistochemical analysis
All immunohistochemical analyses were performed
using horseradish peroxidase-labeled streptavidin. Tissue sections
measuring 4–5 μm were mounted on Poly-L-lysine-coated slides
(Sigma-Aldrich, St. Louis, MO, USA). Following deparaffinization
using a standard protocol, sections were treated with 3%
H2O2 in methanol for 25 min to block the
endogenous peroxidase activity. The slides were subsequently
microwaved in citrate buffer (pH 6.0) for 15 min at 92–98°C, and
cooled to room temperature. After blocking with normal goat serum
(Invitrogen Life Technologies, Carlsbad, CA, USA) for 30 min at
37°C, the slides were incubated with a polyclonal rabbit anti-human
hMSH2 antibody [diluted 1:75 in phosphate-buffered saline (PBS);
SC-22771; Santa Cruz Biotechnology, Dalla, TX, USA] overnight at
4°C in a high-humidity chamber. Oral squamous cell carcinoma (OSCC)
tissue with known hMSH2 expression served as a positive control.
For the negative control, PBS was used instead of a primary
antibody. Subsequent steps were performed using an SP kit (Hebei
Bohai Biology Co., Ltd.), according to the manufacturer’s
instructions. Immunoreactivity was visualized using a solution of
diaminobenzidine as the chromogen, and the nuclei were
counterstained with hematoxylin.
Assessment of staining
All the slides were interpreted by an experienced
pathologist, according to the standards reported by Lo Muzio et
al (18). Positive staining of
hMSH2 was scored in the nucleus and in the plasma with brown
particles. At least five randomly selected high-power fields
(magnification, ×400) were examined. The proportion of
immunoreactive cells in 100 cells per field was calculated, in
which <5% was regarded to show negative expression (−), while
≥5% was considered to indicate positive expression (+).
Statistical analysis
Statistical analysis was performed using the SPSS
17.0 statistical software program (SPSS, Inc., Chicago, IL, USA).
The χ2 or Fisher’s exact tests were used to compare the
differences between groups. Correlation analysis between the
expression of hMSH2 and the clinical characteristics was performed
using the χ2 test. For all analyses, P<0.05 was
considered to indicate a statistically significant difference.
Results
hMSH2 expression
As shown in Table
I, the positive rate of hMSH2 expression in the OLP tissues was
52.94%, which was significantly lower compared with that in the
control group (80%; χ2=7.1993; P<0.05). In the NM
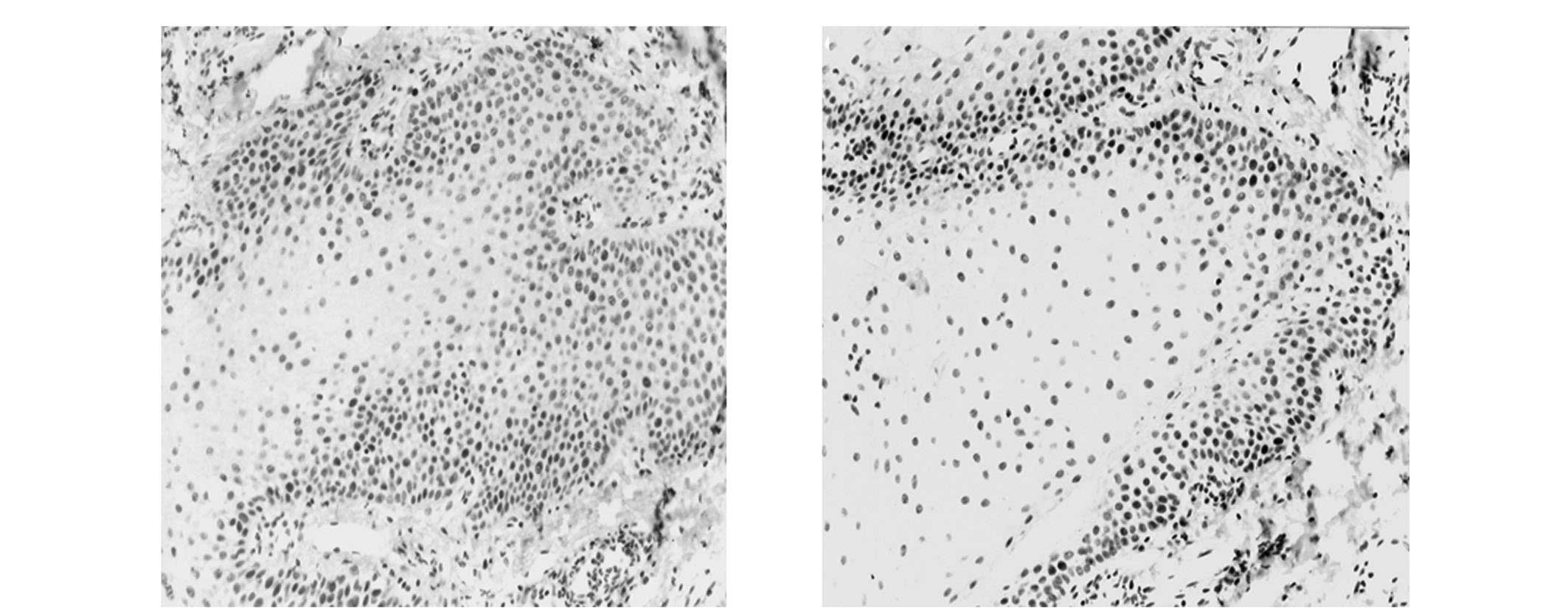
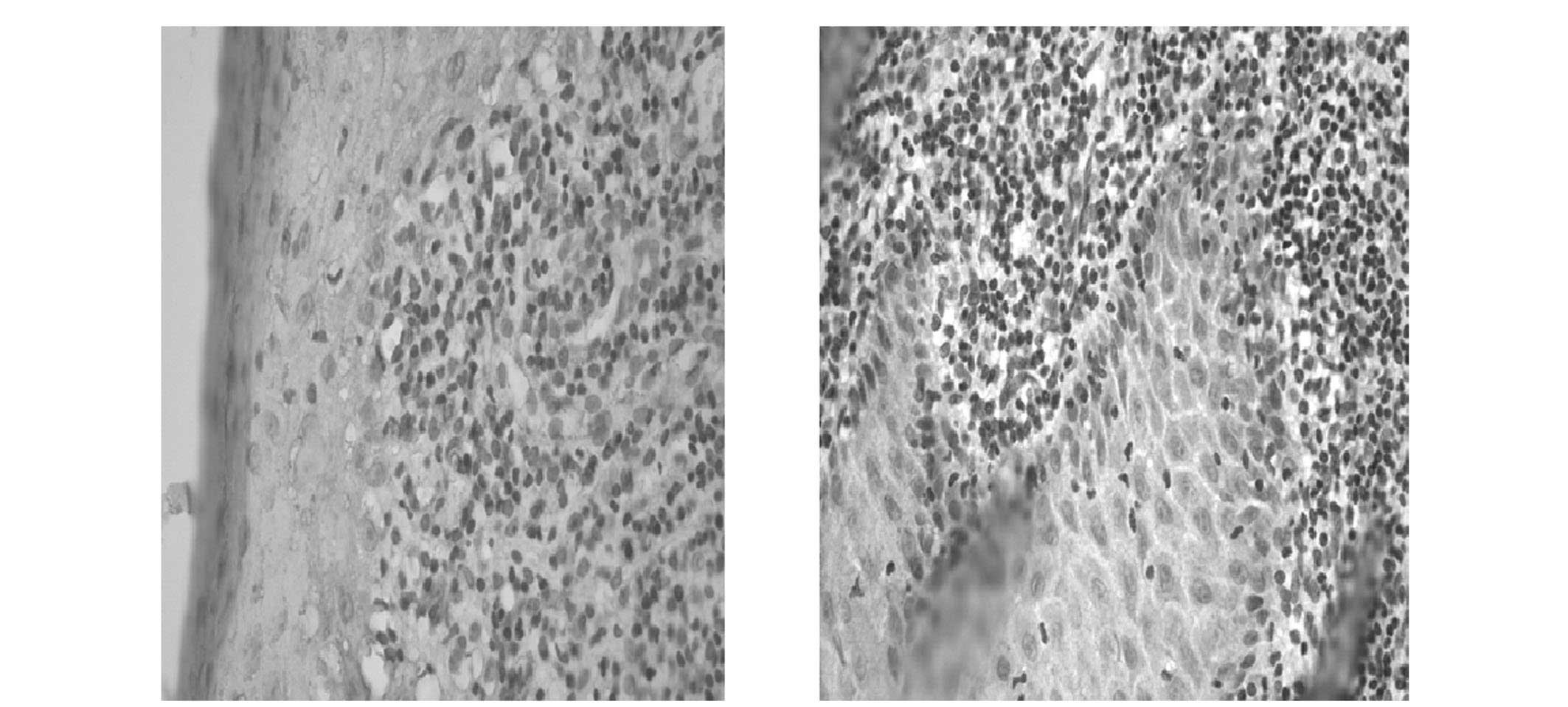
tissue, hMSH2 expression was primarily observed in the cellular
nuclei of the basal and spinous layer (Fig. 1). However, in the OLP tissues,
positive staining of hMSH2 was predominantly observed in the
cytoplasm (Fig. 2).
 | Table IExpression of hMSH2 in patients with
NM and OLP. |
Table I
Expression of hMSH2 in patients with
NM and OLP.
| | hMSH2 (n) | |
|---|
| |
| |
|---|
| Groups | Cases (n) | + | − | Positive rate
(%) |
|---|
| NM | 40 | 32 | 8 | 80.0 |
| OLP | 51 | 27 | 24 | 52.94a |
Associations between hMSH2 expression and
clinicopathological features
In the OLP tissues, the expression of hMSH2 was not
found to significantly correlate with the gender or age of the
patient, or the type of OLP (P>0.05; Table II).
 | Table IIAssociation between hMSH2 expression
and clinicopathological features of patients with OLP. |
Table II
Association between hMSH2 expression
and clinicopathological features of patients with OLP.
| | hMSH2 (n) | | | |
|---|
| |
| | | |
|---|
| Parameters | Cases (n) | + | − | Positive rate
(%) | χ2 | P-value |
|---|
| Gender | | | | | 0.04 | |
| Male | 22 | 12 | 10 | 54.55 | | >0.05 |
| Female | 29 | 15 | 14 | 51.72 | | |
| Age (years) | | | | | 0.14 | |
| <50 | 14 | 8 | 6 | 57.14 | | >0.05 |
| ≥50 | 37 | 19 | 18 | 51.35 | | |
| Types | | | | | 0.92 | |
| Reticular | 24 | 11 | 13 | 45.83 | | >0.05 |
|
Erosive/Atrophic | 27 | 16 | 11 | 59.26 | | |
Discussion
OLP is a common non-infectious disease of the oral
mucosa; however, the pathogenesis remains unknown. Currently, OLP
is considered to be a potential malignant lesion, and the majority
of studies hypothesize that the condition may develop into OSCC
(19,20). However, whether OLP exhibits
cancerization remains controversial (21), with certain studies considering OLP
to be more similar to other benign oral mucosal lesions, and pose
no higher risk of cancerization (22,23).
The aim of the present study was to observe the protein expression
of hMSH2 in the MMR system of OLP, in order to investigate the role
of hMSH2 in the occurrence and development of OLP and assess the
cancerization trend of OLP.
The MMR system is a basic biological mechanism for
maintaining the high fidelity of DNA replication and the stability
of the genetic material by controlling genetic mutations. The
system is an important barrier to prevent the onset of tumors. MMR
proteins always exists in multimeric complex forms to recognize and
specifically repair the mismatched bases during DNA replication,
and may subsequently induce the apoptosis of cells with severely
damaged DNA (24). The
inactivation of MMR genes may present as the activation of
oncogenes or the inactivation of tumor suppressor genes caused by
microsatellite instability, or directly causing mutations in
oncogenes or tumor suppressor genes, thereby inducing
carcinogenesis (25,26).
To date, nine human MMR genes have been identified,
including hMSH2, hMSH6, hMSH5, hMSH4, hMSH3, hMLH1, hPMS1, hPMS2
and hMLH3 (4). hMSH2 was the first
human MMR gene to be isolated, which was isolated from human
hereditary nonpolyposis colorectal cancer (HNPCC). A number of
studies have confirmed that gene mutations of hMSH2 play crucial
roles not only in the pathogenesis of HNPCC (27), but also in the pathogenesis of a
variety of other tumors (28–31),
including cancer of the endometrium, rectal cancer, ovarian cancer,
bladder cancer, as well as OSCC (18). Generally, stronger expression of
the MMR proteins can lead to a stronger repairing ability. Thus,
the positive rate of hMSH2 in non-tumor tissues is significantly
higher compared with that in tumor tissues. For example, the
expression of hMSH2 protein in HNPCC, cervical squamous cell
carcinoma and prostate cancer is significantly lower compared with
that in normal control tissues (32). Lo Muzio et al (18) analyzed the expression of hMSH2 in
OSCC and normal oral mucosal tissues using immunohistochemical
methods, and found that the deletion of hMSH2 protein may be a
potential indicator for OSCC.
In the present study, the positive rate of hMSH2
expression in the OLP tissue was found to be significantly lower
compared with that in the control group. Thus, it was hypothesized
that defects of MMR function may play certain roles in the
occurrence and development of OLP, supporting the hypothesis that
OLP is a potentially malignant lesion that poses a cancer risk.
Pimenta et al (33) also
examined the expression of hMSH2 protein in 26 cases of OLP (12
cases of reticular type and 14 cases of atrophic or erosive type)
using immunohistochemistry. The authors found that the positive
rate of hMSH2 expression was 46.54% in reticular type OLP, 48.79%
in atrophic or erosive type OLP and 61.29% in the normal mucosa,
which is consistent with the observations of the present study.
In conclusion, the present study demonstrated that
hMSH2 expression was significantly reduced in OLP tissues,
resulting in MMR defects in the cells, which may subsequently cause
genetic instability and lead to the onset of OSCC. However, the
exact mechanisms of hMSH2 in the development and carcinogenesis of
OLP should be confirmed by further studies.
References
|
1
|
Reenan RA and Kolodner RD: Isolation and
characterization of two Saccharomyces cerevisiae genes encoding
homologs of the bacterial HexA and Muts mismatch repair proteins.
Genetics. 132:963–973. 1992.PubMed/NCBI
|
|
2
|
Fishel R, Lescoe MK, Rao MR, et al: The
human mutator gene homolog MSH2 and its association with hereditary
nonpolyposis colon cancer. Cell. 77:1661994.
|
|
3
|
Kunkel TA and Erie DA: DNA mismatch
repair. Annu Rev Biochem. 74:681–710. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Manolagas SC and Jilka RL: Bone marrow,
cytokines, and bone remodeling. Emerging insights into the
pathophysiology of osteoporosis. N Engl J Med. 332:305–311. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wagner A, Barrows A, Wijnen JT, et al:
Molecular analysis of hereditary nonpolyposis colorectal cancer in
the United States: high mutation detection rate among clinically
selected families and characterization of an American founder
genomic deletion of the MSH2 gene. Am J Hum Genet. 72:1088–1100.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lipkin SM, Wang V, Jacoby R, et al: MLH3:
a DNA mismatch repair gene associated with mammalian microsatellite
instability. Nat Genet. 24:27–35. 2000. View Article : Google Scholar
|
|
7
|
Hsu HS, Wen CK, Tang YA, Lin RK, Li WY,
Hsu WH and Wang YC: Promoter hypermethylation is the predominant
mechanism in hMLH1 and hMSH2 deregulation and is a poor prognostic
factor in nonsmoking lung cancer. Clin Cancer Res. 11:5410–5416.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Salonen L, Axéll T and Helldén L:
Occurrence of oral mucosal lesions, the influence of tobacco habits
and an estimate of treatment time in an adult Swedish population. J
Oral Pathol Med. 19:170–176. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Carrozzo M: How common is oral lichen
planus? Evid Based Dent. 9:112–113. 2008. View Article : Google Scholar
|
|
10
|
Dissemond J: Oral lichen planus: an
overview. J Dermatolog Treat. 15:136–140. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Carrozzo M and Pellicano R: Lichen planus
and hepatitis C virus infection: an updated critical review.
Minerva Gastroenterol Dietol. 54:65–74. 2008.PubMed/NCBI
|
|
12
|
Lanfranchi-Tizeira HE, Aguas SC and Sano
SM: Malignant transformation of atypical oral lichen planus: a
review of 32 cases. Med Oral. 8:2–9. 2002.(In English and
Spanish).
|
|
13
|
Warnakulasuriya S, Johnson NW and van der
Waal I: Nomenclature and classification of potentially malignant
disorders of the oral mucosa. J Oral Pathol Med. 36:575–580. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bombeccari GP, Guzzi G, Tettamanti M,
Giannì AB, Baj A, Pallotti F and Spadari F: Oral lichen planus and
malignant transformation: a longitudinal cohort study. Oral Surg
Oral Med Oral Pathol Oral Radiol Endod. 112:328–334. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Georgakopoulou EA, Achtari MD, Achtaris M,
Foukas PG and Kotsinas A: Oral lichen planus as a preneoplastic
inflammatory model. J Biomed Biotechnol. 2012:7596262012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
National Centre for Biotechnology
Information. http://www.ncbi.nlm.nih.gov/SNPuri.
Accessed October 16, 2013
|
|
17
|
UniProt Knowledgebase:
UniProtKB/Swiss-Prot. http://www.expasy.ch/sprot/sprot-top.htmluri.
Accessed October 16, 2013
|
|
18
|
Lo Muzio L, Nocini P, Mignogna MD, et al:
Immunocytochemical detection of hMSH2 and hMLH1 expression in oral
SCC. Anticancer Res. 19:933–940. 1999.PubMed/NCBI
|
|
19
|
van der Meij EH, Schepman KP, Smeele LE,
van der Wal JE, Bezemer PD and van der Waal I: A review of the
recent literature regarding malignant transformation of oral lichen
planus. Oral Surg Oral Med Oral Pathol Oral Radiol Endod.
88:307–310. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Scully C, Beyli M, Ferreiro MC, et al:
Update on oral lichen planus: etiopathogenesis and management. Crit
Rev Oral Biol Med. 9:86–122. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Accurso BT, Warner BM, Knobloch TJ,
Weghorst CM, Shumway BS, Allen CM and Kalmar JR: Allelic imbalance
in oral lichen planus and assessment of its classification as a
premalignant condition. Oral Surg Oral Med Oral Pathol Oral Radiol
Endod. 112:359–366. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
O’Flatharta C, Leader M, Kay E, Flint SR,
Toner M, Robertson W and Mabruk MJ: Telomerase activity detected in
oral lichen planus by RNA in situ hybridisation: not a marker for
malignant transformation. J Clin Pathol. 55:602–607. 2002.
View Article : Google Scholar
|
|
23
|
Eisenberg E: Oral lichen planus: a benign
lesion. J Oral Maxillofac Surg. 58:1278–1285. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lynch HT, Lynch JF, Lynch PM and Attard T:
Hereditary colorectal cancer syndromes: molecular genetics, genetic
counseling, diagnosis and management. Fam Cancer. 7:27–39. 2008.
View Article : Google Scholar
|
|
25
|
Amira AT, Mouna T, Ahlem B, et al:
Immunohistochemical expression pattern of MMR protein can
specifically identify patients with colorectal cancer
microsatellite instability. Tumor Biol. 35:6283–6291. 2014.
View Article : Google Scholar
|
|
26
|
Bairwa NK, Saha A, Gochhait S, Pal R,
Gupta V and Bamezai RN: Microsatellite instability: an indirect
assay to detect defects in the cellular mismatch repair machinery.
Methods Mol Biol. 1105:497–509. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang R, Qin W, Xu GL, Zeng FF and Li CX:
A meta-analysis of the prevalence of somatic mutations in the hMLH1
and hMSH2 genes in colorectal cancer. Colorectal Dis. 14:e80–e89.
2012. View Article : Google Scholar
|
|
28
|
González L, Ortiz AP, Suárez EL, et al:
Case-case study of factors associated to hMLH1, hMSH2, and hMSH6
protein expression among endometrial cancer patients of the
University District Hospital of San Juan, Puerto Rico. Int J
Gynecol Cancer. 22:826–829. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lu FI, Gilks CB, Mulligan AM, et al:
Prevalence of loss of expression of DNA mismatch repair proteins in
primary epithelial ovarian tumors. Int J Gynecol Pathol.
31:524–531. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vageli DP, Giannopoulos S, Doukas SG, et
al: Mismatch repair hMSH2, hMLH1, hMSH6 and hPMS2 mRNA expression
profiles in precancerous and cancerous urothelium. Oncol Lett.
5:283–294. 2013.
|
|
31
|
Gu MJ, Bae YK, Kim A, et al: Korean Small
Intestinal Cancer Study Group: Expression of hMLH1, hMSH2 and hMSH6
in small intestinal carcinomas. Hepatogastroenterology.
59:2228–2232. 2012.PubMed/NCBI
|
|
32
|
Ciavattini A, Piccioni M, Tranquilli AL,
Filosa A, Pieramici T and Goteri G: Immunohistochemical expression
of DNA mismatch repair (MMR) system proteins (hMLH1, hMSH2) in
cervical preinvasive and invasive lesions. Pathol Res Pract.
201:21–25. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pimenta FJGS, Pinheiro MDGR and Gomez RS:
Expression of hMSH2 protein of the human DNA mismatch repair system
in oral lichen planus. Int J Med Sci. 1:146–151. 2004. View Article : Google Scholar
|