Introduction
Non-small cell lung cancer (NSCLC) has a high global
cancer-related mortality rate (1).
Two main types of NSCLC, namely adenocarcinoma (AC) and squamous
cell carcinoma (SCC), are the most common subtypes. SCC of the lung
is characterized by a complex pattern of cytogenetic and molecular
genetic changes, and chromosomal aberrations are a hallmark of
cancer cells, occurring at a high prevalence in SCC. Although a
number of studies have been performed to evaluate genetic events
associated with the development and progression of SCC (2,3), the
molecular mechanism remains to be uncovered, and the identification
of predictive markers is crucial.
Genetic alterations in the early stages of cancer
have a close correlation with tumor initiation, and potentially
activate downstream pathways that are implicated in tumor
progression. With the recent advances of computed tomographic
technology, the number of patients diagnosed with stage I lung SCC
has been increasing (3). Although
stage I SCCs are thought to be early-stage diseases and are treated
primarily by surgery without adjuvant therapy, a considerable
fraction of the patients with such SCCs have shown unfavorable
outcomes following surgical treatment. Therefore, to improve the
prognosis of those patients, it is necessary to identify suitable
markers to select patients with poor prognosis, who would benefit
from the use of adjuvant therapy following surgery (4). In particular, the identification of
the genes that are altered during cancer initiation and progression
can be valuable as therapeutic targets or prognostic
indicators.
Studies have previously been performed to evaluate
the genetic events associated with the initiation and progression
of SCCs (5,6). However, little is known about the
specific underlying genes that affect tumorigenesis in early stage
SCCs. Furthermore, the genomic markers that predict aggressive
clinical behavior of SCC remain to be identified. Therefore, in the
present study, high-resolution array-comparative genomic
hybridization (CGH) was conducted to identify early genetic
alterations that define the prognosis of patients with stage I lung
SCC.
Materials and methods
Tumor samples and DNA extraction
A total of 19 stage I SCCs from lung patients
undergoing surgery as primary treatment, without previous radiation
or chemotherapy, were analyzed. The demographic and pathological
data, including age, gender and tumor stage, were obtained by a
review of the medical records. All the patients were classified
according to the World Health Organization classification
histologic typing of lung carcinomas (7). This study has been reviewed and
approved by the Institutional Review Board of the Chungnam National
University Hospital (Daejeon, Korea); informed consent was obtained
from the patients for the use of their tumors in the current
study.
Array-CGH experiment
The MacArray™ Karyo 4000 chips (Macrogen, Seoul,
Korea) used in this study consisted of 4,046 human bacterial
artificial chromosomes (BACs), which were applied in duplicate, and
had a resolution of 1 Mbp (http://www.macrogen.co.kr) (8–11).
Array-CGH was performed as described previously (12). Briefly, hybridizations were
performed in a sealed chamber for 48 h at 37°C. Following the
hybridization, array slides were scanned on a GenePix 4200A
two-color fluorescent scanner (Molecular Devices Corporation,
Sunnyvale, CA, USA) and quantification was performed using GenePix
4200A software (Molecular Devices Corporation). After scanning, the
fluorescent intensities of the red and green channels were saved as
two TIFF image files and the background was subtracted. Locally
weighted scatterplot smoothing (LOWESS) normalization was applied
to adjust for effects due to variation in the intensities between
the red and green dyes. The breakpoint detection and status
assignment of genomic regions was performed using a Gaussian
model-based approach (GLAD) (13).
A low-level copy number gain was defined as a log2 ratio
>0.25, and a copy number loss was defined as a log2
ratio <−0.25. This threshold value was defined empirically as a
value 3-fold that of the standard deviation calculated from
hybridization experiments of 30 normal males to normal females.
Statistical analysis for array-CGH
The Fisher exact test utilized two categories,
normal and abnormal (loss and gain), with the null hypothesis that
the relative proportions of the two categories would be the same in
different groups. A multiple testing correction [Benjamini-Hochberg
false discovery rate (FDR)] was applied to correct for the high
number of false positive calls. R package version 2.2.1 of the
Bioconductor Project (http://www.bioconductor.org) was used for detection of
the frequency of gain or loss and statistical analysis.
Results
Whole genome array analysis of stage I
lung SCC cases
A whole genome array-CGH was conducted to
investigate DNA copy number alterations, and to identify new
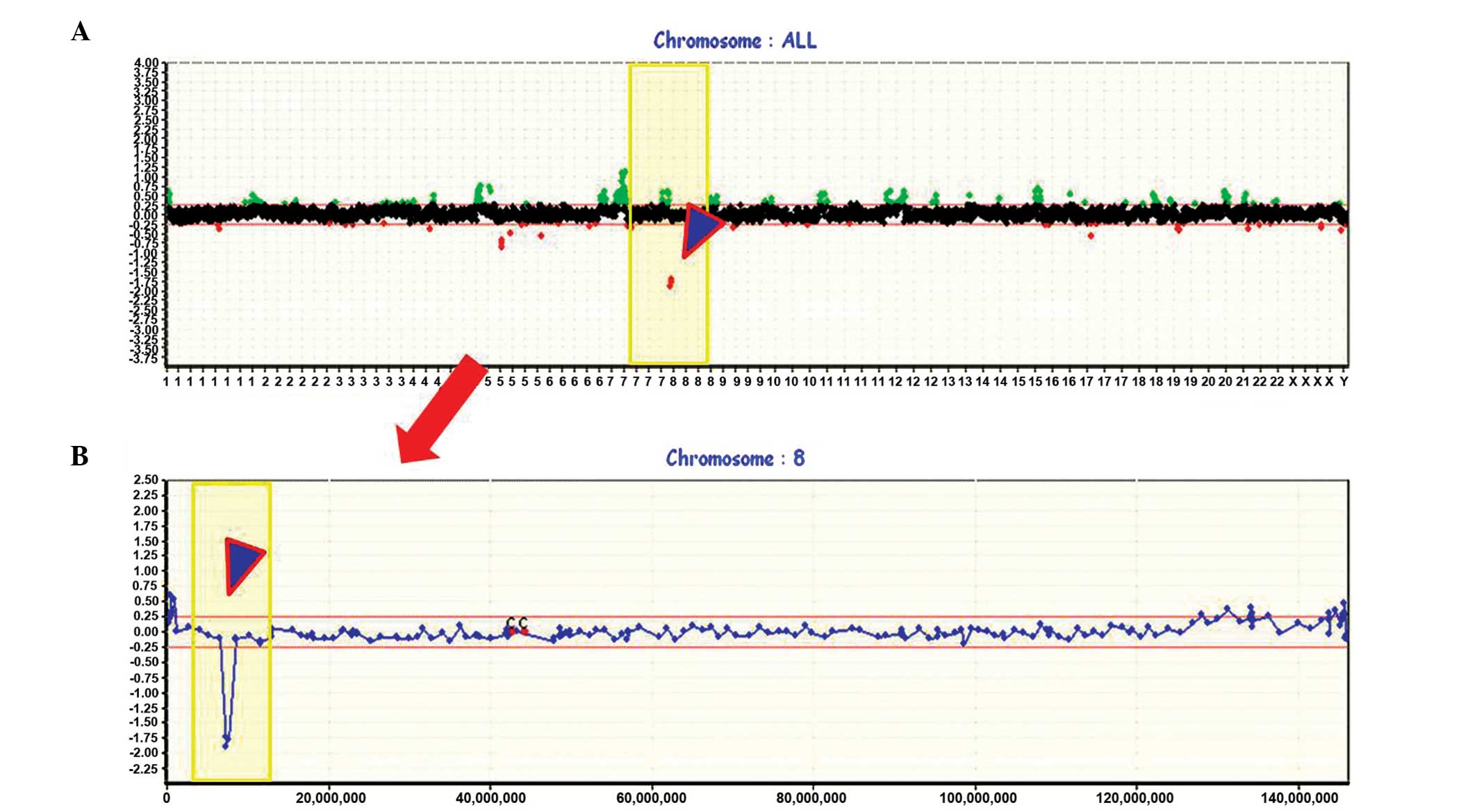
candidate genes in 19 patients with stage I SCC. Frequent gains
were seen on chromosome arms 5p, 7p, 7q, 8q, 11q and 16p (>40%
of patients), and frequent losses on 5q, 8p, 9q, 13q, 14q, 15q, 17p
and 22q (>40% of patients; Fig.
1A). For the first step of the analysis, a decision was made to
focus on chromosome 8p, the most frequently lost (94.7%, 18/19) and
hemizygously deleted (63.2%, 12/19) region in stage I SCCs. More
specifically, three loci of homozygous deletions (HDs) were
displayed in 21.1% (4/19) of the cases. Due to the high frequency
of chromosomal imbalances observed, it was hypothesized that
deletions at 8p must be important and early genetic events in the
pathogenesis of lung SCC, and that 8p may harbor tumor suppressor
genes (TSGs) important for early SCCs. The minimal common region of
chromosome 8p was identified to be located between BAC19_P21 and
BAC181_E17 by genome-wide array-CGH.
Chromosomal alterations of 8p21.1-p23.3
are the most common genetic alterations in stage I lung SCC
cases
The most frequent regions of copy number alterations
in these cases of SCC were defined more clearly, and narrowed down
to 8p21.1-p23.3. A more detailed analysis of chromosome 8p
identified three distinct regions of alterations across the
chromosome. The minimal common region identified by array-CGH was
located between BAC117_I18 and BAC250_C10. The delineation of
8p21.1-p23.3 chromosomal regions and possible target genes of the
SCC cases are shown in Table
I.
 | Table IChromosomal recurrent minimal regions
of genetic alterations on chromosome 8p in 19 stage I SCCs. |
Table I
Chromosomal recurrent minimal regions
of genetic alterations on chromosome 8p in 19 stage I SCCs.
| Regions | Gene contained in
clones | aLoss, n (%) | bHemizygous deletion, n (%) | cHomozygous deletion, n (%) |
|---|
| 8p21.1-p21.3 | EXTL3,
RC74, LOC340414, EXTL3, LOC389642,
NKX3-1, NKX2-6, DPYSL2, DOCK5,
GNRH1, KCTD9, CDCA2, TNFRSF10B,
TNFRSF10C, RHOBTB2, CHMP7, LOC203069,
LOXL2, FLJ10569, XPO7, NPM2,
FGF17, EPB49, RAI16, FLJ22494,
HR, DOK2 | 8/22 (36.4) | 2/22 (9.1) | 3/22 (14.3) |
| 8p22 | LOC392206,
NAT2, DLC1, TUSC3, MTUS1, LOC137012,
NAT1, PDGFRL | 8/22 (36.4) | 5/22 (22.7) | 1/22 (4.8) |
| 8p23.1-p23.3 | MFHAS1,
GATA4, NEIL2, LOC441338, FDFT1,
CTSB, CLDN23, MFHAS1, MSRA,
AGPAT5, DEFB106A, DEFB105A, LOC441316,
FAM90A6P, FAM90A7P, LOC441318, CSMD1,
LOC157693, LOC392169, FBXO25, INM01,
FLJ00290, LOC401441, LOC389607,
LOC157697, LOC442372 | 8/22 (36.4) | 2/22 (9.1) | 3/22 (14.3) |
The first candidate locus spanned 82.0–93.2 kb in
the 8p21.1-p21.3 region. According to the information archived in
the human genome databases (http://genome.ucsc.edu/), it is flanked by the BAC
clones BAC117_I18 and BAC176_L07, and contains 63 possible target
genes (1.8 Mb segment). Notably, a high-frequency of copy number
losses (−0.25>log2 ratio) and hemizygous deletions
(−0.5<log2 ratio <−1) at these region were
detected in 47.4% (9/19) and 10.5% (2/19) of the cases,
respectively.
The second locus spanned 88.6–114.6 kb in the 8p22
region, and demonstrated a high frequency of copy number losses in
7 of 19 cases (36.8%). This locus comprised the putative TSGs of
deleted in liver cancer 1 (DLC1), tumor suppressor candidate
3 (TUSC3) and microtubule-associated tumor suppressor 1
(MTUS1). Furthermore, the following possible target genes
were detected that have not previously been considered to play a
pathogenic role in SCCs: NAT2, LOC392206,
LOC137012, NAT1 and PDGFRL. A high frequency
of hemizygous deletions (−0.5<log2 ratio <11) in
this region was also noted in 26.3% (5/19) of the cases.
The third locus was located distally in the
8p23.1-p23.3 region (87.5–104.7 kb). Notably, a high-frequency of
copy number losses and hemizygous deletions at this region was
detected in 89.5% (17/19) and 52.6% (10/19) of the SCCs,
respectively. More specifically, three HD (−1< log2
ratio) loci at the 8p23.1 region were noted in 21.1% (4/19) of the
cases. The first locus contained clones covering a region of ~112.8
kb, and comprised the CLDN23 and MFHAS1 genes,
occurring in 15.8% (3/19) of the cases. The second locus spanning
~89.6 kb was found to contain the MSRA gene, placing it at
the highest level of HD (21.1%, 4/19). The third locus spanned
~105.3 kb, which contains the DEFB106A, DEFB105A,
FAM90A18P, FAM90A9P, FAM90A10P and
LOC441329 genes, also showing a high-frequency of HDs in the
cases (21.1%, 4/19). The median span of the HDs was 7.5 Mb (range,
105.3–112.8 kb), and all HDs were located between BAC90_M06 and
BAC234_K05. Representative genome profiles of HDs at the 8p23.1
region are presented in Fig. 1.
Whole genome profiles are shown in the upper portion (Fig. 1A), and an individual profile of
chromosome 8, including HDs at the 8p23.1 region, is presented in
more detail below (Fig. 1B). An
example of an individual profile showing HDs in the 8q23.1 region
is presented in Fig. 2, and a
schematic presentation of the cytogenetic bands, as well as map
positions, is provided underneath.
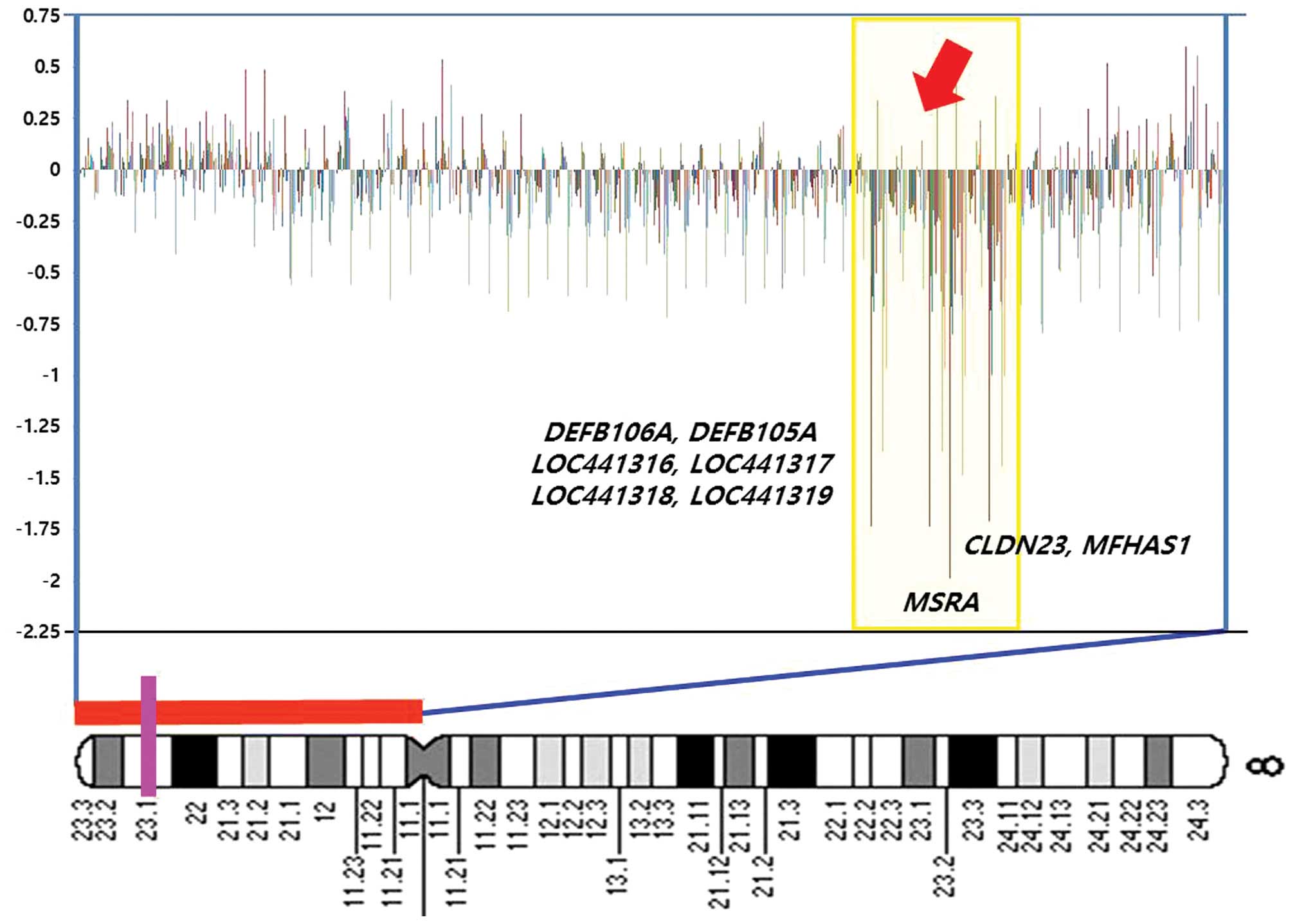 | Figure 2A diagram showing weighted frequencies
(%) of squamous cell carcinoma cases on the short arm of chromosome
8. In the profiles, the y-axis represents the mapped position of
the corresponding clone, and the intensity ratios are assigned to
the x-axis. Cytobands are shown at the bottom of the ideogram.
Vertical lines indicate the lowest locus of chromosome 8 in the
bacterial artificial chromosome (BAC) clone containing the
MSRA, MFHAS1, CLDN23, DEFB106A,
DEFB105A, LOC441316, LOC441317 (FAM90A7P) and
LOC441318 genes. The homozygous deletions (HDs) at 8p23.1
are highlighted in yellow. Log2 ratio <−1 in this BAC
clone, suggesting that homozygous deletions occurred at the
MSRA, MFHAS1, CLDN23, DEFB106A,
DEFB105A, LOC441316, FAM90A7P and
LOC441318 gene loci. Genes contained in clones are shown at
the right. |
Discussion
In this study, whole-genome array-CGH showed that
stage I lung SCCs display non-random patterns of co-occurring gains
and losses. The most striking finding is characterized by a high
frequency of copy number losses and HDs on the short arm of
chromosome 8. Genomic changes on chromosome 8p have long been
considered to be one of the major drivers of cancer progression,
and are suspected to include critical TSGs in lung cancer (14–18).
Previous investigations have focused on identifying somatic genetic
mutations, including deletions and point mutations, of candidate
genes on this region. Yan et al (5) reported that copy number deletions of
chromosome 8p are one of the most prevalent genomic alterations in
SCC of the lung, occurring at an incidence of 46%, and Sy et
al (6) identified a
preferential association of 8p loss with SCC pathogenesis.
Furthermore, Shao et al (19) summarized the loss of heterozygosity
(LOH) of 8p as an early hereditary event during the development of
lung cancer. Allelic losses on 8p are also well described in other
carcinomas, with most studies uncovering a complex pattern that
cannot be reduced to a single minimally deleted region (20). In a study by Moore et al
(21), array-CGH analysis revealed
a high frequency of copy number losses at 8p (38%) in clear cell
renal cell carcinoma, and the finding that the highest frequency of
copy number alterations is on chromosome 8p has also described in
prostate cancer (22). Notably, 8p
allelic losses have also been detected in a relatively early stage
during the pathogenesis of head and neck carcinomas (23). These results and the findings of
the present study suggest that copy number losses on chromosome 8p
are an important and early genetic event in the pathogenesis of
lung SCC, and may harbor gatekeeper TSGs for these cancers
(24).
On genomic analysis, chromosomal aberrations at the
8p21.1-p23.3 regions seem particularly noteworthy, due to the
high-frequency of copy number losses and hemizygous deletions at
this region, detected in 89.5 and 52.6% of the cases, respectively.
Genetic alterations in the distal part of the 8p21.1-p23.3 region
have been reported as early events frequently occurring in lung
cancer. In addition, the size of these alterations, as well as
their frequency, has also been reported to increase during lung
cancer progression (25–27). These regions contain several
interesting TSGs, the most attractive of which is the DCL1
gene on 8p22. It is considered to be one of the prime target genes
on 8p, and its reduced expression or HD has already been described
in connection with various tumors, including lung cancer. Castro
et al (28) reported
DLC1 as the most frequently methylated gene in lung tumors
(50.0%) and described the methylation of this gene as an early
event, associated with early differentiation and stage.
Furthermore, a significant reduction or absence of DLC1 mRNA
expression has been reported in 95% of primary NSCLC, and 58% of
NSCLC cell lines (29). Emerging
data from Kim et al on DLC1, which encodes a RhoA
GTPase-activating protein, indicate that tumor suppressor loss in
NSCLC is associated with genomic deletion or epigenetic silencing
and loss of DLC1 gene transcription (30). Additionally, Pils et al
(31) summarized the epigenetic
events in a frequently deleted region on chromosome 8p22 that
influences the expression of TUSC3, a putative TSG in
ovarian cancer. Similarly, Vaňhara et al (32) suggested that the expression of
TUSC3 epigenetically decreased in epithelial ovarian cancer,
compared with that in benign controls, and provides prognostic
information for patient survival. A recent study by Li et al
(33) reported MTUS1 as a
potential tumor suppressor in gastric cancer. The results of these
studies suggest that these gene changes may occur as early events
in the development of several different types of cancer, and may
also serve as a novel prognostic indicator for lung cancer.
Previous studies have noted that other tumor
suppressors associated with early cancer development are likely to
exist in the 8p21.1-p23.3 regions, in addition to the handful of
identified candidates. In the present study, the following possible
target genes were identified at 8p23.1 in a homozygous deleted
region that was previously not considered to play a pathogenic role
in SCC: MSRA, MFHAS1, CLDN23, DEFB106A,
DEFB105A, LOC441316, FAM90A7P and
LOC441318. Although involvement of these genes in the
pathogenesis of SCC has not been previously mentioned, genetic
mutations of these genes have consistently been reported in
multiple tumor types (31–34). Lei et al (34) reported the MSRA gene as one
of the well-annotated genes significantly downregulated in
hepatocellular carcinoma (HCC), and suggested that it might play a
role in the progression of HCC. Furthermore, Alonso Guervós et
al (17) documented an
association between the loss of the MFHAS1 gene and lymph
node metastases. Additionally, downregulation of the CLDN23
gene in gastric cancer has also been reported (15,16).
These findings support the theory that genetic mutations of these
developmental genes may contribute to lung tumorigenesis at an
early stage, and highlight the value of examining the genomes of
pre-invasive stages of cancer at tiling resolution. Moreover, the
newly identified target genes at the 8p23.1 HD chromosomal sites
should provide important clues with regard to the genetic
mechanisms underlying the initiation and progression of stage I SCC
of the lung. Further investigation is required to validate and
clarify the vital functions of these genes as novel targets for
early SCCs, in larger studies using multiple samples.
In the present study, the previous findings
concerning the 8p chromosome were significantly extended and the
critical regions implicated in early SCC of the lung were firmly
established. Furthermore, genomic analysis allowed the proposition
of novel candidate genes that may be associated with the
pathogenesis of stage I SCC cases. These findings suggest that
genetic alterations on chromosome 8p are the first step in the
initiation of genomic instability at the early onset of SCC and the
newly identified target genes at the 8p23.1 HD chromosomal sites
should provide important information with regard to the genetic
mechanisms of the initiation and progression of stage I SCC.
Acknowledgements
This study was financially supported by the research
fund of Korea Nazarene University in 2014.
References
|
1
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small cell lung cancer: epidemiology, risk
factors, treatment, and survivorship. Mayo Clin Proc. 83:584–594.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kang JU, Koo SH, Kwon KC, Park JW and Kim
JM: Gain at chromosomal region 5p15.33, containing TERT, is the
most frequent genetic event in early stages of non-small cell lung
cancer. Cancer Genet Cytogenet. 182:1–11. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Garnis C, Campbell J, Davies JJ, Macaulay
C, Lam S and Lam WL: Involvement of multiple developmental genes on
chromosome 1p in lung tumorigenesis. Hum Mol Genet. 14:475–482.
2005. View Article : Google Scholar
|
|
4
|
Iwakawa R, Kohno T, Kato M, Shiraishi K,
Tsuta K, Noguchi M, Ogawa S and Yokota J: MYC amplification as a
prognostic marker of early-stage lung adenocarcinoma identified by
whole genome copy number analysis. Clin Cancer Res. 17:1481–1489.
2011. View Article : Google Scholar
|
|
5
|
Yan WS, Song LY, Wei WD, Li A, Liang QW,
Liu JH and Fang Y: Chromosomal imbalance in primary lung squamous
cell carcinoma and their relationship with smoking. Ai Zheng.
24:47–52. 2005.(In Chinese). PubMed/NCBI
|
|
6
|
Sy SM, Wong N, Lee TW, Tse G, Mok TS, Fan
B, Pang E, Johnson PJ and Yim A: Distinct patterns of genetic
alterations in adenocarcinoma and squamous cell carcinoma of the
lung. Eur J Cancer. 40:1082–1094. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mihailovici MS, Danciu M, Teleman S,
Stanciu C, Stan M, Bălan G and Potoroacă A: Diagnosis of gastric
cancer on endobiopsies using the WHO classification. Rev Med Chir
Soc Med Nat Iasi. 106:725–729. 2002.(In Romanian).
|
|
8
|
Hwang KT, Han W, Cho J, Lee JW, Ko E, Kim
EK, Jung SY, Jeong EM, Bae JY, Kang JJ, Yang SJ, Kim SW and Noh DY:
Genomic copy number alterations as predictive markers of systemic
recurrence in breast cancer. Int J Cancer. 123:1807–1815. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Park JJ, Kang JK, Hong S, Ryu ER, Kim JI,
Lee JH and Seo JS: Genome-wide combination profiling of copy number
and methylation offers an approach for deciphering misregulation
and development in cancer cells. Gene. 407:139–147. 2008.
View Article : Google Scholar
|
|
10
|
Choi YW, Choi JS, Zheng LT, Lim YJ, Yoon
HK, Kim YH, Wang YP and Lim Y: Comparative genomic hybridization
array analysis and real time PCR reveals genomic alterations in
squamous cell carcinomas of the lung. Lung Cancer. 55:43–51. 2007.
View Article : Google Scholar
|
|
11
|
Choi YW, Bae SM, Kim YW, Lee HN, Kim YW,
Park TC, Ro DY, Shin JC, Shin SJ, Seo JS and Ahn WS: Gene
expression profiles in squamous cell cervical carcinoma using
array-based comparative genomic hybridization analysis. Int J
Gynecol Cancer. 17:687–696. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kang JU, Koo SH, Kwon KC and Park JW:
Frequent silence of chromosome 9p, homozygous DOCK8, DMRT1 and
DMRT3 deletion at 9p24.3 in squamous cell carcinoma of the lung.
Int J Oncol. 37:327–335. 2010.PubMed/NCBI
|
|
13
|
Willenbrock H and Fridlyand J: A
comparison study: applying segmentation to array CGH data for
downstream analyses. Bioinformatics. 21:4084–4091. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lei KF, Wang YF, Zhu XQ, et al:
Identification of MSRA gene on chromosome 8p as a candidate
metastasis suppressor for human hepatitis B virus-positive
hepatocellular carcinoma. BMC Cancer. 7:1722007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Katoh M: Epithelial-mesenchymal transition
in gastric cancer (Review). Int J Oncol. 27:1677–1683.
2005.PubMed/NCBI
|
|
16
|
Katoh M and Katoh M: CLDN23 gene,
frequently down-regulated in intestinal-type gastric cancer, is a
novel member of CLAUDIN gene family. Int J Mol Med. 11:683–689.
2003.PubMed/NCBI
|
|
17
|
Alonso Guervós M, Alvarez Marcos C,
Llorente JL, Sampedro Nuño A, Suárez C and Hermsen M: Genetic
differences between primary larynx and pharynx carcinomas and their
matched lymph node metastases by multiplex ligation-dependent probe
amplification. Oral Oncol. 45:600–604. 2009. View Article : Google Scholar
|
|
18
|
Kang JU, Koo SH, Kwon KC, Park JW and Kim
JM: Identification of novel candidate target genes, including
EPHB3, MASP1 and SST at 3q26.2-q29 in squamous cell carcinoma of
the lung. BMC Cancer. 9:2372009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shao SJ, Wang Y and Yang PM: Abnormalities
of molecular biology in premalignant lung lesions. Ai Zheng.
23:99–103. 2004.PubMed/NCBI
|
|
20
|
Chitale D, Gong Y, Taylor BS, Broderick S,
Brennan C, Somwar R, Golas B, Wang L, et al: An integrated genomic
analysis of lung cancer reveals loss of DUSP4 in EGFR-mutant
tumors. Oncogene. 28:2773–2783. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Moore LE, Jaeger E, Nickerson ML, et al:
Genomic copy number alterations in clear cell renal carcinoma:
associations with case characteristics and mechanisms of VHL gene
inactivation. Oncogenesis. 1:e142012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheng I, Levin AM, et al: Copy number
alterations in prostate tumors and disease aggressiveness. Genes
Chromosomes Cancer. 51:66–76. 2012. View Article : Google Scholar
|
|
23
|
El-Naggar AK, Coombes MM, Batsakis JG,
Hong WK, Goepfert H and Kagan J: Localization of chromosome 8p
regions involved in early tumorigenesis of oral and laryngeal
squamous carcinoma. Oncogene. 16:2983–2987. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang J, Zheng DL, Qin FS, Cheng N, Chen
H, Wan BB, Wang YP, Xiao HS and Han ZG: Genetic and epigenetic
silencing of SCARA5 may contribute to human hepatocellular
carcinoma by activating FAK signaling. J Clin Invest. 120:223–241.
2010. View
Article : Google Scholar :
|
|
25
|
Kurimoto F, Gemma A, Hosoya Y, Seike M,
Takenaka K, Uematsu K, Yoshimura A, Shibuya M and Kudoh S:
Unchanged frequency of loss of heterozygosity and size of the
deleted region at 8p21-23 during metastasis of lung cancer. Int J
Mol Med. 8:89–93. 2001.PubMed/NCBI
|
|
26
|
Shao S, Sun W, Zhang J, An Q, Xiao T, Yang
P, Cheng S and Gao Y: Fine deletion mapping on chromosome 8p21-8p22
in adenocarcinoma of lung. Zhonghua Yi Xue Za Zhi. 82:740–742.
2002.(In Chinese). PubMed/NCBI
|
|
27
|
Wistuba II, Behrens C, Virmani AK, et al:
Allelic losses at chromosome 8 p21-23 are early and frequent events
in the pathogenesis of lung cancer. Cancer Res. 59:1973–1979.
1999.PubMed/NCBI
|
|
28
|
Castro M, Grau L, Puerta P, et al:
Multiplexed methylation profiles of tumor suppressor genes and
clinical outcome in lung cancer. J Transl Med. 8:862010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yuan BZ, Jefferson AM, et al: DLC-1
operates as a tumor suppressor gene in human non-small cell lung
carcinomas. Oncogene. 23:1405–1411. 2004. View Article : Google Scholar
|
|
30
|
Kim TY, Jackson S, Xiong Y, Whitsett TG,
Lobello JR, Weiss GJ, Tran NL, Bang YJ and Der CJ:
CRL4A-FBXW5-mediated degradation of DLC1 Rho GTPase-activating
protein tumor suppressor promotes non-small cell lung cancer cell
growth. Proc Natl Acad Sci USA. 110:16868–16873. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pils D, Horak P, Vanhara P, Anees M, Petz
M, Alfanz A, Gugerell A, Wittinger M, Gleiss A, Auner V, Tong D,
Zeillinger R, Braicu EI, Sehouli J and Krainer M: Methylation
status of TUSC3 is a prognostic factor in ovarian cancer. Cancer.
119:946–954. 2013. View Article : Google Scholar
|
|
32
|
Vaňhara P, Horak P, Pils D, Anees M, Petz
M, Gregor W, Zeillinger R and Krainer M: Loss of the oligosaccharyl
transferase subunit TUSC3 promotes proliferation and migration of
ovarian cancer cells. Int J Oncol. 1383–1389. 2013.
|
|
33
|
Li X, Liu H, Yu T, Dong Z, Tang L and Sun
X: Loss of MTUS1 in gastric cancer promotes tumor growth and
metastasis. Neoplasma. 61:128–135. 2014. View Article : Google Scholar
|
|
34
|
Lei KF, Wang YF, Zhu XQ, Lu PC, Sun BS,
Jia HL, Ren N, Ye QH, Sun HC, Wang L, Tang ZY and Qin LX:
Identification of MSRA gene on chromosome 8p as a candidate
metastasis suppressor for human hepatitis B virus-positive
hepatocellular carcinoma. BMC Cancer. 7:1722007. View Article : Google Scholar : PubMed/NCBI
|