Introduction
Renal cell carcinoma (RCC) is the second most common
genitourinary tumor, accounting for approximately 3% of all adult
malignancies (1). Worldwide
incidence and mortality rates are rising at a rate of approximately
2–3% every decade (2).
Approximately 70% of patients present with localized diseases and
the malignancy generally resists traditional oncological therapies
including chemotherapy and radiotherapy. Radical or partial
nephrectomy remains the mainstay of curative treatment (3). Although certain environmental and
genetic factors have been identified as being associated with RCC,
the molecular mechanisms involved in the initiation and progression
of the disease are still unclear (4). In addition, more than one-third of
patients may have metastasis when diagnosed, and half of patients
may suffer recurrence even following nephrectomy (5). Therefore, an urgent need to identify
new, sensitive, reliable biomarkers and develop new targeted
therapies is emphasized for RCC.
Currently, one of the most prevalent and progressive
approaches for the molecular characterization of tumors is based on
microRNA (miRNA) expression profiles. miRNAs are endogenous
noncoding 19–23 nucleotide RNAs involved in post-transcriptional
regulation of gene expression (6),
and play significant roles in a variety of biological processes,
including proliferation, migration, differentiation and apoptosis
(7). Mutated or abnormally
expressed miRNAs have been identified as oncogenes or tumor
suppressors in a number of human cancers, including RCC (8–10).
It has been reported that miR-184 is widely dysregulated in various
human cancers, including tongue squamous cell carcinoma (11), neuroblastoma (12), nasopharyngeal carcinoma (13) and hepatocellular carcinoma (HCC)
(14), indicating that miR-184 may
play a significant role in oncogenesis. Previous studies have
demonstrated that miR-184 was downregulated in RCC, which may have
potential significance in the occurrence and development of RCC
(15). However, knowledge on the
mechanism of action of miR-184 in RCC is limited. Therefore, there
is a need to research this function.
The aim of our study was to examine the effects of
miR-184 on proliferation, migration and apoptosis in two RCC cell
lines and to lay the foundation for the further study of the
pathogenesis of renal cell carcinoma.
Materials and methods
Cell culture
This study was approved by the institutional review
board and ethics committee of Peking University Shenzhen Hospital,
Shenzhen, China. Two human renal carcinoma cell lines were used,
namely 786-o and ACHN, purchased from the American Type Culture
Collection (ATCC, Manassas, VA, USA). The cell lines were incubated
in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA,
USA) supplemented with 10% fetal bovine serum (Shanghai ExCell
Biology, Shanghai, China) and 1% antibiotics (100 μg/ml penicillin
and 100 mg/ml streptomycin sulfates) and maintained in a humidified
incubator (5% CO2) at 37°C.
Cell transfection efficiency
The miR-184 mimic or negative control was chemically
synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The
sequences were as follows: miR-184 mimic sense strand
5′-TGGACGGAGAACTGATAA GGGT-3′, and antisense strand
5′-CCTTATCAGTTCTCCGTC CATT-3′; miR-184 negative control sense
strand 5′-TTCTCC GAACGTGTCACGTTT-3′, and antisense strand 5′-ACGTGA
CACGTTCGGAGAATT-3′. ACHN and 786-o cells of 60–80% confluence were
transfected with miR-184 mimic or negative control using
Lipofectamine 2000 reagent (Invitrogen) according to the
manufacturer’s instructions. Transfection efficiency and miR-184
expression changes were confirmed by fluorescence microscopy and
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR). The culture medium was replaced by fresh medium 6 h
after transfection, and the transfection efficiency was observed in
cells which were transfected with green fluorescent protein
Fam-labeled negative control (Shanghai GenePharma Co., Ltd.). The
cells were harvested and total RNAs were extracted for RT-qPCR 24 h
after transfection.
RT-qPCR
Total RNA was extracted from cells using TRIzol
reagent (Invitrogen) and purified with an RNeasy Maxi kit (Qiagen,
Valencia, CA, USA) according to the manufacturer’s instructions.
Only the RNA samples with 260/280 ratios of 1.8–2.0 were used for
further investigation. A total of 1 μg total RNA was reverse
transcribed into cDNA using the miScript reverse transcription kit
(Qiagen) according to the manufacturer’s instructions. The RT-qPCR
reaction of miR-184 was performed in a Lightcycler 480 real-time
PCR system (Roche Diagnostics GmbH, Mannheim, Germany) using a
miScript SYBR-Green PCR kit (Qiagen) according to the instructions
and using U6 as an endogenous control. The 20 μl reaction mixture
contained 10 μl 2X QuantiTect SYBR-Green PCR Master mix, 2 μl 10X
miScript universal primer, 0.4 μl specific miRNA primer, 1 μl cDNA
template and RNase-free water. The forward primer sequence of
miR-184 was 5′-TGGACGGAGAACTGATAAGGGT-3′. Reverse primers were
provided by the miScript SYBR-Green PCR kit. The forward primer of
U6 was 5′-CTCGCTTCGGCAGCACA-3′ and the reverse primer was
5′-ACGCTTCACGAATTTG CGT-3′. PCRs were performed on cDNA of the
mimic and negative control group in triplicate for each set. The
protocol for PCR was 95°C for 15 min, followed by 40 cycles of 94°C
for 15 sec, 55°C for 30 sec and 72°C for 30 sec. The miR-184
expression level was determined using the ΔΔCt method.
Cell proliferation assay
The proliferation potential of cells was measured
using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
bromide (MTT) assay. 786-o and ACHN cells were seeded into 96-well
culture plates at a cell density of 6,000 cells/well in growth
medium and transfected with 10 pmol miR-184 mimic or negative
control. A blank control was set up with medium only. Cell growth
was assayed by the addition of 20 μl MTT (5 mg/ml, Sigma, St.
Louis, MO, USA) to each well, and the plate was incubated for 4 h
at 37°C. The proliferation assay was performed for 3 days and cell
growth was assayed at every 24 h interval. Then, the reaction was
stopped by addition of 150 μl dimethyl sulfoxide (Sigma). After
agitating for 15 min at room temperature, the optical density (OD)
of each sample at the wavelength of 490 nm was measured with an
enzyme immunoassay instrument (Bio-Rad, Hercules, CA, USA). Assays
were repeated at least three times.
Cell migration assay
Cell migration was examined by scratch assay
according to the methods previously described (16). Approximately 500,000 cells (786-o
and ACHN) were seeded in each six-well dish and transfected with
miR-184 mimic (100 pmol) or negative control (100 pmol) 24 h later
using Lipofectamine 2000. After 6 h of transfection, a sterile
200-μl pipette tip and markers were used to make a scratch in the
cell monolayer. After scratching, the cells were washed with
phosphate-buffered saline (PBS) medium three times and incubated at
37°C. Images of the scratches were acquired with a digital camera
system at 0 and 24 h after the scratches were made at the same
points. MIAS-2000 software (Leica Microsystems GmbH, Wetzlar,
Germany) was used to determine the migration distance (μm). The
experiments were performed in triplicate, and repeated at least
three times.
Cell apoptosis assay
The extent of apoptosis was evaluated using an
Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI)
detection kit (Invitrogen). 786-o and ACHN were cultured at 37°C
and 5% CO2 in six-well plates and transfected with
miR-184 mimic or negative control at a confluence of approximately
60%. For apoptosis assays, floating and adherent cells were
collected 48 h after transfection and then combined and washed
twice with pre-chilled PBS and resuspended in in 1X binding buffer
(Invitrogen). In total, 5 μl Alexa Fluor® 488 Annexin V
(Invitrogen) and 3 μl PI (Invitrogen) were added to 500 μl cell
suspension, and the samples were analyzed within 15 min of
staining. The fluorescence was analyzed by flow cytometry (Beckman
Coulter, Miami, FL, USA) using an excitation of 488 nm, according
to the manufacturer’s instructions. Each experiment was performed
at least three times.
Statistical analysis
Statistical significance was determined with the
t-test. Each experiment was repeated at least three times. The
results were expressed as the means ± standard deviation. A
two-tailed P<0.05 was considered to indicate a statistically
significant difference. Statistical analysis was carried out with
the SPSS 16.0 statistical software (SPSS, Inc., Chicago, IL,
USA).
Results
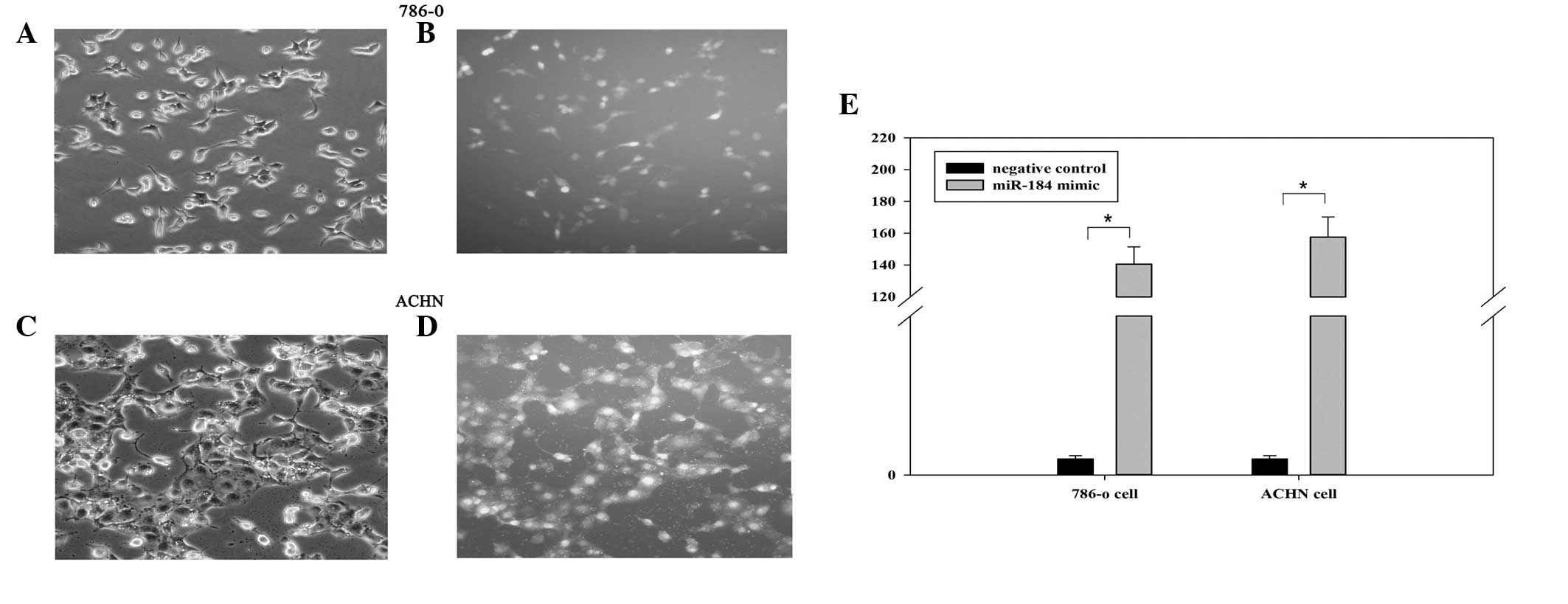
Cell transfection efficiency
To investigate the biological role of miR-184 in
renal cancer, miR-184 mimic and negative control were transfected
into renal cancer cell lines (786-o and ACHN). The Fam-labeled
negative control was transfected into cells, and the transfection
efficiency was analyzed by fluorescence microscopy 6 h after
transfection. As shown in Fig.
1A–D, the transfection efficiency was ~85 and 90% in 786-o and
ACHN cells, respectively. Furthermore, the fold changes of miR-184
determined by RT-qPCR assay in 786-o and ACHN cells were 140.67 and
157.57, respectively (Fig. 1E,
P<0.05).
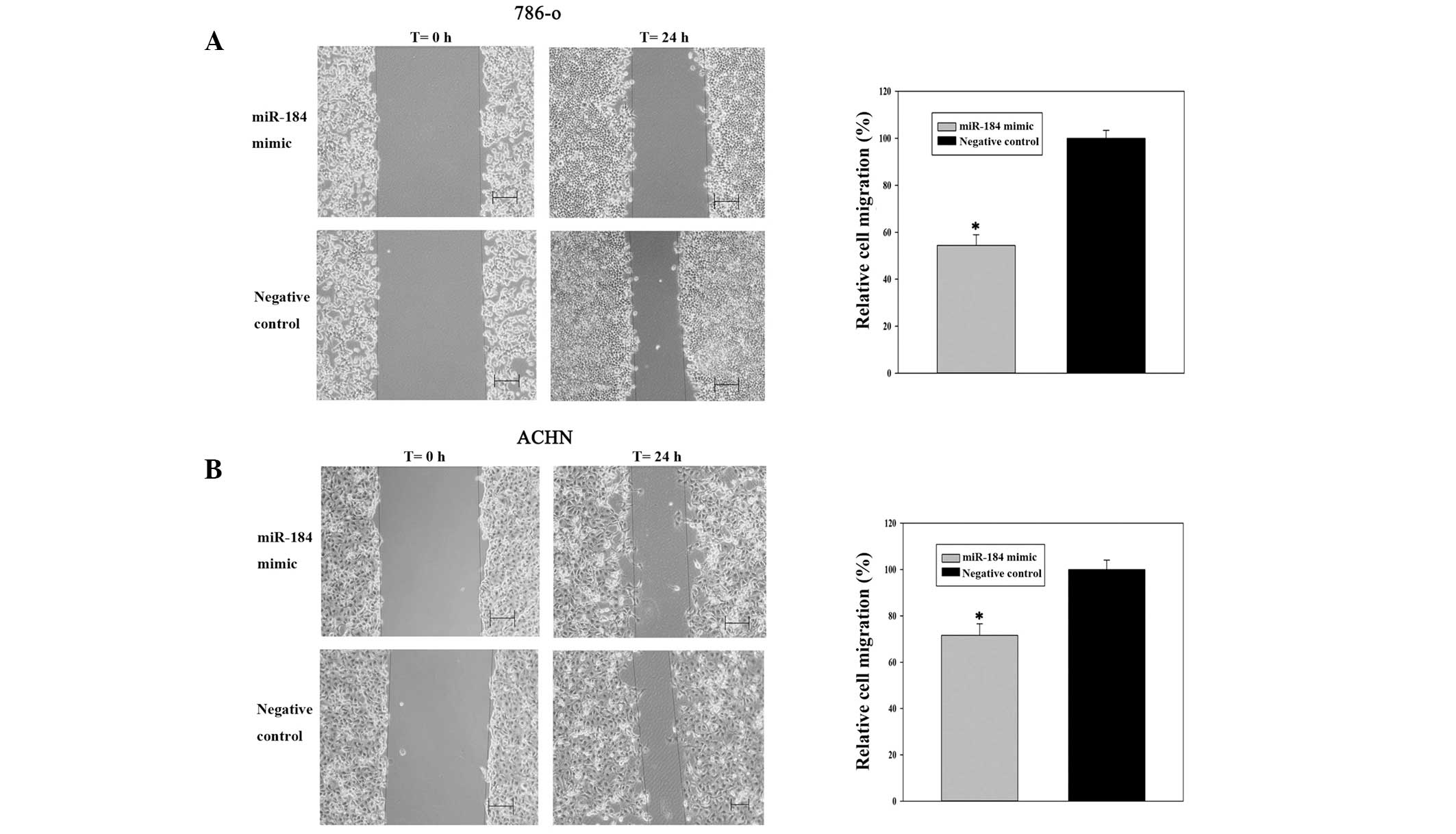
miR-184 mimic inhibits renal cancer cell
migration
Scratch assays were performed to observe the
function of miR-184 in cell migration. As shown in Fig. 2A and B, cell migration was
significantly inhibited in the groups transfected with miR-184
compared with those in the negative control. The inhibition rates
of migration were 45.58% for 786-o cells (P<0.05) and 28.44% for
ACHN cells (P<0.05), indicating that miR-184 mimic inhibited the
migration of renal cancer cells.
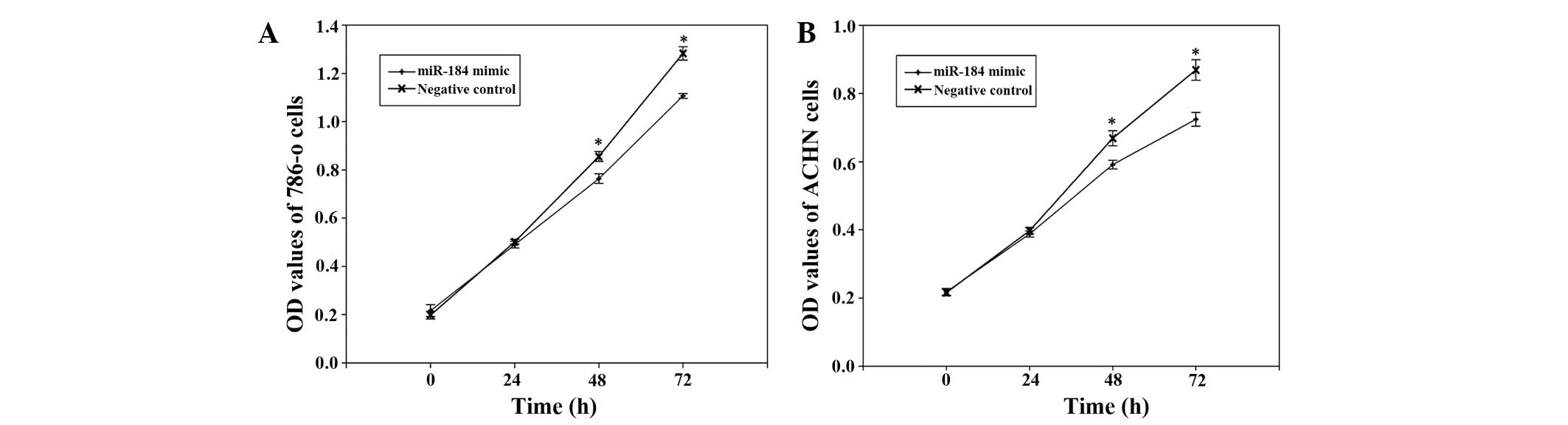
miR-184 mimic suppresses renal cancer
cell proliferation
To determine the potential role of miR-184 on the
proliferation of renal cancer cells, MTT assays were performed. The
miR-184 mimic group and negative control group were measured at 0,
24, 48 and 72 h after transfection. The OD values revealed that
proliferation of 786-o cells was decreased by 2.3% (P>0.05),
10.75% (P<0.05) and 13.72% (P<0.05), while proliferation of
ACHN cells was decreased by 2.4% (P>0.05), 11.57% (P<0.05)
and 16.67% (P<0.05) at 24, 48 and 72 h after transfection,
respectively, suggesting that miR-184 mimic inhibited the growth of
786-o and ACHN compared with the negative control (Fig. 3).
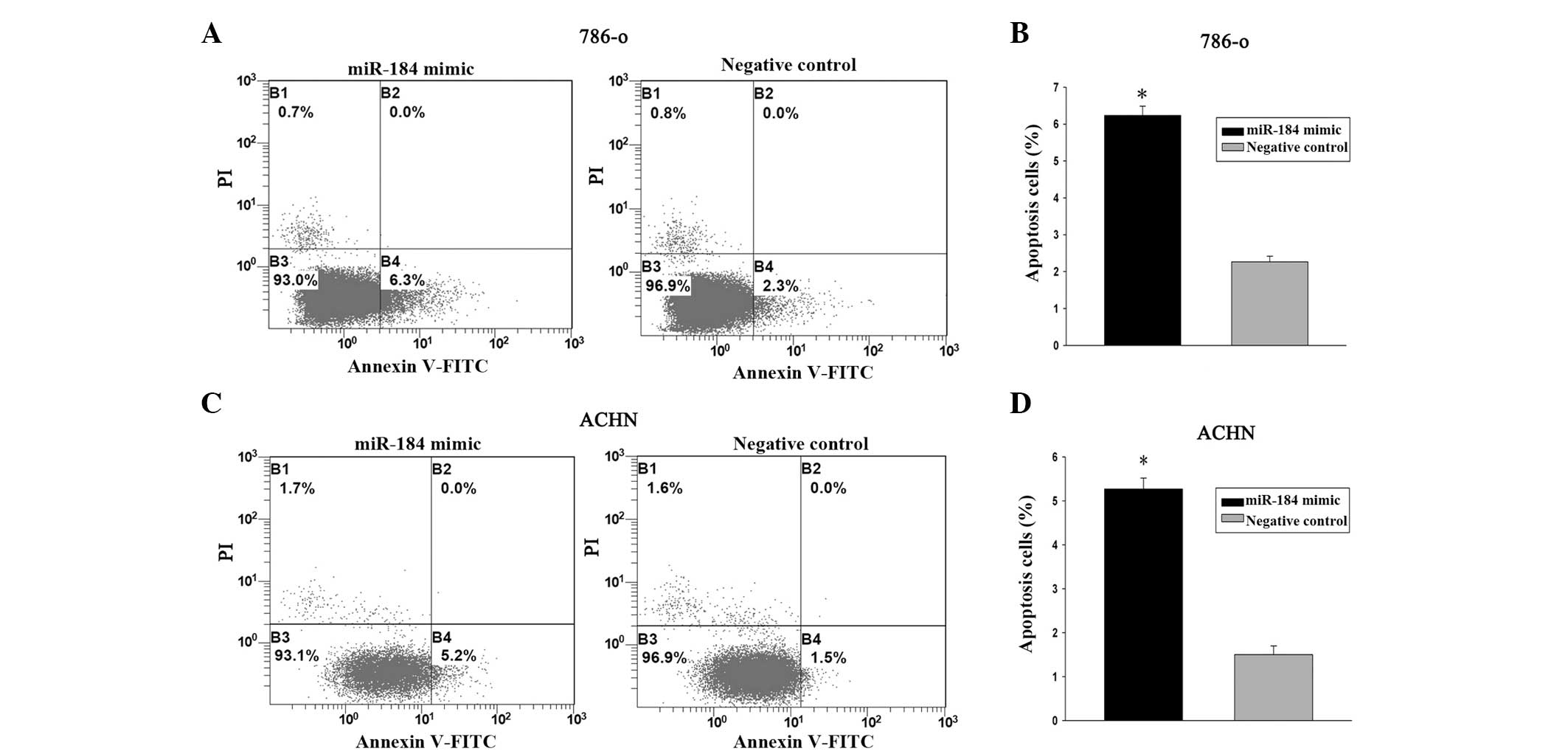
miR-184 mimic induces renal cancer cell
apoptosis
The effects of miR-184 on cell apoptosis were
further evaluated. 786-o and ACHN cells were transfected with
miR-184 mimic and negative control for 48 h. Flow cytometry
analysis demonstrated that the apoptosis rates of 786-o cells
transfected with miR-184 mimic and control were 6.2% versus 2.2%
(P<0.05) while the apoptosis rates of ACHN cells were 5.2%
versus 1.5% (P<0.05). These data demonstrated that miR-184 mimic
promoted renal cancer cell apoptosis (Fig. 4).
Discussion
It is well known that RCC is one of the most common
types of cancers affecting males and females. Despite the great
advances in cancer therapy, major limitations still exist in
managing RCC (17). Traditional
chemotherapy and radiation are not effective in the treatment of
advanced RCC. Therefore, searching for alternative treatment
strategies remains a priority.
miRNAs are a group of endogenous, small, noncoding
RNAs that have been observed in numerous organisms and are involved
in post-transcriptional gene regulation through base pairing to
partially complementary sites, notably in the untranslated region
of mRNA (18). Using microarray
technology, systematic expression analysis of miRNA profiles has
been performed in several human tumors, including breast (19), lung (20) and renal cancer (21). Dysregulation of miRNAs has been
observed between tumors and normal tissues and in distinct stages
of cancer, indicating a possible correlation between miRNAs and
oncogenesis. For example, miR-21, miR-451 and miR-145 have been
identified as being associated with carcinogenesis and development
of cancer by targeting oncogenes or anti-oncogenes (16,22,23).
In previous studies, miRNA expression profiles were
demonstrated to be potential applications for the diagnosis,
prognosis and treatment of tumors (24,25).
These data are consistent with the hypothesis that miRNAs play an
essential role in the development and progression of human cancers.
Among these miRNAs, miR-184 is one of the most frequently studied
in cancer biology.
There is evidence to suggest that aberrant
expression of miRNAs in tumors contributes to human tumorigenesis
by affecting the expression of multiple genes (26). miR-184 has been reported
extensively in human cancers, suggesting that it may function as an
oncogene in a variety of tumors. One comprehensive miRNA profiling
of prostate cancer revealed that miR-184 was upregulated in
high-grade tumors (27).
Inhibition of miR-184 in tongue squamous cell carcinoma cells
reduced cell proliferation and induced apoptosis (11). Furthermore, miR-184 is upregulated
in human HCC cell lines and tissues, and it post-transcriptionally
regulates SOX7 expression and promotes cell proliferation in HCC
(14). Other scholars demonstrated
that miR-184 has a tumor suppressive role in cancers. miR-184
inhibits neuroblastoma cell survival and promotes apoptosis by
targeting AKT2 (12). Taken
together, these studies indicate a possible role of miR-184 in
modulating tumor progression. Our previous studies successfully
identified numerous aberrant expressions of miRNAs in RCC by
massively parallel sequencing technology, and revealed that miR-184
was significantly downregulated in RCC (10); then, researchers observed that the
expression of miR-184 was downregulated in RCC tissues (15). However, the biological role of
miR-184 in RCC is not fully elucidated, and several methods have
been used to study their function.
In the present study, the functions of miR-184 on
cell migration, proliferation and apoptosis were analyzed by
transfecting miR-184 mimic and negative control into renal cancer
786-o and ACHN cells. Our results demonstrated that cells
transfected with miR-184 mimic exhibited less cell migration,
proliferation and more cell apoptosis compared with the negative
control groups. The results provide new insight into the roles and
possible mechanisms of miR-184 in the occurrence and development of
RCC.
Based on the above, the results appear contradictory
in that miR-184 was characterized as an oncogene in certain cancers
and a tumor suppressor in others. This contradiction may be
explained by the ‘imperfect complementarity’ of the interactions
between miRNAs and target genes (28). Researchers observed that miRNAs
post-transcriptionally regulated the expression of more than 30% of
protein coding genes by translational repression, which also
regulated the expression of several putative target genes by
binding to a complementary sequence predominantly in their
untranslated region. However, the bindings are not always
completely complementary, particularly in mammals (29,30).
Moreover, further research should be conducted to determine the
roles and target genes of miR-184 in renal cell carcinoma.
In conclusion, our results revealed that miR-184
dramatically suppressed cell proliferation and migration and
induced cell apoptosis in renal cancer cell lines and plays an
significant role in RCC. In addition, our data suggest that miR-184
may be a promising therapeutic target for renal cancer treatment in
the future. Further research is still needed to explore the roles
and target genes of miR-184.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (no. 81101922), the Medical Scientific
Research Foundation of Guangdong Province of China (no. A2012584
and no. A2013606), the Science and Technology Development Fund
Project of Shenzhen (no. JCYJ20130402114702124) and the funds of
Guangdong and Shenzhen Key medical subject.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wei C, Lai YQ, Li XX and Ye JX:
TGF-beta-activated kinase-1: a potential prognostic marker for
clear cell renal cell carcinoma. Asian Pac J Cancer Prev.
14:315–320. 2013. View Article : Google Scholar
|
|
3
|
Wu S, Lv Z, Wang Y, et al: Increased
expression of pregnancy up-regulated non-ubiquitous calmodulin
kinase is associated with poor prognosis in clear cell renal cell
carcinoma. PLoS One. 8:e599362013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Janzen NK, Kim HL, Figlin RA and
Belldegrun AS: Surveillance after radical or partial nephrectomy
for localized renal cell carcinoma and management of recurrent
disease. Urol Clin North Am. 30:843–852. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wei C, Wu S, Li X, et al: High expression
of FER tyrosine kinase predicts poor prognosis in clear cell renal
cell carcinoma. Oncol Lett. 5:473–478. 2013.PubMed/NCBI
|
|
6
|
Yang XW, Zhang LJ, Huang XH, et al:
miR-145 suppresses cell invasion in hepatocellular carcinoma cells:
miR-145 targets ADAM17. Hepatol Res. 44:551–559. 2014. View Article : Google Scholar
|
|
7
|
Ha TY: MicroRNAs in human diseases: from
cancer to cardiovascular disease. Immune Netw. 11:135–154. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gottardo F, Liu CG, Ferracin M, et al:
Micro-RNA profiling in kidney and bladder cancers. Urol Oncol.
25:387–392. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou L, Chen J, Li Z, et al: Integrated
profiling of microRNAs and mRNAs: microRNAs located on Xq27.3
associate with clear cell renal cell carcinoma. PLoS One.
5:e152242010. View Article : Google Scholar
|
|
11
|
Wong TS, Liu XB, Wong BY, et al: Mature
miR-184 as potential oncogenic microRNA of squamous cell carcinoma
of tongue. Clin Cancer Res. 14:2588–2592. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Foley NH, Bray IM, Tivnan A, et al:
MicroRNA-184 inhibits neuroblastoma cell survival through targeting
the serine/threonine kinase AKT2. Mol Cancer. 9:832010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhen Y, Liu Z, Yang H, et al: Tumor
suppressor PDCD4 modulates miR-184-mediated direct suppression of
C-MYC and BCL2 blocking cell growth and survival in nasopharyngeal
carcinoma. Cell Death Dis. 4:e8722013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu GG, Li WH, He WG, et al: Mir-184
post-transcriptionally regulates SOX7 expression and promotes cell
proliferation in human hepatocellular carcinoma. PLoS One.
9:e887962014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Leng HM, Qian WP, Zhou L, et al: Abnormal
expression and significance of MIR-184 in human renal carcinoma.
Beijing Da Xue Xue Bao. 43:509–513. 2011.(In Chinese). PubMed/NCBI
|
|
16
|
Lu R, Ji Z, Li X, et al: miR-145 functions
as tumor suppressor and targets two oncogenes, ANGPT2 and NEDD9, in
renal cell carcinoma. J Cancer Res Clin Oncol. 140:387–397. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cohen HT and McGovern FJ: Renal-cell
carcinoma. N Engl J Med. 353:2477–2490. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ambros V: MicroRNA pathways in flies and
worms: growth, death, fat, stress, and timing. Cell. 113:673–676.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Iorio MV, Ferracin M, Liu CG, et al:
MicroRNA gene expression deregulation in human breast cancer.
Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yanaihara N, Caplen N, Bowman E, et al:
Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wach S, Nolte E, Theil A, et al: MicroRNA
profiles classify papillary renal cell carcinoma subtypes. Br J
Cancer. 109:714–722. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang A, Liu Y, Shen Y, Xu Y and Li X:
miR-21 modulates cell apoptosis by targeting multiple genes in
renal cell carcinoma. Urology. 78:474 e13–e19. 2011. View Article : Google Scholar
|
|
23
|
Li HY, Zhang Y, Cai JH and Bian HL:
MicroRNA-451 inhibits growth of human colorectal carcinoma cells
via downregulation of Pi3k/Akt pathway. Asian Pac J Cancer Prev.
14:3631–3634. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang GK, Zhu JQ, Zhang JT, et al:
Circulating microRNA: a novel potential biomarker for early
diagnosis of acute myocardial infarction in humans. Eur Heart J.
31:659–666. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guo LJ and Zhang QY: Decreased serum
miR-181a is a potential new tool for breast cancer screening. Int J
Mol Med. 30:680–686. 2012.PubMed/NCBI
|
|
26
|
Cho WC: OncomiRs: the discovery and
progress of microRNAs in cancers. Mol Cancer. 6:602007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Walter BA, Valera VA, Pinto PA and Merino
MJ: Comprehensive microRNA Profiling of Prostate Cancer. J Cancer.
4:350–357. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yu Z, Ni L, Chen D, et al: Identification
of miR-7 as an oncogene in renal cell carcinoma. J Mol Histol.
44:669–677. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kim VN: MicroRNA biogenesis: coordinated
cropping and dicing. Nat Rev Mol Cell Biol. 6:376–385. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yu XY, Zhang Z, Liu J, Zhan B and Kong CZ:
MicroRNA-141 is downregulated in human renal cell carcinoma and
regulates cell survival by targeting CDC25B. Onco Targets Ther.
6:349–354. 2013.PubMed/NCBI
|