Introduction
Human cytoplasmic glutathione S-transferases
(GSTs) are a group of phase II metabolic enzymes, which are
exogenous compounds, and are a supergene family. Glutathione
S-transferase π 1 (GSTP1) has been identified as a member of
the GSTP family. As GSTP1 is present at the highest level in lung
tissue among all GST isoenzymes, it may have an important role in
the detoxification of inhaled carcinogens, such as benzo(a)pyrene
and tobacco carcinogens. Polymorphisms have been detected in exons
5 and 6 of the GSTP1 gene. GSTP1 enzymes with a Val allele in exon
5 have a significantly decreased enzymatic activity, while those
with a Val allele in exon 6 have no significant reduction in
activity. Moreover, GSTP1 exon 5 polymorphisms have been found to
be associated with lung cancer risk, whereas GSTP1 exon 6
polymorphisms have not been found to be correlated (1,2).
Although GSTP1 exon 5 polymorphisms have been
demonstrated to be associated with lung cancer risk, the findings
have not been consistent across studies (2). Moreover, few studies on GSTP1 exon 5
polymorphisms in the Chinese Han population have been reported.
Therefore, the present study attempted to determine the frequency
distribution and characteristics of polymorphic alleles and
genotypes in GSTP1 exon 5 in a Chinese Han population, and to
explore the correlation between GSTP1 exon 5 polymorphisms and
susceptibility to lung cancer using the polymerase chain
reaction-restriction fragment length polymorphism (PCR-RFLP)
technique in a case-control study.
Materials and methods
Subjects
Lung cancer group
Hospitalized patients with primary lung cancer who
received surgical treatment in the Department of Thoracic Surgery
of the Affiliated Hospital of Inner Mongolia Medical University
(Hohhot, China) from May 2006 to October 2008 were diagnosed by
postoperative pathological examination. A total of 150 patients
with lung cancer, including 115 male patients and 35 female
patients, aged 31–76 years (mean, 57.1 years) were enrolled. Of
them, 70 patients had squamous cell carcinoma, 61 had
adenocarcinoma and 19 had other types of lung cancer.
Control group
The control group consisted of 152 healthy
volunteers who received a physical examination in the outpatient
clinics of the Affiliated Hospital of Inner Mongolia Medical
University during the same period, including 117 males and 35
females, aged 33–78 years (mean, 54.7 years). The present study was
approved by the ethics committee of Inner Mongolia Medical
University. All patients provided written informed consent.
Main reagents
The main reagents were GoldView™ DNA dye, 1 ml/tube,
purchased from SBS Genetech (Beijing, China); an UltraPure™ rapid
genomic DNA extraction kit, 50 extractions/kit, purchased from SBS
Genetech; and DNA Marker I, 200 reactions/tube, purchased from
Tiangen Biotech (Beijing, China). In addition, 10 mM dNTP mix, 0.2
ml/tube; 5 U/µl Taq DNA polymerase, 500 U/tube; 10X PCR reaction
buffer, 1 ml/tube; 25 mM MgCl2, 1 ml/tube; and
Restriction endonuclease Alw26I, 1,000 U/tube were purchased
from Fermentas (Pittsburgh, PA, USA).
Key instruments
The key instruments used were an electronic balance
(BP310S; Sartorius AG, Göttingen, Germany); a heated shaking water
bath and fast digital thermostatic tank (HZS-H; Donglian Electronic
& Technology Development Co., Ltd., Harbin, China); a microwave
oven (Guangdong Galanz Microwave Oven and Electrical Appliances
Manufacturing Co., Ltd., Guangdong, China); a constant-voltage
electrophoresis system (Vokam 2541; Bio-Rad, Hercules, CA, USA); a
horizontal electrophoresis tank (Liuyi Instrument Factory, Beijing,
China); a gel scanning image analysis system (Bio-Rad); a UV
spectrophotometer (Smart Spec 3000; Bio-Rad); a 96-well gradient
PCR machine (Hybaid, Thermo Fisher Scientific, Inc., Carlsbad, CA,
USA); a vortexing shaker (WH-851; KOLED, Beijing, China); and a UV
transilluminator (ZF-90; Gucun Electro-optical Instrument Factory,
Shanghai, China).
Genomic DNA extraction
Genomic DNA was extracted from the peripheral venous
blood of the 302 subjects. The blood, which was stored as frozen
EDTA-anticoagulated samples, was thawed at room temperature and
mixed. DNA extraction was performed using 0.5 ml whole blood in
strict accordance with the instructions of the DNA extraction kit.
Then, 5 µl extracted genomic DNA was electrophoresed on 0.8%
agarose gel to determine its purity and yield. In addition, the
purity of the extracted genomic DNA was determined by UV
spectrophotometry, and optical density (OD) values were measured at
260 and 280 nm to calculate OD260:280 nm.
Determination of GSTP1 genotype by
PCR-RFLP
Design and synthesis of primers
A pair of primers for exon 5 of the GSTP1 gene were
designed according to the method described by Ryberg et al
(3) (Table I) and synthesized by SBS
Genetech.
 | Table I.Sequences of the primers for use in
the analysis of GSTP1 by polymerase chain reaction. |
Table I.
Sequences of the primers for use in
the analysis of GSTP1 by polymerase chain reaction.
| Direction | Primer sequence | Product size
(bp) |
|---|
| Forward |
5′-GTAGTTTGCCCAAGGTCAAG-3′ | 433 |
| Reverse |
5′-AGCCACCTGAGGGGTAAG-3′ |
|
PCR amplification
i) PCR system: A 25-µl reaction system consisting of
the following components was used: 2.5 µl 10X PCR reaction buffer,
2 µl 25 mM MgCl2, 2.5 µl 2 mM 4X dNTP, 0.5 µl 20 µM
primer 1, 0.5 µl 20 µM primer 2, 3 µl genomic DNA and 0.3 µl 5 U/µl
Taq DNA polymerase. The solution was made up with double-distilled
water to a total volume of 25 µl. The components were placed in a
tube immersed in an ice water bath, well mixed in a short time in a
vortexing shaker, and centrifuged briefly prior to being placed
into the PCR machine.
ii) PCR conditions: The mixture was amplified under
the following cycling conditions: Initial denaturation at 94°C for
3 min for 1 cycle; denaturation at 94°C for 45 sec and annealing at
60°C for 1 min for 35 cycles; extension at 72°C for 1 min and a
final extension at 72°C for 6 min for 1 cycle
iii) Detection of PCR products: The amplified
fragment length was 433 bp. A 3 µl quantity of PCR products was
electrophoresed on 2% agarose gel (stained with GoldView™ DNA dye,
0.5 µl/10 ml) under 120 V and 60 mA for 20 min, and observed in a
UV transilluminator to determine whether a target gene fragment of
433 bp was obtained.
Restriction enzyme digestion of PCR products
i) Digestion system: A 15 µl quantity of PCR
products was digested with Alw26I in a 20-µl reaction system
comprising the following components: 15 µl PCR products, 2 µl 10X
buffer and 1 µl 10 U/µl Alw26Ia, made up with
double-distilled water to a total volume of 20 µl. The components
were placed in a tube immersed in an ice water bath, well mixed in
a vortexing shaker, centrifuged briefly, and placed into a 37°C
water bath.
ii) Enzyme digestion conditions: The mixtures were
placed in a 37°C water bath for 3 h.
iii) Determination of digestion results: A 10 µl
quantity of the digestion products was electrophoresed on 2%
agarose gel (stained with GoldView™ DNA dye, 0.5 µl/10 ml) under
120 V and 60 mA for 20 min. The digestion results were observed
with a UV transilluminator and recorded.
iv) Determination of GSTP1 genotypes according to
digestion results: No Alw26I restriction site was present at
nucleotide 1578 of the wild-type allele. However, the A→G mutation
created a restriction site at nucleotide 1578. The presence of one
restriction site indicated A/A, and the presence of two and three
restriction sites indicated A/G and G/G, respectively. Genotypes
were determined as follows: A/A: 328 and 105 bp, wild-type
homozygote; A/G: 328, 222, 106 and 105 bp, mutant heterozygote;
G/G: 222, 106 and 105 bp; mutant homozygote.
v) Alw26I restriction sites are: 5′…G T C T C
(N) ↓…3′ and 3′…C A G A G (N)↑ …5′.
Statistical analysis
Data were analyzed using SPSS software, version 10.0
(SPSS, Inc., Chicago, IL, USA). Allele and genotype frequencies
were compared between lung cancer and control groups by Chi-square
test. Lung cancer risk assessment was presented as the odds ratio
(OR) and 95% confidence interval (CI). Logistic regression analysis
was performed to calculate OR and 95% CI. In addition, OR was
adjusted by gender, age, and smoking status based on the actual
statistical data.
Results
PCR amplification of DNA and digestion
of PCR products
PCR amplification of genomic DNA
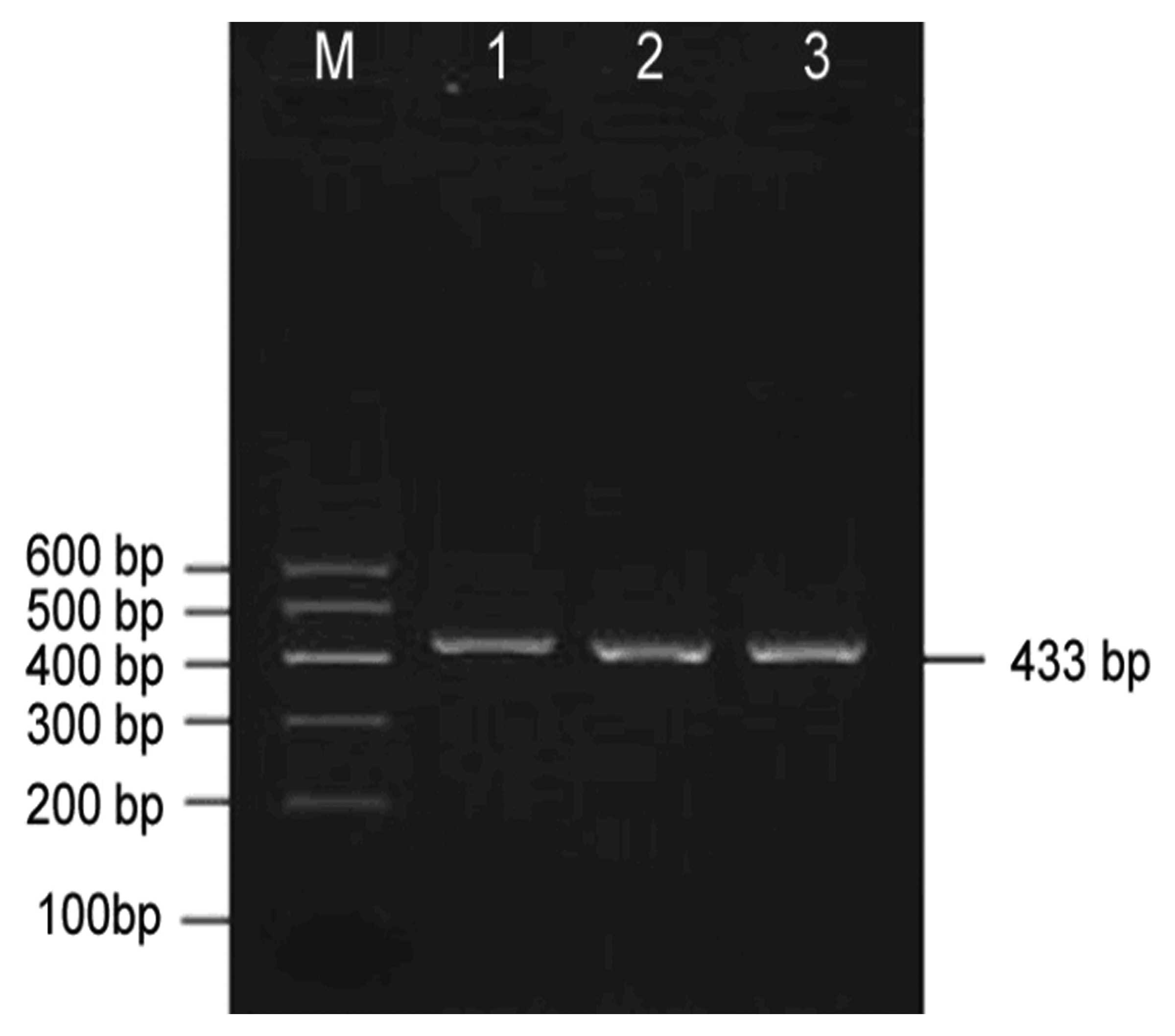
The GSTP1 gene from all study samples was
successfully amplified by PCR. An electrophoretogram of PCR
products is shown in Fig. 1. A
target fragment of 433 bp was observed, which was consistent with
the theoretical length from the primer design. There was a high
yield of PCR products. No non-specific bands were observed.
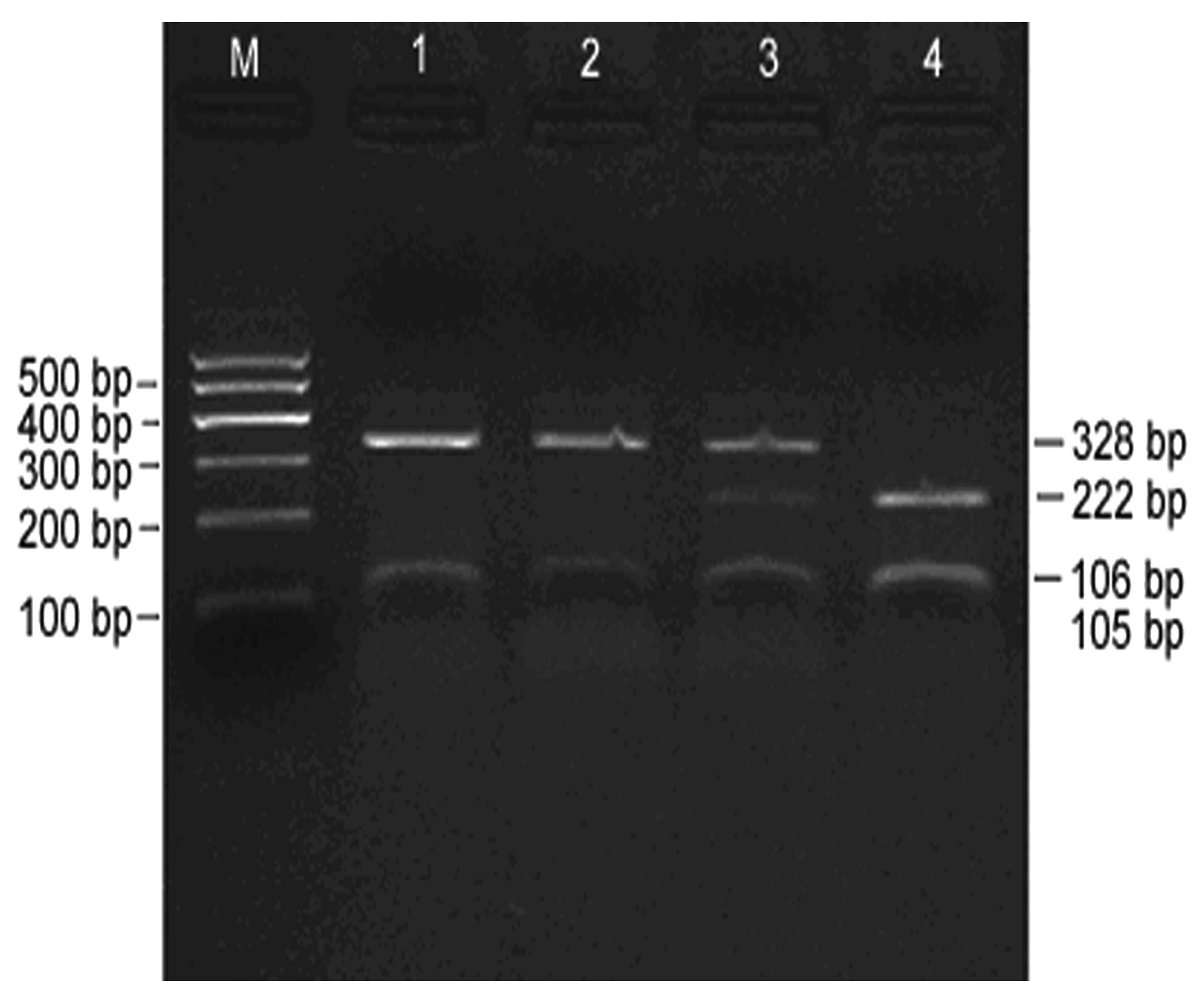
Digestion of PCR products
An electrophoretogram of Alw26I-digested PCR
products is shown in Fig. 2,
indicating effective digestion. As it is sometimes difficult to
distinguish 105 and 106 bp during electrophoresis, they appeared as
a single band. However, this did not affect the determination of
GSTP1 genotypes. Three target fragments with lengths of 328, 222
and 105–106 bp, respectively, were obtained following the digestion
of PCR products, and three different genotypes, A/A, A/G and G/G,
were observed.
Allele and genotype frequencies of
GSTP1
Allele frequencies of GSTP1 in the control and
lung cancer groups
In this study, GSTP1 polymorphisms were analyzed in
302 subjects, including 150 healthy control subjects and 152 lung
cancer patients. The C and G allele frequencies of GSTP1 in the
control and lung cancer groups are shown in Table II. No significant difference was
found between the two groups (P=0.135).
 | Table II.Distribution of GSTP1 alleles in the
control and lung cancer groups. |
Table II.
Distribution of GSTP1 alleles in the
control and lung cancer groups.
|
|
| Alleles of GSTP1, n
(%) |
|
|
|---|
|
|
|
|
|
|
|---|
| Group | No. of alleles | A | G | χ2 | P-value |
|---|
| Control | 304 | 234 (77.0) | 70 (23.0) | 2.229 | 0.135 |
| Lung cancer | 300 | 215 (71.7) | 85 (28.3) |
|
|
Genotype frequencies of GSTP1 in the control and
lung cancer groups
GSTP1 polymorphisms were analyzed in the 302
subjects. The frequencies of three different genotypes, A/A, A/G
and G/G, of GSTP1 in the control and lung cancer groups are shown
in Table III. No significant
difference was observed between the two groups (P=0.223).
 | Table III.Distribution of GSTP1 genotypes in the
control and lung cancer groups. |
Table III.
Distribution of GSTP1 genotypes in the
control and lung cancer groups.
|
|
| Genotypes of GSTP1, n
(%) |
|
|
|---|
|
|
|
|
|
|
|---|
| Group | n | A/A | A/G | G/G | χ2 | P-value |
|---|
| Control | 152 | 86 (56.6) | 62 (40.8) | 4 (2.6) | 3.004 | 0.223 |
| Lung cancer | 150 | 74 (49.3) | 67 (44.7) | 9 (6.0) |
|
|
Goodness of fit of GSTP1 genotype
distribution
All subjects in the lung cancer and control groups
were genotyped for GSTP1 exon 5. The goodness of fit of the GSTP1
genotype distribution in the healthy control group to
Hardy-Weinberg equilibrium was tested. The actual and expected
numbers of healthy controls with the three genotypes are shown in
Table IV. No significant difference
was observed between the actual and expected distributions
(P=0.372). This indicated that GSTP1 genotype frequencies in the
study population fitted the Hardy-Weinberg equilibrium,
demonstrating that the genotype results of this study conform to
this genetic law.
 | Table IV.Actual and expected numbers of GSTP1
genotypes in the control group. |
Table IV.
Actual and expected numbers of GSTP1
genotypes in the control group.
|
| Genotypes of
GSTP1 |
|
|
|---|
|
|
|
|
|
|---|
| Item | A/A | A/G | G/G | χ2 | P-value |
|---|
| Expected | 90 | 54 | 8 | 1.976 | 0.372 |
| Actual | 86 | 62 | 4 |
|
|
Correlation between GSTP1
polymorphisms and lung cancer risk
Since the activity of GST is significantly lower
with the A/G and G/G genotypes than with the A/A genotype, the
former has a decreased ability to detoxify activated carcinogens,
and their carriers thus have an increased susceptibility to lung
cancer (4). Therefore, the A/G and
G/G genotypes were placed in the same category when analyzing the
correlation between GSTP1 polymorphisms and lung cancer risk.
Specifically, the A/G and G/G genotypes represented the non-A/A
genotype. Subjects with the A/A genotype were regarded as a
reference group to evaluate the risk of lung cancer.
Correlation between GSTP1 polymorphisms and
overall lung cancer risk
Overall, 50.7% of the subjects in the lung cancer
group carried the non-A/A genotype of GSTP1, which was higher than
the 43.4% of the control group. The risk of lung cancer in the
subjects with the non-A/A genotype was 1.43-fold higher that that
of the subjects with the A/A genotype; however, no statistical
significance was identified (P=0.138; Table V).
 | Table V.Correlation between genetic
polymorphisms of GSTP1 and susceptibility to lung cancer and
histology of lung cancer. |
Table V.
Correlation between genetic
polymorphisms of GSTP1 and susceptibility to lung cancer and
histology of lung cancer.
|
|
| Genotypes of GSTP1, n
(%) |
|
|
|
|---|
|
|
|
|
|
|
|
|---|
| Group | n | A/A | Non-A/A | ORa | 95% CI | P-value |
|---|
| Control | 152 | 86 (56.6) | 66 (43.4) | 1.00 |
|
|
| Lung cancer | 150 | 74 (49.3) | 76 (50.7) | 1.43 | 0.890–2.306 | 0.138 |
| SQ | 70 | 27 (38.6) | 43 (61.4) | 2.31 | 1.180–4.512 | 0.015 |
| AD | 61 | 34 (55.7) | 27 (44.3) | 1.12 | 0.579–2.180 | 0.730 |
| Others | 19 | 9
(47.4) | 10 (52.6) | 1.71 | 0.636–4.617 | 0.286 |
Correlation between GSTP1 polymorphisms and risk
of different histological types of lung cancer
The lung cancer group was stratified by different
histological types of lung cancer. Comparison with the control
group suggested that the non-A/A genotype was significantly
associated with an increased risk of lung squamous cell carcinoma.
The risk of squamous cell lung carcinoma in subjects with the
non-A/A genotype was 2.31-fold higher than that in those with the
A/A genotype (95% CI: 1.180–4.512, P=0.015). This correlation was
not observed in adenocarcinoma or other types of lung cancer
(Table V).
Correlation between GSTP1 polymorphisms and lung
cancer risk in populations with different smoking statuses
The lung cancer and control groups were stratified
by smoking status and intensity and analyzed. In terms of smoking
status, GSTP1 polymorphisms were found to have no correlation with
susceptibility to lung cancer in never, former or current smokers
(all P>0.05). In terms of smoking intensity, GSTP1 polymorphisms
were found to have no association with susceptibility to lung
cancer in mild and heavy smokers (Table
VI).
 | Table VI.Correlation between genetic
polymorphisms of GSTP1 and susceptibility to lung cancer according
to smoking status. |
Table VI.
Correlation between genetic
polymorphisms of GSTP1 and susceptibility to lung cancer according
to smoking status.
| Item | Control, n (%) | Lung cancer, n
(%) | ORa | 95% CI | P-value |
|---|
| Total subjects | 152 | 150 |
|
|
|
| Non-smokers | 100 | 52 |
|
| 0.102 |
|
A/A | 55 (55.0) | 25 (48.1) | 1.00 |
|
|
|
Non-A/A | 45 (45.0) | 27 (51.9) | 1.85 | 0.884–3.878 |
|
| Ex-smokers | 4 | 12 |
|
| 0.756 |
|
A/A | 1 (25.0) | 4 (33.3) | 1.00 |
|
|
|
Non-A/A | 3 (75.0) | 8 (66.7) | 0.67 | 0.051–8.639 |
|
| Current
smokers | 48 | 86 |
|
| 0.415 |
|
A/A | 30 (62.5) | 45 (52.3) | 1.00 |
|
|
|
Non-A/A | 18 (37.5) | 41 (47.7) | 1.36 | 0.649–2.853 |
|
| PY<30 | 33 | 41 |
|
| 0.347 |
|
A/A | 22 (66.7) | 31 (52.1) | 1.00 |
|
|
|
Non-A/A | 11 (33.3) | 18 (43.9) | 1.61 | 0.598–4.318 |
|
| PY≥30 | 15 | 45 |
|
| 0.766 |
|
A/A | 8 (53.3) | 22 (48.9) | 1.00 |
|
|
| Non-A/A | 7 (46.7) | 23 (51.1) | 1.19 | 0.371–3.852 |
|
Discussion
The GSTP1 gene is located on chromosome 11q13 and
consists of 7 exons and 6 introns, with a length of ~2.8 kb. Board
et al (5) isolated cDNA
clones of the human GSTP1 gene in 1989, and were the first to
report the polymorphisms of the human GSTP1 gene in exons 5
(Ile105→Val) and 6 (Ala114→Val). Exon 5
polymorphism is mutation from A to G at position 1578 of exon 5 of
the GSTP1 gene, turning isoleucine (Ile) into valine (Val) at the
105th amino acid at the protein level. This segment is important
for the biological function of proteins. The substitution of the
105th amino acid decreases the thermal stability and catalytic
activity of the enzyme (2). GSTP1
exon 5 polymorphism has two alleles, A and G, and has three
genotypes, that is, wild-type homozygote A/A (Ile/Ile), mutant
heterozygote A/G (Ile/Val) and mutant homozygote G/G (Val/Val).
Exon 6 polymorphism is mutation from C to T at position 2293 of
exon 6 of the GSTP1 gene, turning alanine (Ala) into valine (Val)
at the 313th amino acid at the protein level. This segment has no
significant impact on the biological function of the protein. The
substitution of the 313th amino acid does not affect the thermal
stability and catalytic activity of the enzyme (2). GSTP1 exon 6 polymorphism also has two
alleles, C and T, and has three genotypes, that is, wild-type
homozygote C/C (Ala/Ala), heterozygote C/T (Ala/Val) and mutant
homozygote T/T (Val/Val). In the present study, the exon 6 GSTP1
gene polymorphism and its correlation with susceptibility to lung
cancer were not investigated in light of the facts that: i) Exon 6
polymorphism does not affect the activity of GSTP1; ii) the
majority of studies have found the exon 6 polymorphism to have no
association with the risk of lung cancer (6,7); iii)
the mutant T allele was not found to be present in exon 6 in a
study on GSTP1 polymorphism in the Japanese population (8); and iv) Zhang et al also did not
find the mutant T allele in exon 6 in a population from Shanghai
(9).
A number of studies have reported the distribution
of GSTP1 exon 5 polymorphisms in different populations (4,10–12). The
results of the present study indicated that the mutant G allele of
the GSTP1 gene occurred at a frequency of 23.0% in a Chinese
population, and that the A/A, A/G, and G/G genotypes of the GSTP1
gene occurred at frequencies of 56.6, 40.8 and 2.6%, respectively.
The frequencies of the GSTP1 exon 5 polymorphism genotypes and the
mutant T allele in populations from different regions reported by
relevant studies are listed in Table
VII. As shown in Table VII,
the mutant G allele is most frequently observed in African
Americans (42%), and occurs least frequently in Chinese (23%);
while the frequency has been found to be 33–36% in European
Americans, British and other Caucasians, and 26% in a Japanese
population (4,10–12).
Although these samples may not be truly representative of their
respective race, these results at least suggest the difference in
the distribution of GSTP1 alleles and genotypes among Chinese,
African Americans and Europeans.
 | Table VII.GSTP1 genotypes and allele
frequencies in different populations. |
Table VII.
GSTP1 genotypes and allele
frequencies in different populations.
|
|
| Genotypes of GSTP1,
n (%) |
|
|
|---|
|
|
|
|
|
|
|---|
| Population | n | A/A | A/G | G/G | G allele | Study |
|---|
| Chinese | 151 | 86
(56.6) | 62
(40.8) | 4 (2.6) | 0.23 | Present study |
| Japanese | 50 | 26
(52.0) | 22
(44.0) | 2 (4.0) | 0.26 | Ishii (8) |
| African
American | 137 | 48
(35.0) | 63
(46.0) | 26 (19.0) | 0.42 | Watson (9) |
| European
American | 287 | 119 (42.0) | 147 (51.0) | 21 (7.0) | 0.33 | Watson (9) |
| English | 151 | 64
(42.4) | 74
(49.0) | 13 (8.6) | 0.33 | Lewis (10)a |
| Caucasian | 151 | 61
(40.4) | 72
(47.7) | 18 (11.9) | 0.36 | Wenzlaff (11) |
Human GSTs detoxify toxins by catalyzing the binding
of toxins to glutathione, thus protecting intracellular
macromolecules from injury induced by cytotoxins and carcinogens
(13). The main human GSTs are GSTA,
GSTM, GSTT and GSTP (or α, µ, θ and π in the old nomenclature).
GSTP1 is an important member of the phase II metabolic enzymes
GSTs, which are exogenous compounds, and is the only identified
member of the GSTP family so far. GST genes, including the GSTP1
gene, have polymorphisms, and the enzymatic activity varies among
different genotypes. The GSTP1 gene is located on human chromosome
11q13, and is widely expressed in human epithelial tissues, such as
lung, kidney, and prostate, but is not expressed or expressed at
low levels in the liver (14). GSTP1
is expressed at the highest level in the lung tissue among all GST
isozymes, accounting for 83% of all GST isozymes (15). Therefore, GSTP1 plays an important
role in the detoxification of inhaled carcinogens, such as
activated polycyclic aromatic hydrocarbons (PAHs), benzo(a)pyrene
and tobacco carcinogens (16). It
has even been suggested to be the most important GST influencing
lung cancer risk (17). The GSTP1
gene has polymorphisms in exon 5. Watson et al (4) reported the association between GSTP1
activity and GSTP1 exon 5 polymorphisms in 1998, and determined the
GSTP1 activity and GSTP1 exon 5 polymorphism genotypes in lung
tissue of 34 subjects. Their results indicated that GSTP1 activity
was significantly higher in subjects with the A/A genotype than in
those with the A/G or G/G genotype (P=0.014 and P=0.05,
respectively), demonstrating that the presence of the mutant G
allele in exon 5 significantly reduced the enzyme activity. Related
studies have also confirmed this finding. It has been demonstrated
that GSTP1 exon 5 Val allele decreases the GSTP activity, and that
higher levels of hydrophobic DNA adducts are present in the lung
tissue in individuals with low activity alleles (17), in addition to increased levels of
PAH-DNA adducts in peripheral blood lymphocytes (18). The GSTP1 exon 5 polymorphisms change
the structure, function and expression level of GSTP1, alter the
body's ability to detoxify carcinogens, and affect individual
susceptibility to lung cancer.
Previous studies have demonstrated that GSTP1
polymorphisms are associated with many tumors, such as rectal
cancer, bladder cancer, prostate cancer, head and neck cancer, and
breast cancer. Although GSTP1 is expressed at a very high level in
human lung tissue, the correlation between GSTP1 polymorphisms and
lung cancer risk has not yet been widely investigated. Case-control
studies found GSTP1 exon 5 polymorphisms to be associated with lung
cancer risk (2). However, other
research results suggested that GSTP1 polymorphisms were not
statistically associated with lung cancer risk (15,17). In
the present study, the non-A/A genotype of GSTP1 occurred at
frequencies of 43.4 and 50.7% in the control and lung cancer
groups, respectively. Although a higher frequency of non-A/A
genotype was observed in the lung cancer group than in the control
group, there was no significant difference between the two groups
(P=0.138). This indicates that GSTP1 exon 5 polymorphisms were not
significantly associated with lung cancer susceptibility. The
stratified analysis by histological type of lung cancer revealed a
significant difference in the frequency of the susceptible GSTP1
genotype, non-A/A, between the control group and lung squamous cell
carcinoma group; and that subjects with the GSTP1 mutant allele had
a higher risk of lung squamous cell carcinoma (OR=2.31, 95% CI:
1.180–4.512, P=0.015), which is consistent with the findings of
Ryberg et al (3). As for lung
adenocarcinoma, GSTP1 exon 5 polymorphisms were not found to be
associated with it in both a previous study (1) and the present study. It is known that
lung squamous cell carcinoma mostly originates from the large
bronchi, while lung adenocarcinoma mostly originates from
peripheral lung tissue. A higher expression level of GSTs has been
observed in bronchial epithelium than in terminal small airway
epithelium (19). The distribution
characteristics of GSTs in the lung account for the association of
GSTP1 polymorphisms with lung squamous cell carcinoma, and its
non-association with lung adenocarcinoma. Furthermore, stratified
analysis by smoking status and intensity in the lung cancer and
control groups indicated non-association of GSTP1 exon 5
polymorphisms with lung cancer risk in nonsmokers and smokers, and
in mild and heavy smokers. This result may be explained by the
findings of Yang and Xian (20),
that the GSTP1 levels were not associated with gender, age or
smoking status, and that certain GSTP1 inducers, such as PAHs and
diet may influence the expression of GSTP1.
In summary, the present study has demonstrated that
GSTP1 exon 5 polymorphisms are associated with lung cancer
susceptibility. However, stratified analysis suggested the
correlation of GSTP1 exon 5 polymorphisms with lung squamous cell
carcinoma risk, and that exon 5 polymorphisms might increase the
risk of lung squamous cell carcinoma. These results suggest that
exon 5 GSTP1 polymorphisms are not a strong influencing factor in
lung cancer risk, but may have a certain effect. These are only
preliminary findings, which can be evaluated in future in-depth
studies; for example, the correlation between GSTP1 polymorphisms
and lung cancer susceptibility can be investigated by measuring DNA
adduct levels on the basis of the present study.
Acknowledgements
This study was supported by a grant from The
Affiliated Hospital of Inner Mongolia Medical University.
References
|
1
|
Gu JD, Hua F, Mei CR, Zheng DJ, Wang GF
and Zhou QH: HapMap-based study on the association between MPO and
GSTP1 gene polymorphisms and lung cancer susceptibility in Chinese
Han population. Acta Pharmacol Sin. 35:636–644. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cote ML, Chen W, Smith DW, et al: Meta-
and pooled analysis of GSTP1 polymorphism and lung cancer: a
HuGE-GSEC review. Am J Epidemiol. 169:802–814. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ryberg D, Skaug V, Hewer A, et al:
Genotypes of glutathione transferase M1 and P1 and their
significance for lung DNA adduct levels and cancer risk.
Carcinogenesis. 18:1285–1289. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Watson MA, Stewart RK, Smith GBJ, et al:
Human glutathione S-transferase P1 polymorphisms: Relationship to
lung tissue enzyme activity and poulation frequency distribution.
Carcinogenesis. 19:275–280. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Board P, Smith S, Green J, Coggan M and
Suzuki T: Evidence against a relationship between fatty acid ethyl
ester synthase and the Pi class glutathione S-transferase in
humans. J Biol Chem. 268:15655–15658. 1993.PubMed/NCBI
|
|
6
|
Yang L, Yang X, Ji W, et al: Effects of a
functional variant c.353T≥C in snai1 on risk of two contextual
diseases. Chronic obstructive pulmonary disease and lung cancer. Am
J Respir Crit Care Med. 189:139–148. 2014.PubMed/NCBI
|
|
7
|
Fathy M, Hamed M, Youssif O, Fawzy N and
Ashour W: Association between environmental tobacco smoke exposure
and lung cancer susceptibility: modification by antioxidant enzyme
genetic polymorphisms. Mol Diagn Ther. 18:55–62. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nishina T, Takano Y, Denda T, et al: A
phase II clinical study of mFOLFOX6 plus bevacizumab as first-line
therapy for Japanese advanced/recurrent colorectal cancer patients.
Jpn J Clin Oncol. 43:1080–1086. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang CS, Tan Z, Lu L, et al: Polymorphism
of Prodynorphin promoter is associated with schizophrenia in
Chinese population. Acta Pharmacol Sin. 25:1022–1026.
2004.PubMed/NCBI
|
|
10
|
Ishii T, Teramoto S and Matsuse T: GSTP1
affects chemoresistance against camptothecin in human lung
adenocarcinoma cells. Cancer Lett. 216:89–102. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lewis AD, Forrester LM, Hayes JD, et al:
Glutathione S-transferase isoenzymes in human tumours and tumour
derived cell lines. Br J Cancer. 60:327–331. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wenzlaff AS, Cote ML, Bock CH, et al:
CYP1A1 and CYP1B1 polymorphisms and risk of lung cancer among never
smokers: a population-based study. Carcinogenesis. 26:2207–2212.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Buratti FM, Scardala S, Funari E and
Testai E: Human glutathione transferases catalyzing the conjugation
of the hepatoxin microcystin-LR. Chem Res Toxicol. 24:926–933.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chan QK, Khoo US, Chan KY, et al: Promoter
methylation and differential expression of pi-class glutathione
S-transferase in endometrial carcinoma. J Mol Diagn. 7:8–16. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Oguztüzun S, Aydin M, Demirag F, et al:
The expression of GST isoenzymes and p53 in non-small cell lung
cancer. Folia Histochem Cytobiol. 48:122–127. 2010.PubMed/NCBI
|
|
16
|
Haroun RA, Zakhary NI, Mohamed MR,
Abdelrahman AM, Kandil EI and Shalaby KA: Assessment of the
prognostic value of methylation status and expression levels of
FHIT, GSTP1 and p16 in non-small cell lung cancer in Egyptian
patients. Asian Pac J Cancer Prev. 15:4281–4287. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kalaivani P, Saranya S, Poornima P, et al:
Biological evaluation of new nickel(II) metallates: Synthesis,
DNA/protein binding and mitochondrial mediated apoptosis in human
lung cancer cells (A549) via ROS hypergeneration and depletion of
cellular antioxidant pool. Eur J Med Chem. 82:584–599. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shantakumar S, Gammon MD, Eng SM, et al:
Residential environmental exposures and other characteristics
associated with detectable PAH-DNA adducts in peripheral
mononuclear cells in a population-based sample of adult females. J
Expo Anal Environ Epidemiol. 15:482–490. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Aynacioglu AS, Nacak M, Filiz A, Ekinci E
and Roots I: Protective role of glutathione S-transferase P1
(GSTP1) Val105Val genotype in patients with bronchial asthma. Br J
Clin Pharmacol. 57:213–217. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang Y and Xian L: The association between
the GSTP1 A313G and GSTM1 null/present polymorphisms and the
treatment response of the platinum-based chemotherapy in non-small
cell lung cancer (NSCLC) patients: a meta-analysis. Tumour Biol.
35:6791–6799. 2014. View Article : Google Scholar : PubMed/NCBI
|