Introduction
Over recent decades, the Enterobacter cloacae
complex (consisting of Enterobacter cloacae, Enterobacter
asburiae, Enterobacter hormaechei, Enterobacter kobei, Enterobacter
ludwigii and Enterobacter nimipressuralis) has taken on
clinical significance, with its species emerging as nosocomial
pathogens, particularly in intensive care units (ICUs). E.
cloacae is the most representative species of the E.
cloacae complex (1). This
species can cause severe opportunistic infections in hospital
patients, including lower respiratory, urinary tract and wound
infections, as well as hospital-acquired sepsis (2). The increasing prevalence of E.
cloacae is due predominantly to a high level of resistance to
antimicrobial agents. SHV-type extended-spectrum β-lactamases
(ESBLs), encoded by the blaSHV gene confer
significant resistance to β-lactam antibiotics (3). SHV-type ESBLs have >140 variants,
and the SHV-12-type ESBL is one of the most prevalent types in Asia
(4,5).
SHV-12 ESBL can be located on the chromosome, a
plasmid or both (6) and can be
transmitted horizontally and/or vertically. The location of the
SHV-12 ESBL gene and the pathway of transmission vary by region,
hospital and time span (7,8).
Bacteria continuously adjust gene expression in
response to environmental changes. Currently, central venous
catheters, malignant tumors, history of antibiotic usage and Pitt
bacteremia score are regarded as the risk factors for bacterial
infection (9); however, associated
risk factors are distinct based on differences in hospital
settings, practitioners, usage of medications and the condition of
the patient, suggesting that additional external factors may be at
play in the evolution of antibiotic resistance for E.
cloacae (10–12).
Currently, the association between SHV-12 ESBL and
the antibiotic resistance of E. cloacae has not been
reported in the Guangdong region of China. In this study,
therefore, the prevalence of E. cloacae and the correlation
between the antibiotic resistance of E. cloacae and the
prevalence status of SHV-12 ESBL were analyzed, a transmission
model was delineated, and external factors that affect the location
and transmission of SHV-12 ESBL were investigated. By analyzing the
association between the SHV-12 ESBL gene and the antibiotic
resistance of E. cloacae with the combined effect of both
internal and external factors, the aim was to provide evidence for
the proper usage of antibiotics to help control the spread of
drug-resistant genes.
Materials and methods
Bacterial isolates
Unique isolates of E. cloacae (n=100) were
collected from randomly selected inpatients (62 men and 38 women)
from the Affiliated Hospital of Guangdong Medical College
(Zhanjiang, China) and the Taiping People's Hospital of Dongguan
(Dongguan, China) between June 2012 and July 2013. All patients
provided informed consent. Bacteria were identified using the BD
Phoenix Automated Microbiology System (BD Pheonix™ 100; BD
Biosciences, Franklin Lakes, NJ, USA) and the corresponding
identification card. Escherichia coli ATCC 25922, E.
coli C600 and SHV-12 ESBL-negative and -positive strains were
from the Laboratory of Pathogenic Biology of Guangdong Medical
College.
Antimicrobial susceptibility
Antimicrobial susceptibility testing was performed
on 100 isolates by agar dilution, as recommended by the Clinical
and Laboratory Standards Institute (13). The antibiotics were β-lactams
[ampicillin (AM), cefazolin, cefuroxime (CXM), ceftazidime (CAZ),
cefepime, cefoxitin (FOX), aztreonam (AZT), sulbactam +
cefoperazone (SCF), tazobactam + piperacillin (TZP), imipenem
(IMP), fluoroquinolone [ciprofloxacin (CIP) and levofloxacin
(OFL)], aminoglycosides [amikacin (AMK) and gentamycin (GEN)] and a
sulfonamide [sulfamethoxazole (SXT)]. All antibiotics were
purchased from the National Institute for Food and Drug Control
(Beijing, China)
Sequencing of SHV-12 ESBLs
The plasmid was eliminated using the
variable-temperature-sodium dodecyl sulfate plasmid elimination
method as described previously (14). Plasmid removal was verified using
agarose gel electrophoresis. Chromosomal DNA was prepared using the
FastPure™ DNA kit [Takara Biotechnology (Dalian) Co., Ltd., Dalian,
China] and plasmid DNA was prepared using the Plasmid Mini kit
(Qiagen, Hilden, Germany) following the manufacturers'
instructions. The purified DNA was used as a template for PCR using
an upstream primer pair, SHV-12-U (5′-GGTTATGCGTTATATTCGCC-3′), and
a downstream primer pair, SHV-12-D (5′-TTAGCGTTGCCAGTGCTC-3′),
which were synthesized in Sangon Biotech (Shanghai) Co., Ltd.
(Shanghai, China). The desired polymerase chain reaction (PCR)
product of the SHV-12 ESBL gene was 865 bp. The PCR product was
purified using a PCR purification kit [Takara Biotechnology
(Dalian) Co., Ltd.] and cloned into the pMDTM 18-T vector.
Recombinant plasmids were transformed into E. coli DH5α
using T-A cloning; the transformants were selected using PCR and
were then subjected to DNA sequencing. The PCR cycle conditions
were as follows: Initial denaturation at 95°C for 5 min;
denaturation at 95°C for 1 min, annealing at 52°C for 30 min and
extension at 72°C for 1 min, repeated for 30 cycles; final
extension at 72°C for 5 min. The resulting sequences were aligned
with available GenBank data using the Basic Local Alignment Search
Tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Genotyping of enterobacteria
Enterobacterial repetitive intergenic consensus
(ERIC)-PCR was used to genotype the clinical isolates. ERIC1R
(5′-ATGTAAGCTCCTGGGGATTCAC-3′) and ERIC2R
(5′-AAGTAAGTGACTGGGGTGAGCG-3′) primers that were complementary to
ERIC sequences of E. cloacae genomic DNA were used for
ERIC-PCR, which was performed as described by Stumpf et al
(15).
Conjugation assay
Mueller-Hinton agar (National Institute for Food and
Drug Control) containing rifampicin (200 µg/ml) and ceftriaxone (5
µg/ml) was used to select the E. cloacae.
Rifampicin-resistant E. coli C600 (LacZ−
Nalr F− Rifr) was used as the
recipient strain, and the ungrown C600 was used as the donor
strain. E. coli transconjugants were selected on
Mueller-Hinton agar plates containing rifampicin (200 µg/ml) and
ceftriaxone (5 mg/l), and bacterial colonies with rifampicin and
ceftriaxone-resistance were inoculated onto Eosin-Methylene blue
agar plates (containing the same concentration of rifampicin and
ceftriaxone). Colorless bacterial colonies on Eosin-Methylene blue
agar plates were considered as transconjugants. PCR amplification
of the SHV-12 ESBL gene was performed using plasmid DNA of the
transconjugant as a template and C600 donor strain as a negative
control.
Statistical analysis
An RxC contingency table for the χ2 test,
2×2 contingency table for the χ2 test and Fisher's exact
probability were used to assess whether statistical difference
could be found in the antibiotic resistance rates between the
positive and negative isolates, as well as among the three gene
locations for positive isolates.
SHV-12 ESBL-positive and -negative isolates were
designated as groups 1 and 2, respectively. Group 3 consisted of
E. cloacae Types A, B and C, as typed by ERIC-PCR, while
group 4 consisted of other types. Group 5 consisted of isolates
that tested positive in the conjugation assay, while group 6
consisted of isolates that tested negative. For the comparison
between groups 1 and 2, and groups 3 and 4, variables in these
groups with P-values <0.1 in univariate analyses were separately
entered into a multiple logistic regression model to identify the
independent risk for E. cloacae carrying SHV-12 ESBLs and
vertical spread, respectively. Yates' χ2 test or exact
probability was used to analyze the difference between groups 5 and
6. All comparisons were performed using SPSS 17 software (SPSS,
Inc., Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference for all tests.
Results and Discussion
Association between the SHV-12 ESBL
gene and the resistance of E. cloacae to antibiotics
The SHV-12 ESBL gene was detected in all 58 E.
cloacae isolates expressing ESBLs (Fig. 1). This percentage is higher than that
reported in France (16) and Taiwan
(9), but lower than that in Tunisia
(17). A total of 34.48% (20 out of
58 isolates) of the SHV-12 ESBL genes were located in the bacterial
chromosome, 48.28% (28 out of 58 isolates) in plasmids and 17.24%
(10 out of 58 isolates) in both (Tables
I and II).
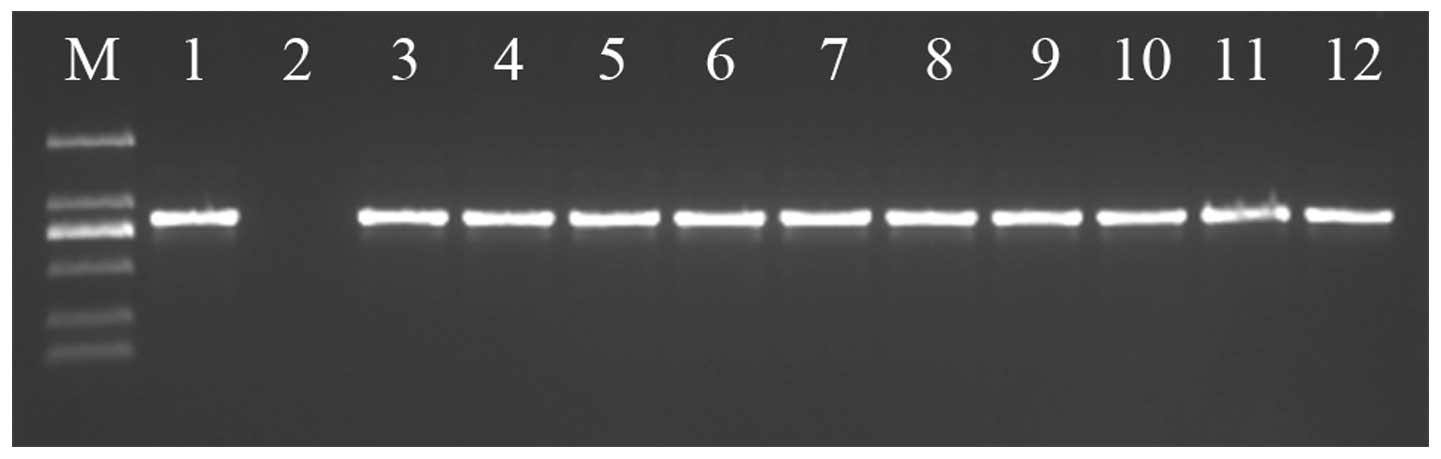 | Figure 1.Polymerase chain reaction
amplification of the SHV-12 extended-spectrum β-lactamase gene in
experimental isolates. M, DNA ladder (from top to bottom: 2,000,
1,000, 750, 500, 250 and 100 bp); 1, positive control; 2, negative
control; 3-l2, experimental isolates. |
 | Table I.Antibiotic resistance and SHV-12
ESBL. |
Table I.
Antibiotic resistance and SHV-12
ESBL.
|
| Resistant isolates,
n (%) |
|
|
|
|---|
|
|
|
|
|
|
|---|
| Antibiotic |
SHV-12-positive |
SHV-12-negative | χ2 | P-value | Total resistance
rate, % |
|---|
| AM | 58
(100) | 42
(100) |
|
| 100 |
| CEF | 58
(100) | 42
(100) |
|
| 100 |
| CXM | 43 (74) | 30 (71) |
0.09 | >0.05 | 73 |
| CAZ | 39 (67) | 20 (48) |
3.88 | <0.05 | 59 |
| FEP | 18 (31) | 10 (24) |
1.59 | >0.05 | 28 |
| FOX | 55 (95) | 27 (64) | 15.40 | <0.05 | 83 |
| TZP | 8
(14) | 3 (7) |
1.10 | >0.05 | 11 |
| SCF | 27 (47) | 12 (29) |
3.31 | >0.05 | 39 |
| AZT | 37 (57) | 14 (24) |
9.04 | <0.05 | 51 |
| IMP | 0 (0) | 0 (0) |
0.00 |
| 0 |
| CIP | 22 (38) | 8
(19) |
4.14 | <0.05 | 30 |
| OFL | 24 (41) | 18 (43) |
0.02 | >0.05 | 42 |
| AMK | 9
(16) | 3 (7) |
1.62 | >0.05 | 12 |
| GEN | 24 (41) | 9
(21) |
4.39 | <0.05 | 33 |
| SXT | 31 (53) | 11 (26) |
7.43 | <0.05 | 42 |
 | Table II.Antibiotic resistance and gene
location of SHV-12 ESBL. |
Table II.
Antibiotic resistance and gene
location of SHV-12 ESBL.
|
|
No. of
resistant isolates |
|---|
|
|
|
|---|
| Gene location (no.
of isolates) | AM | CEF | CXM | CAZ | CPM | FOX | TZP | SCF | AZT | IMP | CIP | OFL | AMK | GEN | SXT |
|---|
| Chromosome
(20) | 20 | 20 | 20 | 7 | 8 | 18 | 2 | 7 | 12 | 0 | 5 | 4 | 1 | 4 | 15 |
| Plasmid (28) | 28 | 28 |
17a |
22a | 5 | 27 | 4 | 11 | 18 | 0 | 12 | 13 | 7 | 10 |
10a |
| Chromosome +
plasmid (10) | 10 | 10 |
6a |
10a | 5 | 10 | 2 |
9a,b | 7 | 0 | 5 |
7a | 1 |
8a, b | 6 |
As shown in Table I,
SHV-12 ESBL-positive E. cloacae isolates demonstrated
significant differences in antibiotic resistance against β-lactam
antibiotics (CAZ, FOX, AZT), fluoroquinolone antibiotics (CIP),
aminoglycoside antibiotics (GEN) and sulfonamides (SXT) compared
with SHV-12 ESBL-negative isolates (P<0.05). It is known that
ESBL-expressing E. cloacae carry resistance against
fluoroquinolone antibiotics (18).
Additionally the highly prevalent class I integron contains
antibiotic resistance genes, mainly for aminoglycoside antibiotics
and trimethoprim (19,20). Similar results have been reported in
isolates from Korean hospitals; however, SHV-12 ESBL-positive and
-negative isolates did not show significant differences in
antibiotic resistance to the β-lactam antibiotics (21). Other mechanisms may, therefore, also
contribute to β-lactam antibiotic resistance.
The AMK-resistance rate in the SHV-12 ESBL
expression strains was found to be 16% in the present study, which
is between that in Mexico (25%) and Taiwan (0%) (22,23). The
AM resistance rate, however, was 100%, which is higher than that in
Mexico (13%) (22). The fact that
the antibiotic-resistance rate of E. cloacae in the current
region is different from that in other areas may derive from the
difference in drug usage and disease type.
SHV-12 ESBL gene location affects
antibiotic resistance
The present results showed that all three types of
gene location of SHV-12 ESBL could be found in the Guangdong
region: A total of 34.48% (20 out of 58 isolates) of the SHV-12
ESBL genes were located in the bacterial chromosome, 48.28% (28 out
of 58 isolates) in plasmids and 17.24% (10 out of 58 isolates) in
both (Table II). The proportion of
each type of location of SHV-12 ESBL in E. cloacae was
similar to that of SHV ESBLs in E. coli in India (24), but was different from the chromosomal
location of SHV-12 ESBL in Klebsiella pneumoniae in Egypt
(6), indicating that the gene
location of SHV-12 ESBL may be distinct in different regions or
different strains.
In the present study, it was found that the three
types of gene locations exhibited significant differences in
antibiotic resistance rates for CXM, CAZ, SCF, OFL, GEN and SXT
(Table II). The resistance rate for
CXM, for example, was higher in the E. cloacae isolates
carrying the chromosomal SHV-12 ESBL gene (100%) than that in
isolates carrying both plasmid and chromosomal SHV-12 ESBL genes
(60%). The opposite was true for CAZ; however, certain antibiotics,
such as TZP, were unaffected by the gene location of SHV-12 ESBL,
indicating that the mechanism of antibiotic resistance in E.
cloacae is complicated and may be compounded by both internal
and external factors.
EPIC-PCR typing
A total of 100 E. cloacae isolates were
divided into 82 different chromosomal gene types via ERIC-PCR. The
isolates were divided into three groups: A, B and C. Type A (9
isolates) and type B (6 isolates) all tested positive for the
SHV-12 ESBL gene. Type C isolates (of which there were 6) tested
negative for the SHV-12 ESBL gene. The remaining 79 isolates
belonged to an orphan clone (Figs. 2
and 3). Of type A, 6 isolates
carried chromosome-coded SHV-12 ESBL, while 3 isolates carried both
chromosomal and plasmid-coded SHV-12 ESBL. Of type B, 4 isolates
carried the chromosomal-coded gene, 1 carried the plasmid-coded
gene and 1 carried both. The differential location of the SHV-12
ESBL gene within and between different bacterial gene types
indicated that E. cloacae carrying SHV-12 ESBL could be
transmitted via both vertical and horizontal transmission.
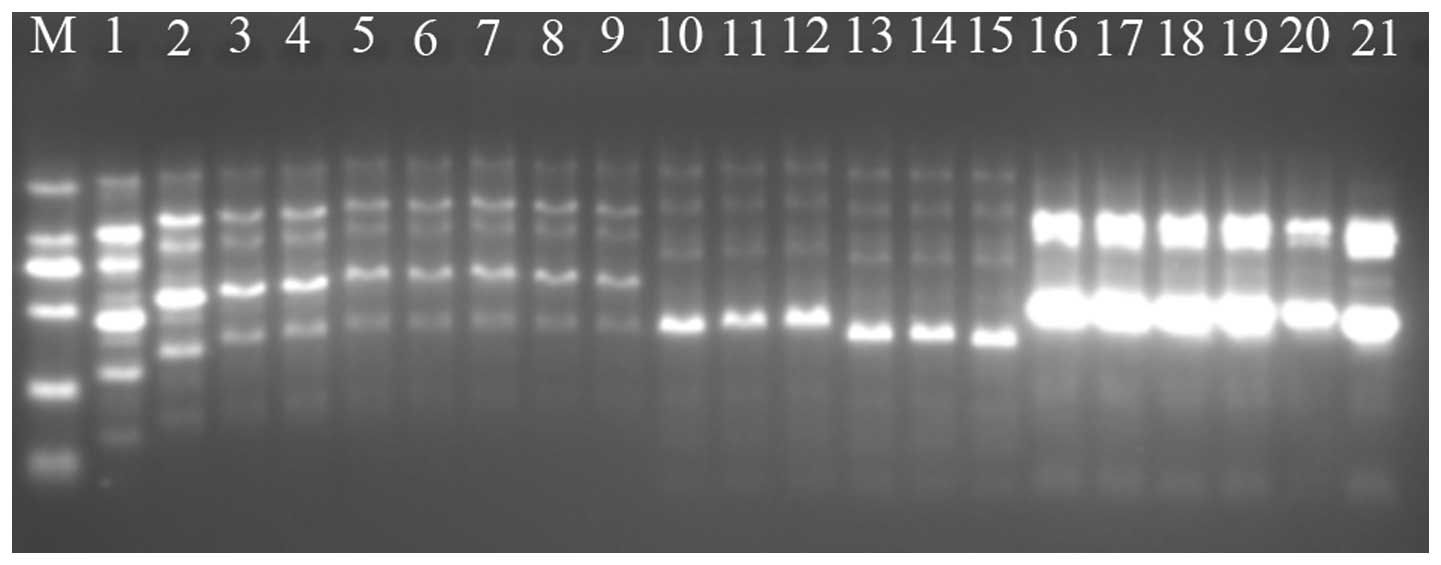 | Figure 2.Enterobacterial repetitive intergenic
consensus-polymerase chain reaction for type A, B and C isolates.
M, DNA ladder (from top to bottom: 2,000, 1,000, 750, 500, 250 and
100 bp); 1–9, type A isolates; 10–15, type B isolates; 16–21, type
C isolates. |
The ERIC-PCR result showed that E. cloacae of
the same gene type could be detected throughout different clinical
departments, suggesting an inter-departmental spread of E.
cloacae (data not shown). Vertical spread, even outbreaks of
E. cloacae carrying SHV-12 ESBLs, has also been reported in
various regions of the world (7,17,25). In
this study, type C E. cloacae carrying no SHV-12 ESBL were
also found to spread between different clinical departments,
implying that other factors could also contribute to the spreading
of antibiotic-resistant strains in hospital.
Transconjugation experiment
A total of 31 E. cloacae isolates carrying
rifampicin (200 µg/ml) plus ceftriaxone (5 mg/l) resistance were
selected for the transconjugation experiment. Of these isolates
52%, carried the SHV-12 ESBL gene. In 94% of the cases, antibiotic
resistance of E. cloacae could be transferred into E.
coli, which indicated that plasmid-mediated resistance was
common in E. cloacae and that antibiotic resistance genes
could be easily spread via horizontal transmission. The positive
rate of transconjugation in the Taiwan region, however, has been
shown to be only 70% (26). In 84%
(26/31) of the transconjugation cases of the present study, only
partial antibiotic resistance genes could be transferred,
demonstrating that certain antibiotic resistance genes could not be
transferred horizontally. It was also reported that antibiotic
resistance genes in Aeromonas spp. could not be transferred
horizontally. In 10% of the cases in the present study, E.
coli acquired complete resistance from E. cloacae, which
indicated that the antibiotic resistance plasmid in E.
cloacae could mediate complete antibiotic resistance (Table III).
 | Table III.Conjugation assays. |
Table III.
Conjugation assays.
|
|
| Transconjugants, n
(%) |
|---|
|
|
|
|
|---|
| SHV-12 location
type (n) | Positive test, n
(%) | SHV-12
positive | Acquired partial
resistance | Acquired complete
resistance |
|---|
| Chromosome (6) | 5
(83) | 0 (0) | 5
(83) | 1
(17) |
| Plasmid (9) |
9 (100) | 6
(67) | 7
(78) | 2
(22) |
| Chromosome and
plasmid (2) |
2 (100) | 1
(50) |
2 (100) | 0 (0) |
| SHV-12 negative
(14) | 13 (93) | 0 (0) | 12 (92) | 1 (8) |
| Total (31) | 29 (94) | 7
(23) | 26 (84) | 3
(10) |
In 64% of the cases, the SHV-12 ESBL gene could be
transconjugated successfully from E. cloacae to E.
coli, which meant that horizontal transmission was a major
method for the spread of SHV-12. A high prevalence of
plasmid-encoded SHV-12 ESBL producers has been observed in the
Sichuan province of China (27) and
Tunisia (16).
Through the Yates' χ2 or Fisher's exact
probability test, it was found that the factors that improved
transconjugation rates in patients were male gender, use of
antibiotics in the last 30 days, hospital stays lasting >14 days
and Charlson comorbidity index (CCI) >2 (P<0.05). This result
also indicated that these factors could affect the horizontal
transmission of antibiotic resistance in clinical niches.
Association between external factors
and E. cloacae antibiotic resistance
The E. cloacae strains were isolated from 7
types of samples collected from 18 clinical departments (Table IV), which meant that E.
cloacae could cause various infections. In the ICU and
respiratory department, E. cloacae had the highest (30%) and
second highest (11%) positive rates, respectively. In Taiwan, the
ICU is also the most common department in which E. cloacae
strains have been isolated (9). In
the region analyzed in the present study, the majority of the E.
cloacae strains were from the sputum; this is similar to the
situation in Tunisia and southern Brazil (17), but different from France and Taiwan,
where the urinary tract is the most common infection site for E.
cloacae (28).
 | Table IV.External factors and antibiotic
resistance. |
Table IV.
External factors and antibiotic
resistance.
|
| Isolates, n(%) |
|---|
|
|
|
|---|
| Factor | Group 1 (n=58) | Group 2 (n=42) | Group 3 (n=21) | Group 4 (n=79) | Group 5 (n=72) | Group 6 (n=3) |
|---|
| Source of
isolates |
|
|
|
|
|
|
|
Sputum | 28 (48) | 21 (50) | 16 (76) | 33 (42) | 35 (78) |
3 (100) |
|
Urine | 10 (17) | 12 (29) | 3
(14) | 19 (24) | 13 (18) | 0 (0) |
| Other
sources1 | 20 (35) | 9
(21) | 2
(10) | 27
(34)b | 24 (33) | 0 (0) |
| Clinical
department |
|
|
|
|
|
|
|
Intensive Care Unit | 27 (47) | 3
(7)a | 11 (52) | 19
(24)b | 27 (38) | 0 (0) |
|
Respiratory Medicine | 6
(10) | 5
(12) | 3
(14) | 8 (10) | 8
(11) | 0 (0) |
| Other
departments2 | 25 (43) | 34
(81)a | 7
(33) | 52
(66)b | 37 (51) |
3 (100) |
| Age in years |
|
|
|
|
|
|
|
1–14 | 7
(12) | 2 (5) | 0 (0) | 9
(11) | 7
(10) | 1
(33) |
|
15–59 | 24 (41) | 13 (31) | 5
(24) | 32 (41) | 28 (39) | 2
(33) |
|
≥60 | 27 (47) | 27 (64) | 16 (76) | 38
(48)b | 37 (51) | 0 (0) |
| Gender |
|
|
|
|
|
|
|
Male | 40 (69) | 22 (52) | 11 (52) | 51 (65) | 50 (69) | 0 (0) |
|
Female | 18 (31) | 20 (48) | 10 (48) | 28 (35) | 22 (31) |
3 (100) |
| Other factors |
|
|
|
|
|
|
|
Invasive procedure | 44 (76) | 20
(48)a | 16 (76) | 48 (61) | 49 (68) | 2
(67) |
|
Presence of any underlying
illnesses and/or comorbidities | 56 (97) | 34
(81)a | 21
(100) | 69 (87) | 64 (89) | 2
(67) |
| Use of
antibiotics in the last 30 days | 53 (91) | 39 (93) | 21
(100) | 71 (90) | 67 (93) | 0 (0)c |
|
Hospitalization >14
days | 53 (91) | 31
(74)a | 21
(100) | 63 (80) | 60 (83) | 0 (0)c |
| CCI
>2 | 51 (88) | 29
(69)a | 19 (90) | 61 (77) | 60 (83) | 0 (0)c |
|
Hospital patients | 57 (98) | 42
(100) | 21
(100) | 77 (97) | 71 (99) |
3 (100) |
|
Mortality | 5 (9) | 0 (0) | 4
(19) | 1 (5) | 4 (6) | 0 (0) |
Respiratory tract infection caused by E.
cloacae occurs in different age groups. Patients aged ≥60 years
had the highest E. cloacae infection rate (54%), and male
patients were more likely to be infected by E. cloacae than
female patients (62 male vs. 38 female patients; male to female
ratio, 1.63:1). A similar effect of age and gender on E.
cloacae infection rate has also been reported in other regions
of the world (16).
In total, >50% of the patients infected with
E. cloacae were undergoing invasive treatment, had
underlying diseases and/or complications, had been taking
antibiotics for >30 days, were hospitalized for >14 days or
had a CCI >2, which further illustrated that infection with
E. cloacae is closely associated with external factors; this
is consistent with other investigations (10–12).
Notably, antibiotic abuse has been considered an
essential reason for antibiotic resistance and the wide spread of
antibiotic-resistant strains; however, no significant effect of
antibiotic usage for >30 days was found in groups 1–4. A
possible reason could be the 3-year application of antibiotic usage
policy in Guangdong, where the samples were collected. In this
study, a higher positive rate of ESBLs was found compared with
other regions of China. One possible explanation is that the
antibiotic resistance gene is preserved in the strain for long
periods of time and spread once it is captured by mobile genetic
elements, such as integrons (29).
As analyzed by multiple logistic regressions, the
risk factors for infection by SHV-12-producing E. cloacae
were age, clinical department and having an underlying disease or
complication (P<0.05). In a previous study, the risk factors for
suffering bloodstream infections of E. cloacae
carrying ESBLs were disease severity, category of
healthcare-associated infection and prior use of antibiotics or a
ventilator (23). This difference
may be associated with the method of drug administration used by
the doctors and the sample source.
E. cloacae isolated from the bile had the
highest antibiotic resistance rate compared with those isolated
from the other sources (data not shown). Although the positive rate
for E. cloacae was low in the pus, secretions, bile and
chest water, the majority of the E. cloacae detected were
positive for SHV-12 ESBL, which is noteworthy for clinicians.
E. cloacae isolated from ICU samples
had the highest positive rate for SHV-12 ESBLs. This may have been
due to the fact that those patients in the ICU had an increased
incidence of underlying diseases or complications. Additionally,
ICU patients often have undergone prior stays in other departments
or have used broad-spectrum antibiotics, which may increase the
possibility of acquiring SHV-12 ESBLs for E. cloacae
(28).
Multiple logistic regression analysis results showed
that the risk factors for vertical transmission were infection
site, age and clinical department. The horizontal spread of SHV-12
ESBLs was the most common between E. cloacae strains
separated from the sputum (76%), the elderly (76%) and ICU patients
(52%), which is consistent with the most common sample type,
clinical department and patient age in this region. Horizontal
transmission of E. cloacae was reported in an ICU in
Croatia, and was common in respiratory tract infections and the
elderly (30).
In conclusion, the expression pattern, gene location
and transmission pathways of SHV-12 ESBL and other external factors
have a close correlation with the antibiotic resistance of E.
cloacae. To prevent the wide spread of SHV-12 ESBL, as well as
outbreaks of multi-drug-resistant strains, we propose that more
attention be given to the clinical investigation of SHV-12
ESBL-expressing E. cloacae to better monitor its spread and
provide evidence for the proper usage of antibiotics.
Acknowledgements
The authors would like to acknowledge the financial
support from the Science and Technology Key Projects Initiative of
Zhanjiang City (grant no. 2012C3106022), the Special Fund for
Science and Technology, Treasury Department of Zhanjiang City
(grant no. 2013A01007), the General Projects of Guangdong Medical
College (grant no. M2012005), the Science and Technological Program
for Dongguan's Higher Education, Science and Research, and Health
Care Institutions (grant no. 01310515000270), as well as the
Medical Research Fund Project of Guangdong Province (grant no.
A2015329).
References
|
1
|
Mezzatesta ML, Gona F and Stefani S:
Enterobacter cloacae complex: Clinical impact and emerging
antibiotic resistance. Future Microbiol. 7:887–902. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xiao YH, Shen P, Wei ZQ, Chen YB, Kong HS,
Yang Q, Zhang WL, Chen X and Li LJ: Mohnarin report of 2011:
Monitoring of bacterial resistance in China. Zhong Hua Yi Yuan Gan
Ran Xue Za Zhi. 22:4946–4952. 2012.(In Chinese).
|
|
3
|
Bradford PA: Extended-spectrum
beta-lactamases in the 21st century: Characterization, epidemiology
and detection of this important resistance threat. Clin Microbiol
Rev. 14:933–951. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hawkey PM: Prevalence and clonality of
extended-spectrum beta-lactamases in Asia. Clin Microbiol Infect.
14(Suppl 1): 159–165. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chia JH, Chu C, Su LH, Chiu CH, Kuo AJ,
Sun CF and Wu TL: Development of a multiplex PCR and SHV
melting-curve mutation detection system for detection of some SHV
and CTX-M beta-lactamases of Escherichia coli, Klebsiella
pneumoniae and Enterobacter cloacae in Taiwan. J Clin
Microbiol. 43:4486–4491. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Newire EA, Ahmed SF, House B, Valiente E
and Pimentel G: Detection of new SHV-12, SHV-5 and SHV-2a variants
of extended spectrum beta-lactamase in Klebsiella pneumoniae
in Egypt. Ann Clin Microbiol Antimicrob. 12:162013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Juhász E, Jánvári L, Tóth A, Damjanova I,
Nobilis A and Kristóf K: Emergence of VIM-4- and SHV-12-producing
Enterobacter cloacae in a neonatal intensive care unit. Int
J Med Microbiol. 302:257–260. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tansawai U, Boonkerd N, Polwichai P,
Dejsirilert S and Niumsup PR: SHV-12 extended spectrum
beta-lactamase associated with high-level ceftazidime resistance in
Enterobacter cloacae isolated from Thailand. Southeast Asian
J Trop Med Public Health. 40:148–154. 2009.PubMed/NCBI
|
|
9
|
Lee CH, Su LH, Li CC, Chien CC, Tang YF
and Liu JW: Microbiologic and clinical implications of bacteremia
due to extended-spectrum-beta-lactamase-producing Klebsiella
pneumoniae with or without plasmid-mediated AmpC beta-lactamase
DHA-1. Antimicrob Agents Chemother. 54:5395–5398. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee CC, Lee NY, Yan JJ, Lee HC, Chen PL,
Chang CM, Wu CJ, Ko NY, Wang LR, Chi CH and Ko WC: Bacteremia due
to extended spectrum-beta-lactamase-producing Enterobacter
cloacae: Role of carbapenem therapy. Antimicrob Agents
Chemother. 54:3551–3556. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu CP, Wang NY, Lee CM, Weng LC, Tseng
HK, Liu CW, Chiang CS and Huang FY: Nosocomial and
community-acquired Enterobacter cloacae bloodstream
infection: Risk factors for and prevalence of SHV-12 in
multiresistant isolates in a medical centre. J Hosp Infect.
58:63–77. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rodríguez-Baño J, Miró E, Villar M, Coelho
A, Gozalo M, Borrell N, Bou G, Conejo MC, Pomar V, Aracil B, et al:
Colonisation and infection due to Enterobacteriaceae producing
plasmid-mediated AmpC β-lactamases. J Infect. 64:176–183. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
CLSI: Performance Standards for
Antimicrobial Susceptibility Testing; Twenty-Second Informational
Supplement. CLSI document M100-S22. Clinical and Laboratory
Standards Institute. (Wayne, PA). 2013.
|
|
14
|
Ni YX: Plasmid annihilation. Basic and
Clinical Microbial Drug Resistance. Zhang ZR, Xia MY and Ni YX:
(Beijing). People's Medical Publishing House. 240–242. 2006.
|
|
15
|
Stumpf AN, Roggenkamp A and Hoffmann H:
Specificity of Enterobacterial repetitive intergenic consensus and
repetitive extragenic palindromic polymerase chain reaction for the
detection of clonality within the Enterobacter cloacae
complex. Diagn Microbiol Infect Dis. 53:9–16. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Biendo M, Manoliu C, Laurans G, Castelain
S, Canarelli B, Thomas D, Hamdad F, Rousseau F and Eb F: Molecular
typing and characterization of extended-spectrum TEM, SHV and CTX-M
beta-lactamases in clinical isolates of Enterobacter
cloacae. Res Microbiol. 159:590–594. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lahlaoui H, Anis BH, Mohamed K and Mohamed
BM: Emergence of SHV-12 extended spectrum beta-lactamase among
clinical isolates of Enterobacter cloacae in Tunisia. Microb
Pathog. 53:64–65. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Miró E, Segura C, Navarro F, Sorlí L, Coll
P, Horcajada JP, Alvarez-Lerma F and Salvadó M: Spread of plasmids
containing the bla(VIM-1) and bla(CTX-M) genes and the qnr
determinant in Enterobacter cloacae, Klebsiella
pneumoniae and Klebsiella oxytoca isolates. J Antimicrob
Chemother. 65:661–665. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu J, Li GM, Zhao Y, Hu XH, Yang WQ and
Yang JR: Study on the resistance genes of avriable region of class
I integron in Enterobacter cloacae. Zhong Guo Kang Sheng Su
Za Zhi. 36:543–547. 2011.(In Chinese).
|
|
20
|
Liu J, Li GM, Zhao Y, Hu XH, Yang WQ and
Yang JR: Correlation among integron positioning, ESBL-production
and resistance in Enterobacter cloacae. Guang Dong Yi Xue.
32:195–198. 2011.(In Chinese).
|
|
21
|
Ko KS, Lee MY, Song JH, Lee H, Jung DS,
Jung SI, Kim SW, Chang HH, Yeom JS, Kim YS, et al: Prevalence and
characterization of extended-spectrum beta-lactamase-producing
Enterobacteriaceae isolated in Korean hospitals. Diagn Microbiol
Infect Dis. 61:453–459. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Garza-González E, Mendoza Ibarra SI,
Llaca-Díaz JM and Gonzalez GM: Molecular characterization and
antimicrobial susceptibility of extended-spectrum
{beta}-lactamase-producing Enterobacteriaceae isolates at a
tertiary-care centre in Monterrey, Mexico. J Med Microbiol.
60:84–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen CH and Huang CC: Risk factor analysis
for extended-spectrum β-lactamase-producing Enterobacter
cloacae bloodstream infections in central Taiwan. BMC Infect
Dis. 13:4172013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sharma J, Sharma M and Ray P: Detection of
TEM & SHV genes in Escherichia coli & Klebsiella
pneumoniae isolates in a tertiary care hospital from India.
Indian J Med Res. 132:332–336. 2010.PubMed/NCBI
|
|
25
|
Nogueira Kda S, Paganini MC, Conte A, Cogo
LL, de Messias Taborda Reason I, da Silva MJ and Dalla-Costa LM:
Emergence of extended-spectrum β-lactamase producing
Enterobacter spp. in patients with bacteremia in a tertiary
hospital in southern Brazil. Enferm Infecc Microbiol Clin.
32:87–92. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yan JJ, Ko WC, Chuang CL and Wu JJ:
Metallo-beta-lactamase-producing Enterobacteriaceae isolates in a
university hospital in Taiwan: Prevalence of IMP-8 in
Enterobacter cloacae and first identification of VIM-2 in
Citrobacter freundii. J Antimicrob Chemother. 50:503–511.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu G, Ling BD, Zeng Y, Lin L, Xie YE and
Lei J: Molecular characterization of extended-spectrum
beta-lactamases produced by clinical isolates of Enterobacter
cloacae from a teaching hospital in China. Jpn J Infect Dis.
61:286–289. 2008.PubMed/NCBI
|
|
28
|
Biendo M, Manoliu C, Laurans G, Castelain
S, Canarelli B, Thomas D, Hamdad F, Rousseau F and Eb F: Molecular
typing and characterization of extended-spectrum TEM, SHV and CTX-M
beta-lactamases in clinical isolates of Enterobacter
cloacae. Res Microbiol. 159:590–594. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Machado E, Ferreira J, Novais A, Peixe L,
Cantón R, Baquero F and Coque TM: Preservation of integron types
among Enterobacteriaceae producing extended-spectrum
beta-lactamases in a Spanish hospital over a 15-year period (1988
to 2003). Antimicrob Agents Chemother. 51:2201–2204. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Novak A, Goic-Barisic I, Andrasevic AT,
Butic I, Radic M, Jelic M, Rubic Z and Tonkic M: Monoclonal
outbreak of VIM-1-carbapenemase-producing Enterobacter
cloacae in intensive care unit, University Hospital Centre
Split, Croatia. Microb Drug Resist. 20:399–403. 2014. View Article : Google Scholar : PubMed/NCBI
|