Introduction
Systemic inflammatory response syndrome (SIRS),
which occurs following systemic infection (sepsis), burns, shock
and major surgery, is a type of cytokine storm, in which there is
an abnormal upregulation of various cytokines (1). SIRS may lead to septic shock, multiple
organ dysfunction syndrome (MODS) and an increased risk of
mortality (1,2). A prospective multicenter analysis of
intensive care units (ICU) was previously conducted to assess the
epidemiology of sepsis, which demonstrated that the incidence and
mortality rates of sepsis were high, and that the annual cost of
hospital care for patients with sepsis was expensive (3,4). The
incidence of sepsis has been shown to be affected by numerous
factors, which involve disruption of inflammatory and immune
responses (1,2,5).
The Toll-like receptor (TLR) family has an important
role in the immune response, and recognizes pathogen-associated
molecular patterns (PAMPs) in order to activate inflammatory
signaling pathways (6,7). Lipopolysaccharide (LPS), which is
released by gram-negative bacteria and is a type of PAMP, is
recognized by TLR4, which subsequently initiates a signaling
cascade that leads to the upregulation of specific cytokines that
have previously been associated with the occurrence and development
of sepsis (7–9). Previous studies reported that the risk
of sepsis, and its mortality rate, was higher in European
populations, as compared with other regional populations (7,8). Single
nucleotide polymorphisms (SNPs) have been shown to affect the
development of immune and inflammatory reactions (7–10), and
there is an issue regarding the significance and distribution of
TLR4 genetic polymorphisms among various ethnicities (7,8,10).
In order to investigate whether TLR4 and
Toll/interleukin-1 receptor (TIR)-associated protein (TIRAP)
genetic polymorphisms, and/or their expression levels, influence
the susceptibility of an individual to sepsis, the present study
analyzed the sequences and expression levels of TLR4 and TIRAP in
106 patients from the ICU of our hospital. In particular, the
following genetic polymorphisms were analyzed: The Ser180Leu allele
of the TIRAP gene, and the Asp299Gly and Thr399I1e alleles of the
TLR4 gene.
Materials and methods
Subjects
A total of 106 patients with sepsis were enrolled in
the present study. The patients were admitted to the medical ICU
between October 2012 and May 2013. At the time of enrollment, the
acute physiology and chronic health evaluation (APACHE II) scores
were recorded for each patient (11). On the basis of the APACHE II scores,
all patients were divided into either sepsis group A (APACHE II
score <20; male, 43; female, 37) or sepsis group B (APACHE II
score >20; male, 14; female, 12). The ratio of male to female
participants was 1.5 to 1, and the mean age was 60-years-old
(range, 44–80 years). All patients satisfied the diagnostic
criteria of the International Sepsis Conference of Washington,
December 2001 (12). In addition,
100 healthy individuals, including 58 males and 42 females (age
range, 40–75 years), from the Medical Examination Center in our
hospital were enrolled in a control group. All individuals in the
control group were without disease or a history of recent
infection. There were no significant differences in age and gender
between the sepsis groups and the control group. The present study
was conducted in accordance with the Declaration of Helsinki, and
with approval from the Ethics Committee of Inner Mongolia Medical
University (Baotou, China). Written informed consent was obtained
from all participants.
Collection of specimens
Venous blood samples (4 ml) were collected from all
patients, and 2 ml blood was added to anticoagulant tubes
containing 2% ethylenediaminetetraacetic acid, which were
subsequently stored at −80°C. Another 2 ml blood was centrifuged at
685 × g for 5–7 min, and the separated serum was stored in tubes at
−80°C. The storage method used was the gradient
temperature-reduction method (13).
Extraction of genomic DNA and
polymerase chain reaction (PCR)
Genomic DNA samples were extracted from peripheral
blood samples using the Blood Gen Mini kit (Beijing ComWin Biotech,
Co., Ltd., Beijing, China), according to the manufacturer's
protocol. The TIRAP (Ser180Leu) and TLR4 (Asp199Gly, Thr399I1e)
genes were amplified using the Veriti™ 96 PCR Amplifier (Applied
Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA), in
which the primers used were as follows: TLR4 (Asp299Gly) forward,
5′-ATACTTAGACTACTACCTCCATG-3′ and reverse, 5′-TTGTTGGAAGTGAAAGTA
AG-3′; TLR4 (Thr399I1e) forward, 5′-TGTTATCAAAGTGATTTTGGGAGAA-3′
and reverse, 5′-AGGTAAATGAGGTTTCTGAGTGATAGG-3′; and
TIRAP/MyD88-adaptor-like (Mal) (Ser180Leu): forward,
5′-AGTGCTGTACCATCGACCTGCTG-3′ and reverse
5′-TTCCCCTTCTCCCTCCTGTAGTAG-3′. All primers were synthesized by
Sangon Biotech Co. Ltd. (Shanghai, China). Each amplification
reaction was conducted in a total volume of 50 µl reaction mixture,
including 16 µl genomic DNA, 2 µl forward primer and 2 µl reverse
primer, 25 µl Taq 2X PCR Master Mix (Beijing Tiangen
Biotech, Co., Ltd., Beijing, China) and 5 µl nuclease-free water.
Following initial denaturation at 94°C for 3 min, the genomic DNA
was amplified by 35 cycles of PCR (94°C for 30 sec, 58°C for 45
sec, and 72°C for 1 min), which was followed by a final 5 min
extension at 72°C.
The PCR products (30 µl) were separated by 2%
agarose gel electrophoresis, and were visualized by staining with
0.5 µg/ml ethidium bromide (Thermo Fisher Scientific, Inc.) in an
Electrophoresis Meter (100 V; 50 A; 30 min; Wide Mini-Sub® Cell GT;
Bio-Rad Laboratories, Inc., Hercules, CA, USA). Images were then
captured under ultraviolet (UV) light. DNA Marker I (Beijing ComWin
Biotech, Co., Ltd.) was used as a standard. Subsequently, the PCR
products were purified using the Midi Purification kit (Beijing
Tiangen Biotech, Co., Ltd.).
Restriction fragment length
polymorphism (RFLP)
Purified Asp299Gly (TLR4), Thr399I1e (TLR4) and
Ser180Leu (TIRAP) alleles were digested using the restriction
enzymes NcoI, HinfI and Eam1105I, respectively
(Thermo Fisher Scientific, Inc.). The enzyme digestion reaction
mixture consisted of 2 µl 10X FastDigest Green Buffer (Thermo
Fisher Scientific, Inc.), 2 µl restriction enzyme (NcoI,
HinfI or Eam1105I), 10 µl PCR products and 6 µl
nuclease-free water. The reaction occurred at 37°C for 5 min, after
which the genotypes were separated by 2% agarose gel
electrophoresis (labclinics, Barcelona, Spain), and visualized
using ethidium bromide staining, UV illumination and image
capturing. DNA Marker 20 (Takara Bio, Inc., Otsu, Japan) was used
as an internal standard.
PCR products (20 µl) prior to enzyme digestion and
10 µl each of the forward and reverse primers for TLR4 (Asp299Gly,
Thr399I1e) and TIRAP (Ser180Leu) were sequenced by the Beijing
Genomics Institute (BGI; Beijing, China). Sequencing results were
investigated using Chromas 2.31 software (Technelysium, Pty Ltd.,
South Brisbane, Australia). TA repeat sequences upstream of the
A(TA)nTAA polymorphism in the promoter region of the UDP
glucuronosyltransferase 1 family gene were analyzed using CLC
Sequence Viewer software (version 4.6.1; CLC bio, Waltham, MA, USA)
to conduct sequence alignments with template sequences in the
GenBank database (Human TLR4, NC_000009; Human TIRAP, NC_000011;
http://www.ncbi.nlm.nih.gov/genbank/).
Enzyme-linked immunosorbent assay
(ELISA)
The protein expression levels of TLR4 and TIRAP in
peripheral blood samples were detected using the Human TLR4 and
TIRAP ELISA kits (R&D Systems, Inc., Minneapolis, MN, USA), in
accordance with the manufacturer's protocol (Thermo Scientific
Multiskan MK3; Thermo Fisher Scientific, Inc.).
Statistical analysis
Statistical analyses were conducted using the
statistical SPSS17.0 software package (SPSS, Inc., Chicago, IL,
USA). Quantitative variables were expressed as the mean ± standard
deviation. Comparisons of continuous variables between two groups
were conducted using one-way analysis of variance (ANOVA) for
parameters with a normal distribution, and the rank-sum test for
parameters with an abnormal distribution. Three-way ANOVA was used
in order to compare differences among the three groups. In
addition, the genetic frequencies test was used to assess the
Hardy-Weinberg equilibrium. Differences in genotype distributions
among the groups were analyzed using the χ2 test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
PCR products
The PCR products were separated by agarose gel
electrophoresis, and were visualized by staining with ethidium
bromide under UV light. (Fig. 1).
Each band on the electrophoretogram represents a single PCR
product, including TLR4 (Asp299Gly; 218 bp), TLR4 (Thr399I1e; 405
bp) and TIRAP (Ser180Leu; 161 bp).
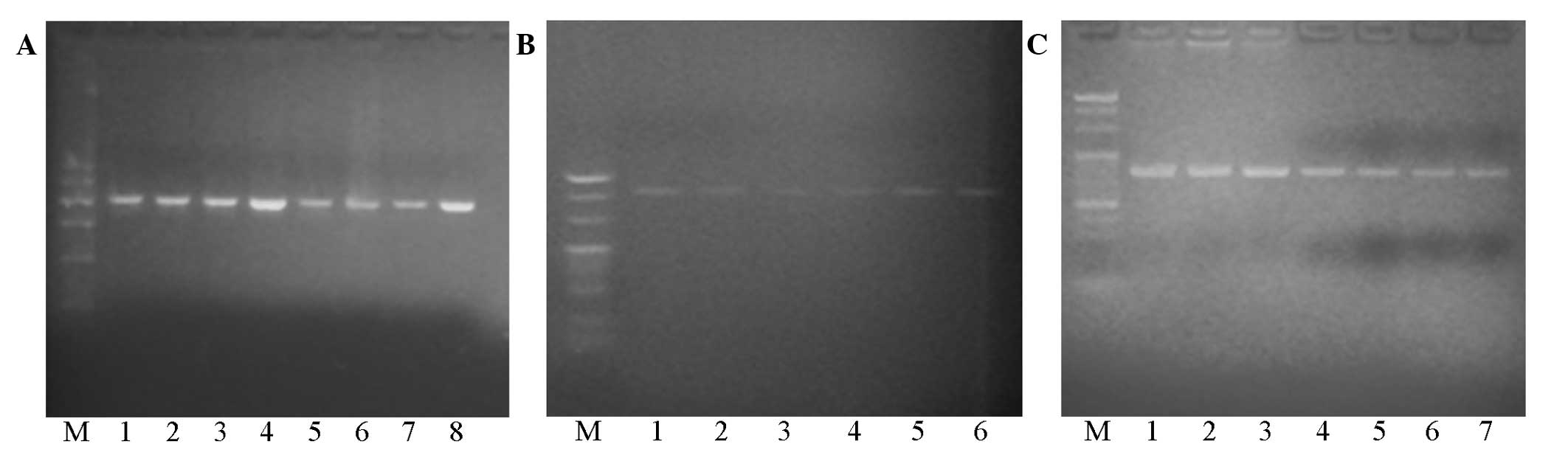
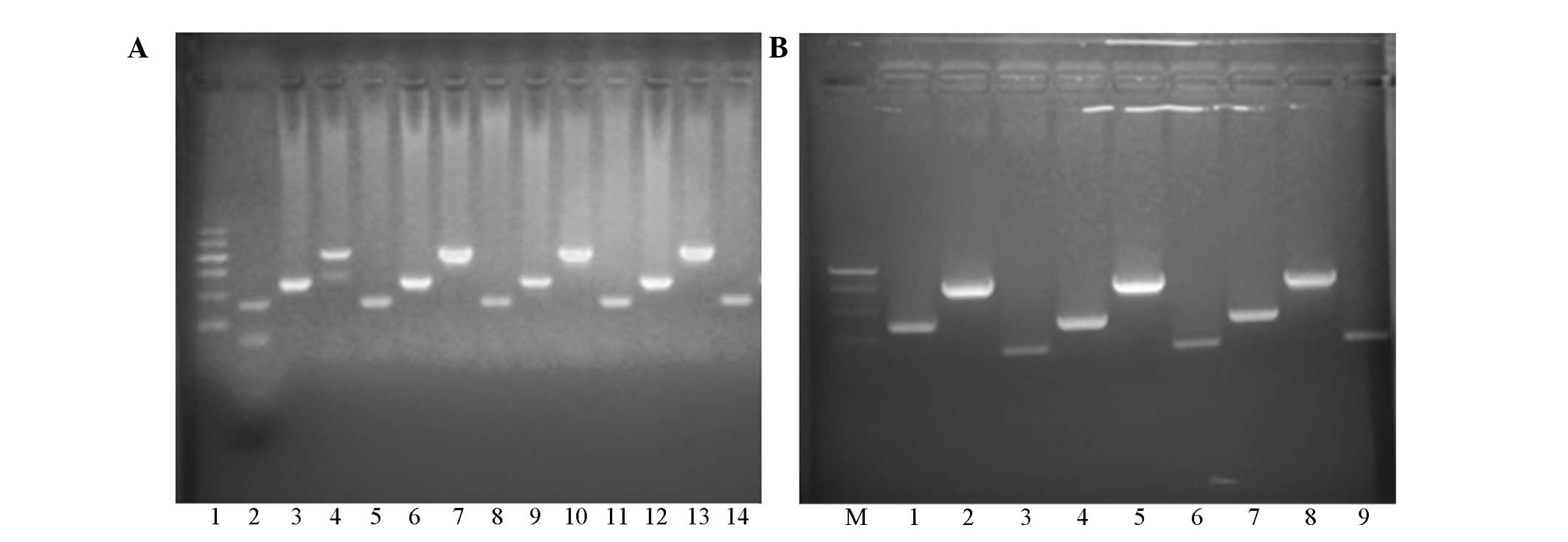 | Figure 1.(A) Electrophoresis of polymerase
chain reaction (PCR) products. Lane 1, DNA Marker I (100, 200, 300,
400, 500 and 600 bp, from top-to-bottom); lane 2, positive control;
lanes 3, 6, 9 and 12, Asp299Gly (TLR4) gene (218 bp); lanes 4, 7,
10 and 13, Thr399Ile (TLR4) gene (405 bp); lanes 5, 8, 11 and 14,
Ser180Leu (TIRAP/Mal) gene (161 bp). (B) Electrophoresis of
purified PCR products. Lane M, DNA Marker 20 (200, 300, 400 and 500
bp, from top-to-bottom); lanes 1, 4 and 7, Asp299Gly (TLR4) gene
(218 bp); lanes 2, 5 and 8 Thr399Ile (TLR4) gene (405 bp); lanes 3,
6 and 9 Ser180Leu (TIRAP/Mal) gene (161 bp). |
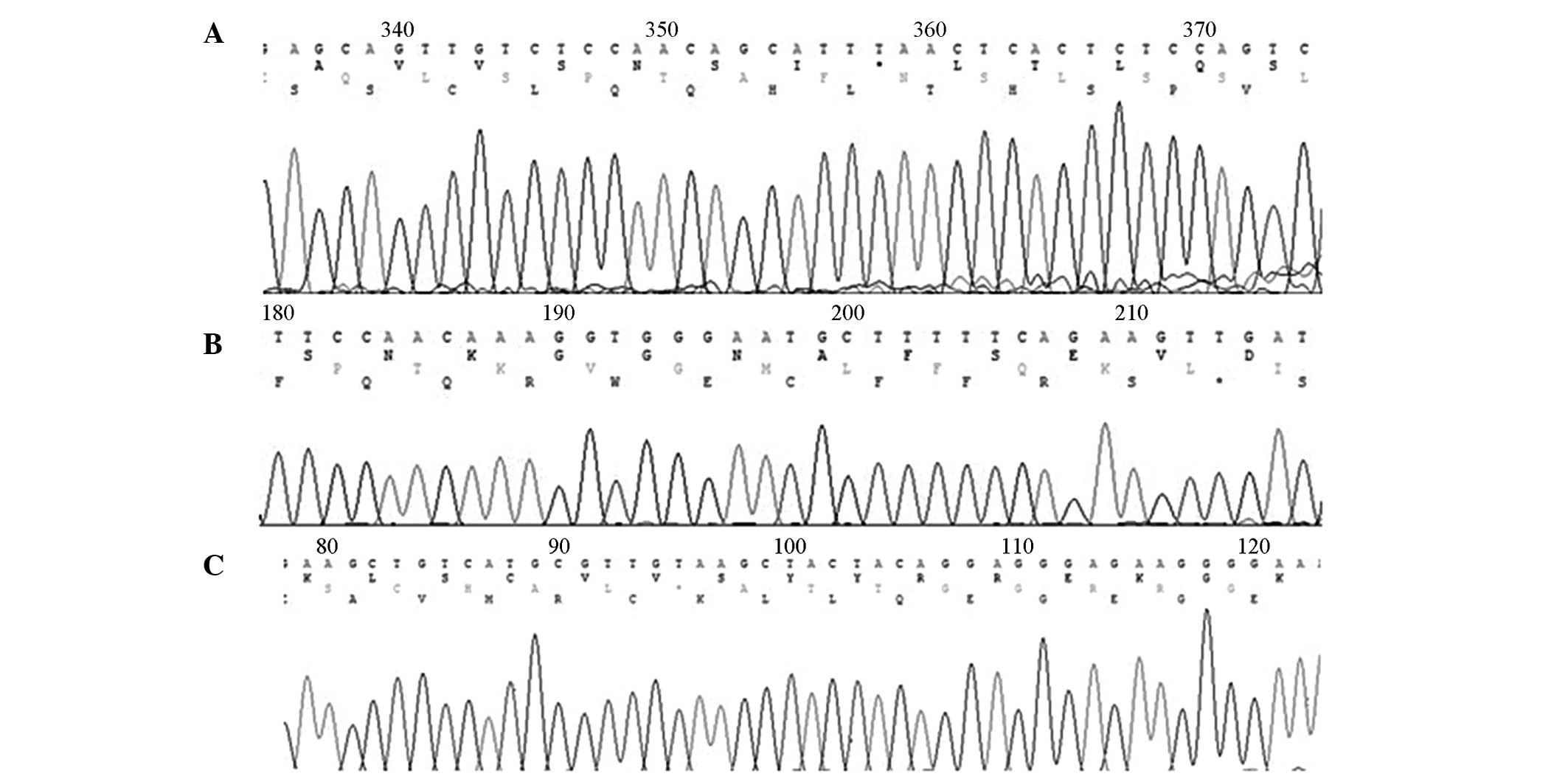
Confirmation of enzyme digestion
Purified PCR products were digested using
restriction enzymes and were separated by agarose gel
electrophoresis (Fig. 2). A single
band corresponding to the TLR4 Asp299Gly allele occurred at 218 bp
following digestion by the NcoI restriction enzyme; thus
indicating the presence of the Asp/Asp genotype only (Fig. 2A). In addition, a single band
corresponding to the TLR4 Thr399I1e allele occurred at 405 bp
following digestion by the HindI restriction enzyme; thus
indicating the presence of the Thr/Thr genotype only (Fig. 2B). Furthermore, a single band
corresponding to the TIRAP Ser180Leu allele occurred at 161 bp
following digestion by the Eam1105I restriction enzyme,
which suggested that only the Ser/Ser genotype was present
(Fig. 2C). These results indicated
that the purified TLR4 and TIRAP genes were not digested by the
restriction enzymes; thus suggesting that there were no genetic
polymorphisms in the TLR4 and TIRAP genes in the sepsis groups, as
compared with the control group.
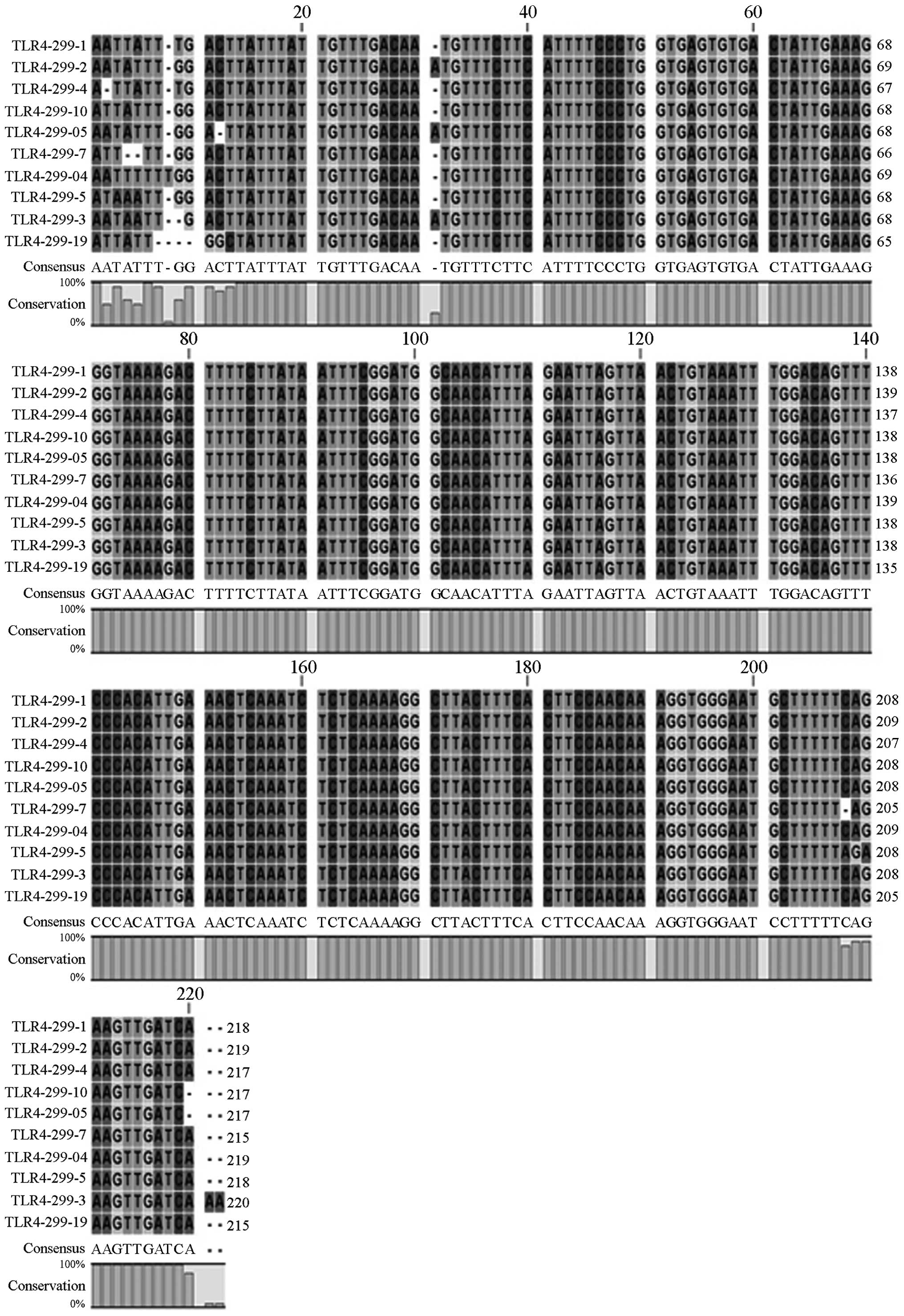
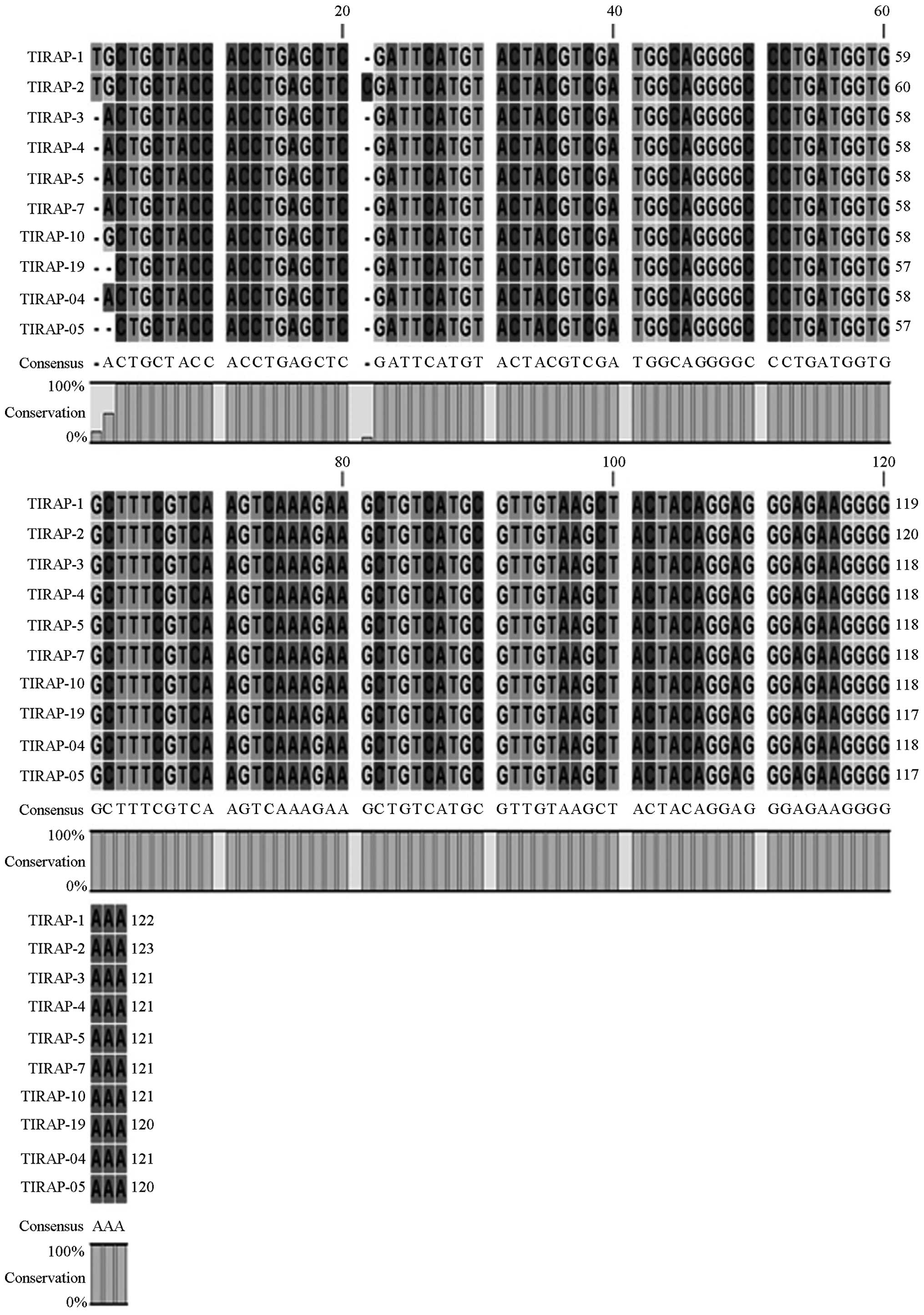
Gene sequencing
Following enzyme digestion, all PCR products were
sequenced by the BGI, which confirmed the gene sequences and
mutations (Fig. 3). Sequencing
results demonstrated that the three gene maps were almost identical
to the template sequences in Genbank. Using the Chromas 2.31
software, sequence alignments of ten samples demonstrated that
there were no mutations in the TLR4 (Asp299Gly, Thr399I1e) and
TIRAP (Ser180Leu) genes in the sepsis group, as compared with the
control group (Figs. 4, 5 and 6).
TLR4 and TIRAP protein expression
levels
TLR4 and TIRAP protein expression levels were
significantly different between the control, sepsis A and sepsis B
groups (TLR4: F=116.550, P<0.01; TIRAP: F=94.950, P<0.01;
Table I).
 | Table I.Protein expression levels of TLR4 and
TIRAP (ng/ml; n=206). |
Table I.
Protein expression levels of TLR4 and
TIRAP (ng/ml; n=206).
|
| Control group | Sepsis group A
(APACHE II≤20) | Sepsis group B
(APACHE II>20) | F value | P-value |
|---|
| TLR4 |
0.886±0.058a,b |
2.253±0.379c | 4.494±0.709 | 116.550 | <0.001 |
| TIRAP |
5.216±0.410a,b |
9.540±2.294c | 19.206±1.755 |
94.950 | <0.001 |
An APACHE II score of 20 was used as a baseline in
order to differentiate sepsis severity. Pearson analysis
demonstrated that TLR4 and TIRAP protein expression levels were
positively correlated with sepsis severity (TLR4: r=0.931,
P<0.05; TIRAP: r=0.972, P<0.05; Table I). Furthermore, the TLR4 protein
expression levels were positively correlated with the TIRAP protein
expression levels (r=0.936; P<0.05; Table I).
Discussion
SNPs are the most common type of human genetic
variant, accounting for 2–3 cm of the human genome, and occurring
at a higher density than microsatellites (14). In addition, SNPs that exhibit a high
genetic stability may directly affect protein expression or
structure and therefore underlie numerous genetic disorders (~30%
SNPs have been associated with disease) (15). Furthermore, SNPs have previously been
associated with the metabolism of numerous drugs. Therefore, the
analysis of SNPs has been widespread in biomedical research and
drug development (15).
In previous studies, specific genetic polymorphisms
have been associated with the susceptibility to, and clinical
features and prognosis of, sepsis (9,16). In
addition, it has been suggested that genetic variation may
influence sepsis development and increase the risk of MODS during
severe infections (16).
Furthermore, TLR-mediated signaling pathways have a critical role
in the mechanisms underlying the development of sepsis, and
alterations in TLR gene structures have previously been associated
with susceptibility to sepsis and sepsis severity (17).
The majority of sepsis cases (>50%) are caused by
gram-negative bacteria, which are predominantly recognized by TLR4
(18). Previous studies reported
that C3H/HeJ TLR4 gene knock-out mice exhibited low sensitivity to
LPS (19,20). In addition, Hagberg et al
(21) reported that C3H/HeJ mice
exhibited significantly increased susceptibility to gram-negative
bacteria in an experimental model of urinary tract infections.
However, TLR4 variation was not associated with the development of
organ injury or cellular stress responses in severe sepsis.
Therefore, there exists a controversy regarding the effects of TLR4
genetic polymorphisms on the susceptibility to and development of
sepsis.
Various mutations in the extracellular domain of
TLR4 have been identified, including Asp299Gly and Thr399I1e
substitution mutations (22). Arbour
et al (23) demonstrated that
the Asp299Gly mutation was able to interrupt TLR4-mediated LPS
signaling, whereas Schröder and Schumann (24) were unable to identify a direct
correlation between Asp299Gly genetic polymorphisms and
susceptibility to infections caused by gram-negative bacteria. It
is possible that the Asp299Gly and Thr399I1e alleles exhibit
co-segregation, including the wild-type/wild-type,
Asp299Gly/wide-type, Thr399I1e/wide-type and Asp299Gly/Thr399I1e
haplotypes. A previous study demonstrated that the
Asp299Gly/wild-type haplotype was associated with increased
LPS-induced release of tumor necrosis factor (TNF)-α (25); however, there was no significant
difference between the other haplotypes and the release of TNF-α.
In addition, as the Asp299Gly/wild-type polymorphism is uncommon in
the human population, there is no confidence that the association
between this haplotype and susceptibility to gram-negative bacteria
is relevant to humans (25).
The incidence and development of sepsis may be
associated with polygenic polymorphisms. The present study enrolled
106 patients previously diagnosed with sepsis, and sequenced the
Asp299Gly and Thr399I1e alleles of TLR4; however, mutations in
these alleles were not identified. These results are consistent
with a previous study which reported that the Asp299Gly allele was
associated with sepsis in the European population, but not in the
Asian population, and that the Asp299Gly and Thr399I1e TLR4 genetic
polymorphisms are uncommon in the Asian population (25). In addition, Okayama et al
(26) were unable to detect the
Asp299Gly allele of the TLR4 gene in specimens from Japanese
patients, and Lin et al (27)
analyzed DNA samples from healthy individuals and patients with
shock in the Chinese Han population, and were unable to identify
Asp299Gly polymorphisms.
TLR-mediated signaling involves at least four
intracellular signaling adaptor molecules, including TIRAP, which
is also known as Mal (28).
TIRAP/Mal acts as a bridging adaptor, which has an important role
in downstream inflammatory responses mediated by TLR2 and TLR4
(29). In addition, genetic
polymorphisms in TIRAP/Mal have been shown to affect the incidence
and development of diseases (30).
Kumpf et al (30) suggested
that the TIRAP Ser180Leu (rs8177374) allele may increase
susceptibility to infections. Furthermore, previous studies
reported that TIRAP (rs8177374 and rs7932766) polymorphisms may
influence the production of inflammatory cytokines and have
important consequences for the susceptibility to and severity of
infections (31). However, the
present study was unable to identify the rs8177374 polymorphism,
which was consistent with previous studies that demonstrated that
the frequencies of the rs8177374 and rs7932766 alleles were higher
in European populations, as compared with Asian populations, and
were in fact rarely detected in Asian populations (31,32).
TLR4 and TIRAP have important roles in TLR-mediated
inflammatory signaling pathways in severe immune responses
(33). In addition, alterations in
the expression levels of TLR4 and TIRAP have previously been
associated with the extent of inflammatory reactions (33). Tsujimoto et al (34) reported that the serum TLR4 protein
expression levels increased following infection of patients with
pathogens, and that the protein expression levels of TLR4 increased
markedly in patients with sepsis. Numerous studies have previously
demonstrated that the severity of acute lung injury was dependent
on systemic inflammatory reactions (35,36). As
TIRAP activates intracellular signaling pathways via TLR-mediated
recognition of various types of pathogen, the TIRAP adaptor protein
has an integral role in the development of acute lung injury. TIRAP
is essential for MyD88-dependent signaling downstream of TLR2 and
TLR4 (its expression was shown to increase following stimulation of
TLR2 and TLR4), initiating a signaling cascade that culminates in
the nuclear localization of nuclear factor-κB and the activation of
the pro-inflammatory response (37).
In the present study, the protein expression levels
of TLR4 and TIRAP were significantly different among the control
group, sepsis group A (APACHE II<20) and sepsis group B (APACHE
II≥20). An APACHE II score of 20 was used as a baseline in order to
differentiate sepsis severity, and Pearson analysis demonstrated
that the protein expression levels of TLR4 and TIRAP were
positively correlated with sepsis severity. In addition, the TLR4
protein expression levels were positively correlated with the TIRAP
protein expression levels. These results suggested that the
inflammatory response and severity of sepsis were associated with
serum TLR4 and TIRAP protein expression levels.
The present study was unable to detect specific
polymorphisms in the TIRAP (Ser180Leu) and TLR4 genes (Asp299Gly,
Thr399I1e), which may have been associated with susceptibility to
sepsis and sepsis severity. However, the protein expression levels
of TLR4 and TIRAP in peripheral blood samples were positively
correlated with sepsis severity, and were shown to have synergistic
effects. It is possible that other previously described
polymorphisms in the TLR4 and TIRAP genes may be associated with
susceptibility to sepsis, and these should be the focus of future
studies.
References
|
1
|
Fry DE: Sepsis, systemic inflammatory
response and multiple organ dysfunction: The mystery continues. Am
Surg. 78:1–8. 2012.PubMed/NCBI
|
|
2
|
Maloney PJ: Sepsis and septic shock. Emerg
Med Clin North Am. 31:583–600. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lopez-Bushnell K, Demaray WS and Jaco C:
Reducing sepsis mortality. Med Surg Nurs. 23:9–14. 2014.
|
|
4
|
Mayr FB, Yende S and Angus DC:
Epidemiology of severe sepsis. Virulence. 5:4–11. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Martin GS: Sepsis, severe sepsis and
septic shock: Changes in incidence, pathogens and outcomes. Expert
Rev Anti Infect Ther. 10:701–706. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wiersinga WJ, Leopold SJ, Cranendonk DR
and van der Poll T: Host innate immune responses to sepsis.
Virulence. 5:36–44. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Esposito S, Molteni CG, Zampiero A, Baggi
E, Lavizzari A, Semino M, Daleno C, Groppo M, Scala A, Terranova L,
et al: Role of polymorphisms of toll-like receptor (TLR) 4, TLR9,
toll-interleukin 1 receptor domain containing adaptor protein
(TIRAP) and FCGR2A genes in malaria susceptibility and severity in
Burundian children. Malar J. 11:1962012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Plantinga TS, Ioana M, Alonso S, Izagirre
N, Hervella M, Joosten LA, van der Meer JW, de la Rúa C and Netea
MG: The evolutionary history of TLR4 polymorphisms in Europe. J
Innate Immun. 4:168–175. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Namath A and Patterson AJ: Genetic
polymorphisms in sepsis. Crit Care Clin. 25:835–856. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang H, Wei C, Li Q, Shou T, Yang Y, Xiao
C, Yu M, Li M, Yang Z, Zhang J and Zheng B: Association of TLR4
gene non-missense single nucleotide polymorphisms with rheumatoid
arthritis in Chinese Han population. Rheumatol Int. 33:1283–1288.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sekulic AD, Trpkovic SV, Pavlovic AP,
Marinkovic OM and Ilic AN: Scoring systems in assessing survival of
critically ill ICU patients. Med Sci Monit. 21:2621–2629. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Levy MM, Fink MP, Marshall JC, Abraham E,
Angus D, Cook D, Cohen J, Opal SM, Vincent JL and Ramsay G:
SCCM/ESICM/ACCP/ATS/SIS: 2001 SCCM/ESICM/ACCP/ATS/SIS International
Sepsis Definitions Conference. Crit Care Med. 31:1250–1256. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sunna A, Gibbs MD, Chin CW, Nelson PJ and
Bergquist PL: A gene encoding a novel multidomain
beta-1,4-mannanase from Caldibacillus cellulovorans and
action of the recombinant enzyme on kraft pulp. Appl Environ
Microbiol. 66:664–670. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jin SB, Zhang XF, Lu JG, Fu HT, Jia ZY and
Sun XW: Genetic analysis of QTL for eye cross and eye diameter in
common carp (Cyprinus carpio L.) using microsatellites and
SNPs. Genet Mol Res. 14:3557–3569. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen HY, Huang W, Leung VH, Fung SL, Ma
SL, Jiang H and Tang NL: Functional interaction between SNPs and
microsatellite in the transcriptional regulation of insulin-like
growth factor 1. Hum Mutat. 34:1289–1297. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Papathanassoqlou ED, Giannakopoulou MD and
Bozas E: Genomic variations and susceptibility to sepsis. AACN Adv
Crit Care. 17:394–422. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Salomao R, Brunialti MK, Gomes NE, Mendes
ME, Diaz RS, Komninakis S, Machado FR, da Silva ID and Rigato O:
Toll-like receptor pathway signaling is differently regulated in
neutrophils and peripheral mononuclear cells of patients with
sepsis, severe sepsis, and septic shock. Crit Care Med. 37:132–139.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Angus DC, Linde-Zwirble WT, Lidicker J,
Clermont G, Carcillo J and Pinsky MR: Epidemiology of severe sepsis
in the United States: Analysis of incidence, outcome and associated
costs of care. Crit Care Med. 29:1303–1310. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Akira S, Takeda K and Kaisho T: Toll-like
receptors: Critical proteins linking innate and acquired immunity.
Nat Immunol. 2:675–680. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Janeway CA Jr and Medzhitov R: Innate
immune recognition. Annu Rev Immunol. 20:197–216. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hagberg L, Hull R, Hull S, McGhee JR,
Michalek SM and Svanborg Edén C: Difference in susceptibility to
gram-negative urinary tract infection between C3H/HeJ and C3H/HeN
mice. Infect Immun. 46:839–844. 1984.PubMed/NCBI
|
|
22
|
Song Z, Yin J, Yao C, Sun Z, Shao M, Zhang
Y, Tao Z, Huang P and Tong C: Variants in the Toll-interacting
protein gene are associated with susceptibility to sepsis in the
Chinese Han population. Crit Care. 15:R122011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Arbour NC, Lorenz E, Schutte BC, Zabner J,
Kline JN, Jones M, Frees K, Watt JL and Schwartz DA: TLR4 mutations
are associated with endotoxin hyporesponsiveness in humans. Nat
Genet. 25:187–191. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schröder NW and Schumann RR: Single
nucleotide polymorphisms of Tol-llike receptors and susceptibility
to infectious disease. Lancet Infect Dis. 5:156–164. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ferwerda B, McCall MB, Alonso S,
Giamarellos-Bourboulis EJ, Mouktaroudi M, Izagirre N, Syafruddin D,
Kibiki G, Cristea T, Hijmans A, et al: TLR4 polymorphisms,
infectious diseases, and evolutionary pressure during migration of
modern humans. Proc Natl Acad Sci USA. 104:16645–16650. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Okayama N, Fujimura K, Suehiro Y, Hamanaka
Y, Fujiwara M, Matsubara T, Maekawa T, Hazama S, Oka M, Nohara H,
et al: Simple genotype analysis of the Asp299Gly polymorphism of
the toll-like receptor-4 gene that is associated with
lipopolysaccharide hyporesponsiveness. J Clin Lab Anal. 16:56–58.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lin YC, Chang YM, Yu JM, Yen JH, Chan JG
and Hu CJ: Toll-like receptor 4 gene C119A but not Asp299Gly
polymorphism is associated with ischemic stroke among ethnic
Chinese in Taiwan. Atherosclerosis. 180:305–309. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hall C, Flores MV, Chien A, Davidson A,
Crosier K and Crosier P: Transgenic zebrafish reporter lines reveal
conserved Toll-like receptor signaling potential in embryonic
myeloid leukocytes and adult immune cell lineages. J Leukoc Biol.
85:751–765. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Couture LA, Piao W, Ru LW, Vogel SN and
Toshchakov VY: Targeting Toll-like receptor (TLR) signaling by
Toll/interleukin-1 receptor (TIR) domain-containing adapter
protein/MyD88 adapter-like (TIRAP/Mal)-derived decoy peptides. J
Biol Chem. 287:24641–24648. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kumpf O, Giamarellos-Bourboulis EJ, Koch
A, Hamann L, Mouktaroudi M, Oh DY, Latz E, Lorenz E, Schwartz DA,
Ferwerda B, et al: Influence of genetic variations in TLR4 and
TIRAP/Mal on the course of sepsis and pneumonia and cytokine
release: An observational study in three cohorts. Crit Care.
14:R1032010. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ferwerda B, Alonso S, Banahan K, McCall
MB, Giamarellos-Bourboulis EJ, Ramakers BP, Mouktaroudi M, Fain PR,
Izagirre N, Syafruddin D, et al: Functional and genetic evidence
that the Mal/TIRAP allele variant 180L has been selected by
providing protection against septic shock. Proc Natl Acad Sci USA.
106:10272–10277. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hamann L, Kumpf O, Schuring RP, Alpsoy E,
Bedu-Addo G, Bienzle U, Oskam L, Mockenhaupt FP and Schumann RR:
Low frequency of the TIRAP S180L polymorphism in Africa, and its
potential role in malaria, sepsis, and leprosy. BMC Med Genet.
10:652009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yew KH, Carpenter C, Duncan RS and
Harrison CJ: Human cytomegalovirus induces TLR4 signaling
components in monocytes altering TIRAP, TRAM and downstream
interferon-beta and TNF-alpha expression. PLoS One. 7:e445002012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tsujimoto H, Ono S, Efron PA, Scumpia PO,
Moldawer LL and Mochizki H: Role of toll-like receptors in the
development of sepsis. Shock. 29:315–321. 2008.PubMed/NCBI
|
|
35
|
Du L, Zhou J, Zhang J, Yan M, Gong L, Liu
X, Chen M, Tao K, Luo N and Liu J: Actin filament reorganization is
a key step in lung inflammation induced by systemic inflammatory
response syndrome. Am J Respir Cell Mol Biol. 47:597–603. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bhatia M, Zemans RL and Jeyaseelan S: Role
of chemokines in the pathogenesis of acute lung injury. Am J Respir
Cell Mol Biol. 46:566–572. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Song Z, Tong C, Sun Z, Shen Y, Yao C,
Jiang J, Yin J, Gao L, Song Y and Bai C: Genetic variants in the
TIRAP gene are associated with increased risk of sepsis-associated
acute lung injury. BMC Med Genet. 11:1682010. View Article : Google Scholar : PubMed/NCBI
|