Introduction
Bronchial asthma is a chronic inflammatory airway
disease involving many inflammatory cells and mediators. T cells,
particularly T helper (Th)1 and Th2. have a crucial role in the
induction of airway inflammation in asthmatic patients (1). Complicated immune responses are capable
of inducing Th1 deficiency and Th2 hyperactivity, which results in
a Th1/Th2 imbalance. This imbalance promotes immunoglobulin (Ig)E
secretion and sensitizes mastocyte and eosinophils via altered
cytokine secretion and causes allergic inflammation and
hyper-responsiveness of the airway (2,3).
Interferon (IFN)-γ and interleukin (IL)-4 are typical cytokines of
Th1 and Th2, respectively.
In patients with asthma, persistent airway
inflammation is initiated by antigen presenting cells (APC), which
integrate various allergens into a signal for T cells and prime the
subsequent immune responses (4,5).
Activation of T cells requires signals which are initiated via the
TCR complex and cluster of differentiation (CD)28. Mature dendritic
cells (mDCs) express high levels of the co-stimulatory molecules
CD80 and CD86, which provide the signal that is required for
triggering T cell activation, expansion and differentiation via
interaction with CD28 (6). Previous
studies have demonstrated that CD80 and CD86 levels are elevated in
patients with asthma (7,8).
In previous studies investigating asthmatic models,
mDCs have been shown to induce Th2 polarization, upregulate IL-4
secretion, downregulate IFN-γ production and induce eosinophilic
inflammation (9,10). However, studies investigating the
effects of the knockdown of CD80 and CD86 in DCs on the
differentiation of and cytokine secretion by T helper cells in
murine models of asthma are lacking.
In the present study, co-stimulatory T-cell
activation signals were blocked by the suppression of CD80 and CD86
molecule expression on DCs using small interfering RNA (siRNA), and
the effects of CD80 and CD86 knockdown on the expression levels of
the Th1/Th2 typical cytokines IFN-γ and IL-4 were evaluated. Thus,
the potential of CD80 and CD86 as targets for the application of
RNA interference (RNAi) in the therapeutic targeting of asthma were
investigated.
Materials and methods
Animals
A total of 20 healthy specific-pathogen-free grade
BALB/c mice (6–8 weeks; mean weight, 18±2 g) were purchased from
the Center of Experimental Animals of Sun Yat-Sen University
(Guangzhou, China). Experiments were performed according to
protocols approved by the Animal Studies Committee of Sun Yat-Sen
University.
Asthma models
A total of 20 mice were randomly assigned to two
groups: i) the asthmatic group, and ii) the normal control group.
In the asthmatic group, each mouse was sensitized to ovalbumin
(OVA; Sigma-Aldrich, St. Louis, MO, USA) intraperitoneally on days
1, 14 and 21 with 100 µg OVA that was emulsified in 20 mg alum
(Guangzhou Chemical Reagent Co., Guangzhou, China). Subsequently,
the mice were exposed to an aerosol challenge of 5% OVA for 30
min/day in airtight containers (with dimensions of 50×50×50 cm) on
days 28–34. In the normal control group, mice were sensitized and
challenged as above with an equivalent amount of saline solution
instead of the OVA protein solution. At 24 h after the last
challenge, mice were sacrificed by an approved cervical dislocation
procedure conducted by skilled and fully trained personnel. The
lungs were removed and then fixed in 10% ethanol for 24 h.
Specimens were dehydrated, embedded in paraffin, and stained with
hematoxylin and eosin as previously described (11). Pathological changes in bronchial and
lung tissues were assessed under a Nikon Eclipse Ti light
microscope (Nikon Corporation, Tokyo, Japan).
Separation of bone marrow-derived
DCs
All mice were sacrificed by cervical dislocation 24
h after the final challenge. Bone marrow was flushed from the
femurs and tibiae with RPMI-1640 culture medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA). Following
centrifugation at 250 × g for 5 min, cells were treated with red
blood cell (RBC) lysis buffer (CWBio Co., Ltd., Beijing, China),
washed with phosphate-buffered saline (PBS), centrifuged at 250 × g
for 5 min and cultured in RPMI-1640 supplemented with recombinant
mouse granulocyte macrophage colony-stimulating factor (rmGM-CSF;
Peprotech, Inc., Rocky Hill, NJ, USA; 10 ng/ml), and rmIL-4
(Peprotech; 10 ng/ml) were used in turn. After 6 days of culture, 1
µg/ml lipopolysaccharide (LPS; Sigma-Aldrich) was added, and the
non-adherent mDCs were harvested on day 7. DCs were stained at 4°C
for 30 min with fluorescein isothiocyanate (FITC)-conjugated
hamster anti-CD11c (5 µg/ml; 11–0114), phycoerythrin (PE)-
conjugated hamster anti-CD80 (5 µg/ml; 12–0801), FITC-conjugated
rat anti-CD86 (5 µg/ml; 11–0862) and PE-conjugated rat anti-major
histocompatibility complex (MHC) II (5 µg/ml; 12–5322; all
eBioscience, Inc., San Diego, CA, USA) monoclonal antibodies, and
were subsequently analyzed by flow cytometry (BD FACSVerse; BD
Biosciences, Franklin Lakes, NJ, USA) in order to determine the
positive expression rate of the labeled antigen expression.
siRNA and transfection
The specific siRNA sequences (Table I) targeting CD80 and CD86 were
designed and selected according to the methods of Gu et al
(12). All siRNA was purchased from
Shanghai GenePharma Co., Ltd. (Shanghai, China). The transfection
step was performed according to the manufacturer's protocol for
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
Briefly, DCs were cultured in a 24-well tissue culture plate at a
density of 1×105 cells/well on the day prior to transfection. To
prepare lipid-siRNA complexes, 3 µl Lipofectamine 2000 was
incubated in 50 µl Opti-MEM (Gibco; Thermo Fisher Scientific, Inc.)
at room temperature for 5 min, and 12 µl of the indicated siRNA was
concurrently combined with 50 µl Opti-MEM. The diluted
Lipofectamine 2000 and siRNA were subsequently mixed and incubated
for a further 20 min at room temperature for complex formation.
Subsequently, the complex was incubated with the DCs in a 24-well
plate at 37°C in a 5% humidified CO2 in air atmosphere
for 6 h. When cotransfection was performed, equivalent amounts of
CD80 siRNA:Lipofectamine 2000 and CD86 siRNA:Lipofectamine 2000
complexes were added to each well. FAM-scrambled-siRNA was used as
the negative control in order to determined the transfection
efficiency. Three groups of transfected DCs were established: In
the non-siRNA group, only Lipofectamine 2000 was added, without any
siRNA being added to the DCs. In the siRNA group, mDCs were
cotransfected by CD80- and CD86-specific siRNA. In the negative
siRNA group, mDCs were transfected by non-specific non-targeting
FAM-siRNA, which has no homology with the targeted RNAs.
Experiments were performed in triplicate for each sample.
Transfection efficiency was determined using fluorescence
microscopy (Nikon Eclipse Ti, Nikon Corporation) and detected by
flow cytometry.
 | Table I.Sequences of siRNA. |
Table I.
Sequences of siRNA.
| siRNA | Sense (5′–3′) | Antisense
(5′–3′) |
|---|
| CD80 siRNA |
GGAAAGAGGAACGUAUGAAdTdT |
UUCAUACGUUCCUCUUUCCdTdT |
| CD86 siRNA |
CAGAGAAACUUGAUAGUGUdTdT |
ACACUAUCAAGUUUCUCUGdTdT |
| FAM siRNA |
UUCUCCGAACGUGUCACGUTT |
ACGUGACACGUUCGGAGAATT |
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
To evaluate CD80 and CD86 mRNA expression levels
following transfection, RT-qPCR was performed. Primer sequences
(Table II) were designed according
to GenBank and synthesized by DaAn Gene Co., Ltd. of Sun Yat-Sen
University (Guangzhou, China). At 24 h post-transfection, the total
RNA of 1×106 DCs was extracted using TRIzol reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) and reverse transcribed and
amplified using QuantiTect SYBR Green RT-PCR kit (Qiagen GmbH,
Hilden, Germany) in a Roche LightCycler 480 (Roche Diagnostics,
Basel, Switzerland). The amplifications were carried out according
to the manufacturer's protocol for the QuantiTect SYBR Green RT-PCR
kit (Takala, Japan). Amplification conditions were 40 cycles of
93°C for 3 min, 93°C for 30 sec, 55°C for 45 sec and 72°C for 45
sec. Every sample was administered to each of three wells. Relative
gene expression levels were calculated using the quantification
cycle (Cq) method with normalization to glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) as the reference gene using 2−∆∆Cq
(13).
 | Table II.Primer sequences of mRNA. |
Table II.
Primer sequences of mRNA.
| mRNA | Forward
(5′–3′) | Reverse
(5′–3′) |
|---|
| CD80 |
CTGGGAAAAACCCCCAGAAG |
TGACAACGATGACGACGACTG |
| CD86 |
CATGGGCTTGGCAATCCTTA |
AAATGGGCACGGCAGATATG |
| GAPDH |
CGTGTTCCTACCCCCAATGT |
TGTCATCATACTTGGCAGGTTTCT |
Flow cytometry
To detect the positive expression rates of CD80 and
CD86 on the DCs following transfection, flow cytometry was
performed on the MHC II/CD11c gate for CD80 and CD86. Six hours
after transfection, DCs were washed twice with PBS and incubated
with fluorescently-labeled antibody at 4°C for 30 min.
Subsequently, the cells were washed again with PBS and fixed with
10 g/l paraformaldehyde. The following anti-mouse monoclonal
antibodies were used: PE-anti-MHC II, FITC-anti-CD11c,
PE-anti-CD80, and FITC-anti-CD86, as mentioned above. All flow
cytometric analyses were performed using IgG isotypic controls.
T-cell separation
Spleens were removed after the mice had been
euthanized by cervical dislocation. T cells were separated using
Mouse Lymphocyte Separation Medium according to the manufacturer's
protocol (Dakewe Biotech Co., Ltd., China). The cell density was
adjusted to 1×109/l prior to further processing.
Mixed lymphocyte reaction (MLR)
The asthmatic murine bone marrow-derived DCs
(1×104/well) and healthy T cells (1×105/well) were co-cultured in
96-well plates at a 1:10 ratio. The co-culture systems were divided
into three groups: i) The non-siRNA group, ii) the siRNA group, and
iii) the negative siRNA group, with DCs from the corresponding
groups as described above. Next, OVA was added to each well to a
final concentration of 10 mg/l in a total volume of 200 µl. The
cells were incubated at 37°C in a 5% humidified CO2 in
air atmosphere for 72 h.
Enzyme-linked immunosorbent assays
(ELISAs)
After 3 days of co-culture, the supernatant was
collected. IFN-γ and IL-4 levels were analyzed using ELISA kits
specific for IFN-γ and IL-4 (Dakewe Biotech Co., Ltd.) according to
the manufacturer's instructions. Absorbance values were read at 450
nm using a Multiskan MK3 (Thermo Fisher Scientific, Inc.).
Statistical analysis
Statistical analyses were performed with SPSS
software, version 13.0 (SPSS, Inc., Chicago, IL, USA). Data are
presented as the mean ± standard deviation (SD). Examinations were
carried out in triplicate for each mouse. Statistical comparisons
between groups were performed using one-way analysis of variance,
and comparisons within a group were performed using Student's
t-test. Differences among groups were considered statistically
significant when P<0.05.
Results
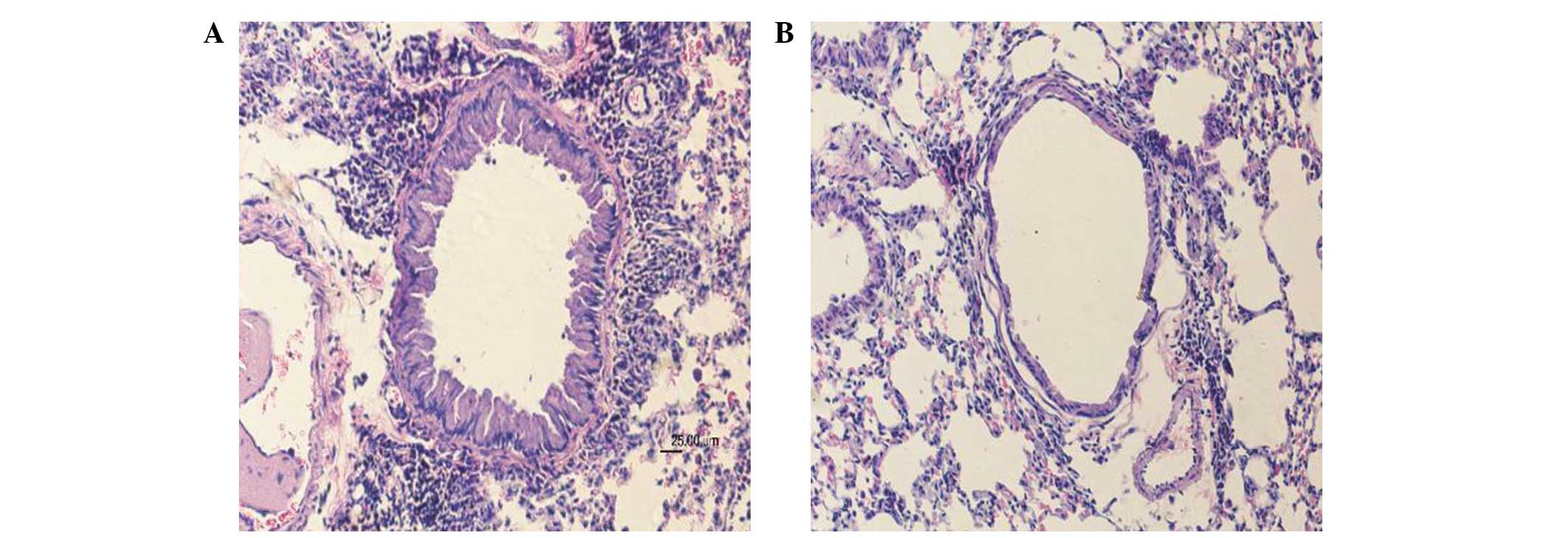
Asthmatic model
The 20 healthy SPF-grade BALB/c mice were assigned
to asthmatic and normal control groups, with 10 mice in each. All
mice were evaluated in the final analysis without experimental
animal loss. According to the lung tissue pathology, lung sections
from the OVA-immunized mice showed clear airway inflammation with
peribronchiolar and perivascular infiltrates. These infiltrates
consisted predominantly of eosinophils and lymphocytes, and
sections showed bronchial mucosa and smooth muscle thickening,
increased mucus secretion, airway epithelial cell shedding, airway
stenosis, and inflammatory cells scattered in the lung
interstitium. No significant pathological changes were observed in
lung sections from the normal control group. Representative
histopathological data are shown in Fig.
1.
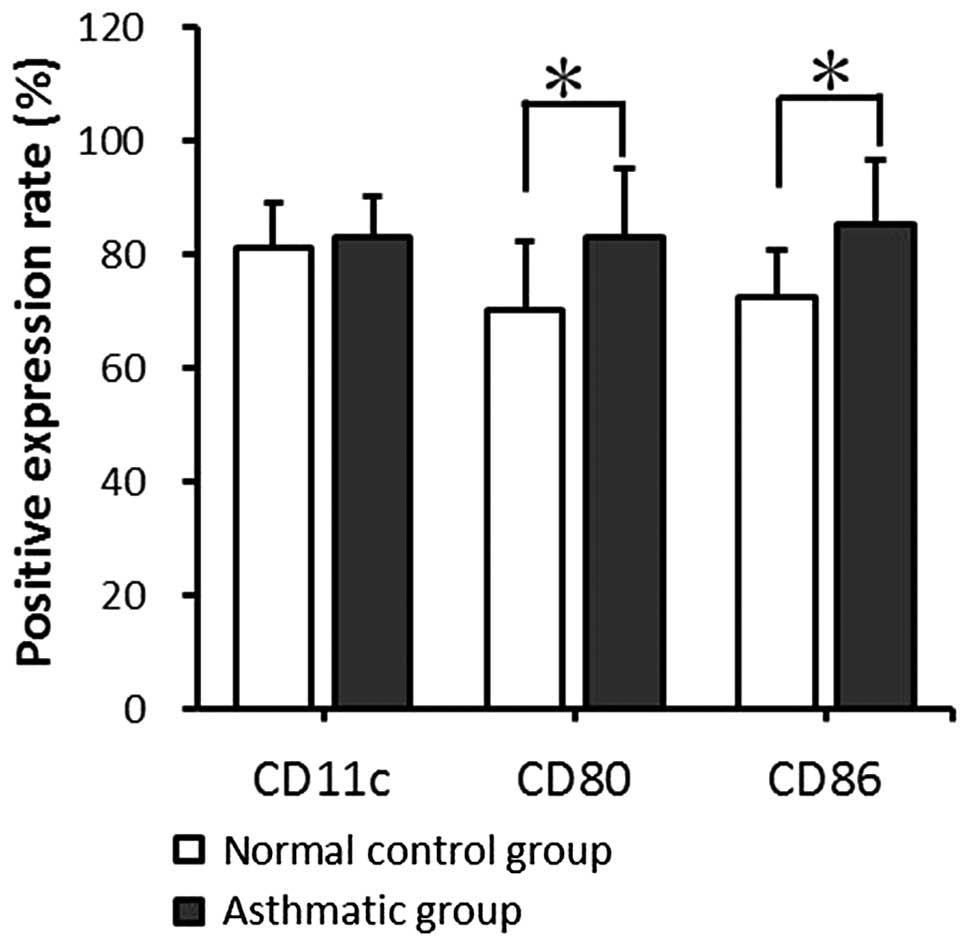
Cell surface molecule expression by
mDCs
The expression levels of CD11c, CD80 and CD86 on the
mDC surfaces were detected by fluorescence-activated cell sorting
(FACS). mDCs from the asthmatic group expressed CD11c at a level
comparable with that in the normal control group; no significant
difference was found between the two groups (P>0.05). However,
in comparison with the normal control group, the asthmatic group
showed significantly higher CD80 and CD86 expression levels
(P<0.05; Fig. 2).
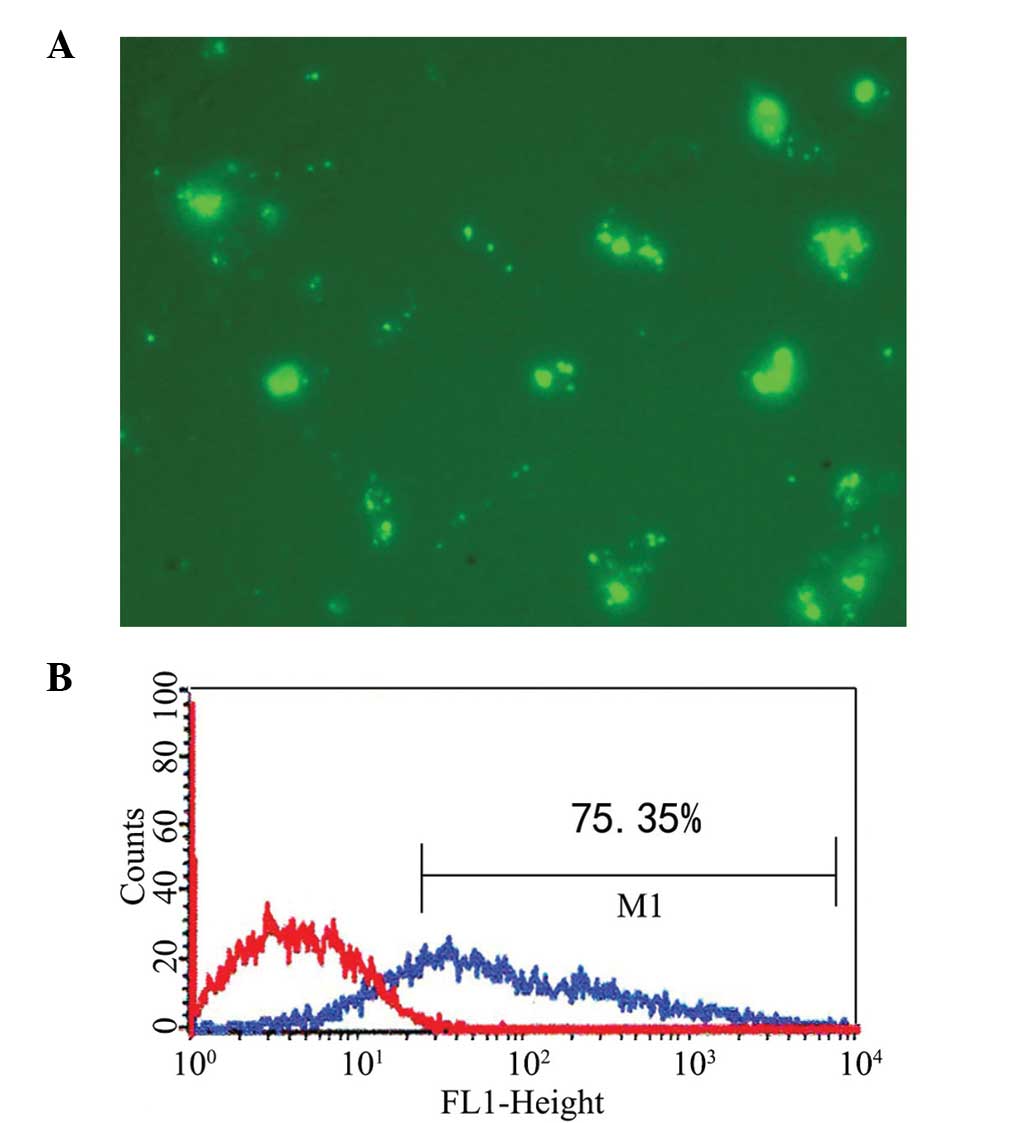
Transfection of mDCs
The CD80- and CD86-specific siRNA constructs were
successfully transferred into the mDCs. The transfected mDCs were
observed under a fluorescence microscope 6 h after transfection.
The transfection efficiency of siRNA detected by FACS was ~75%
(Fig. 3).
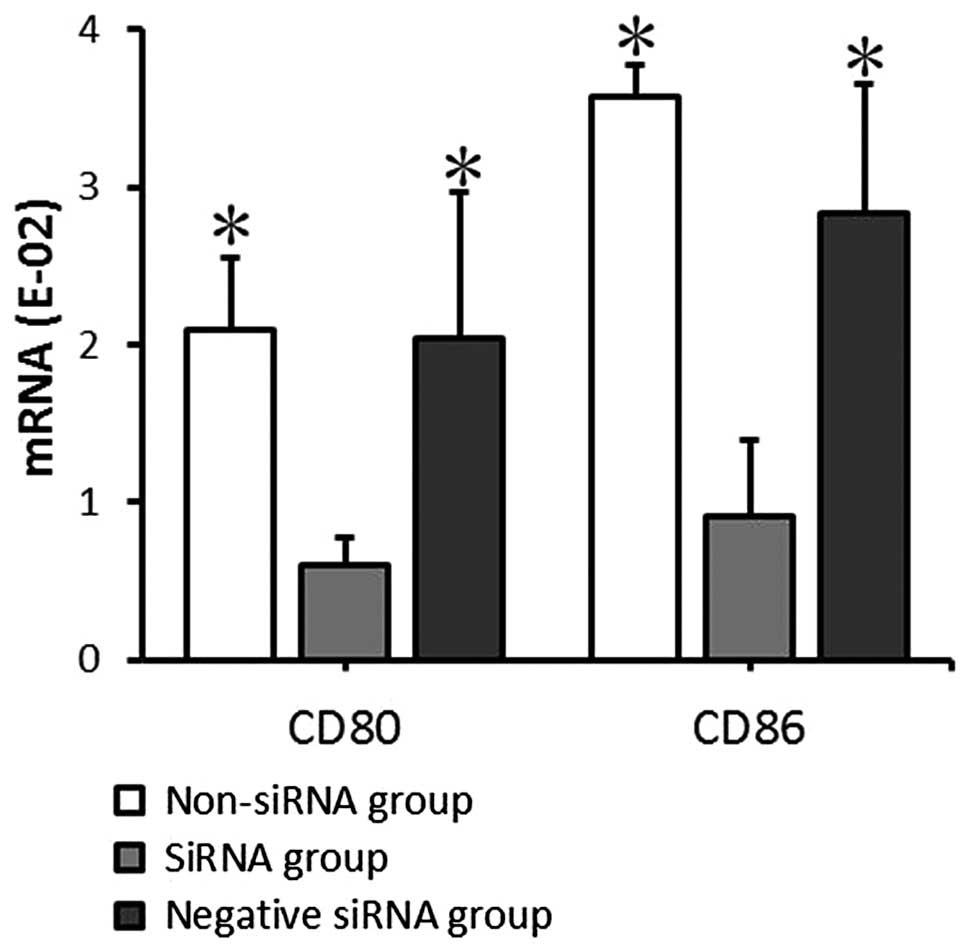
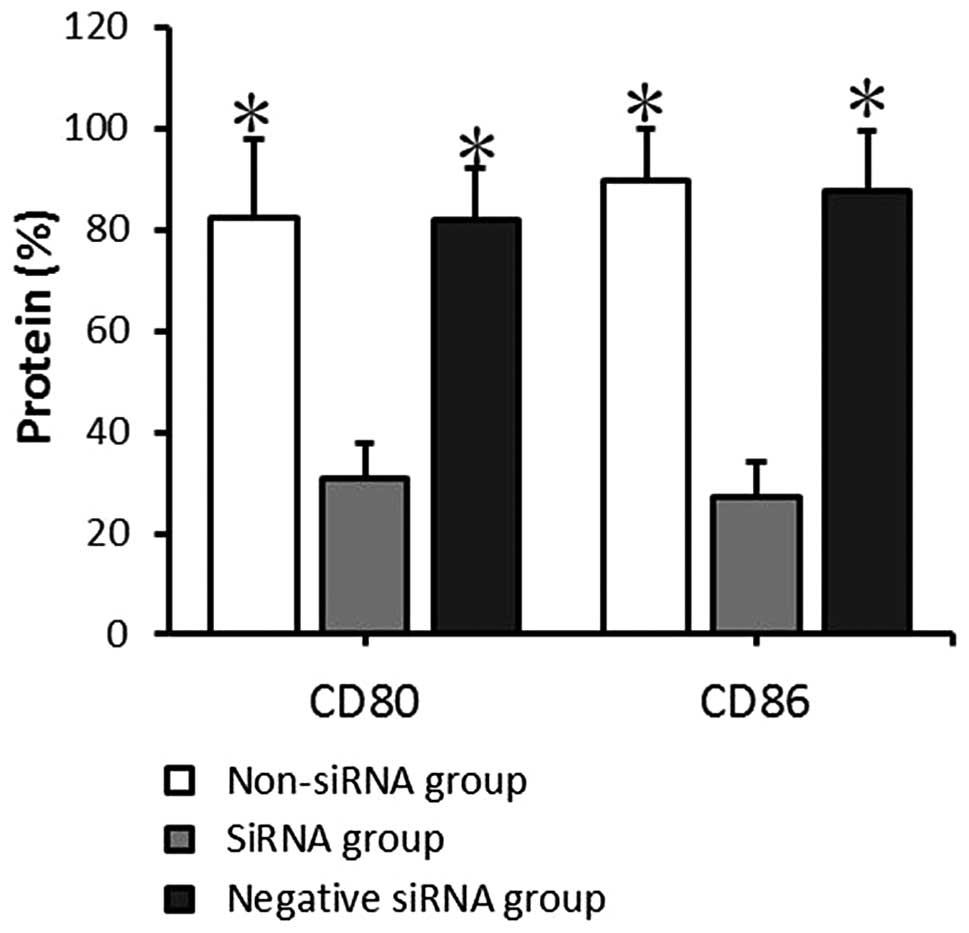
mRNA and protein expression of CD80
and CD86 by mDCs following transfection
The mRNA and protein expression levels of CD80 and
CD86 in the transfected mDCs were detected by RT-qPCR and FACS
analysis at 24 and 72 h, respectively, after transfection. The CD80
and CD86 mRNA expression levels detected by RT-qPCR indicated that
CD80 mRNA expression levels in the non-siRNA, siRNA and negative
siRNA groups were 2.09±0.46, 0.60±0.17, and 2.04±0.93,
respectively, and the CD86 mRNA expression levels were 3.58±0.20,
0.91±0.48, and 2.83±0.83, respectively. The data provided by FACS
indicated that the CD80 protein positive expression rates in the
non-siRNA, siRNA and negative siRNA groups were 82.45±15.80,
30.79±7.07 and 81.83±10.07%, respectively, and the CD86 protein
positive expression rates were 89.45±10.22, 27.29±6.99, and
87.66±11.74%, respectively. The mRNA expression level and protein
positive expression rates exhibited comparable results. There was
no significant significance between the non-siRNA group and the
negative siRNA group (P>0.05). CD80 and CD86 expression at the
mRNA and protein levels in the siRNA group decreased significantly
compared with that in the non-siRNA and negative siRNA groups
(P<0.05; Figs. 4 and 5). This demonstrates that siRNA-targeted
interference can significantly suppress CD80 and CD86 mRNA and
protein expression levels.
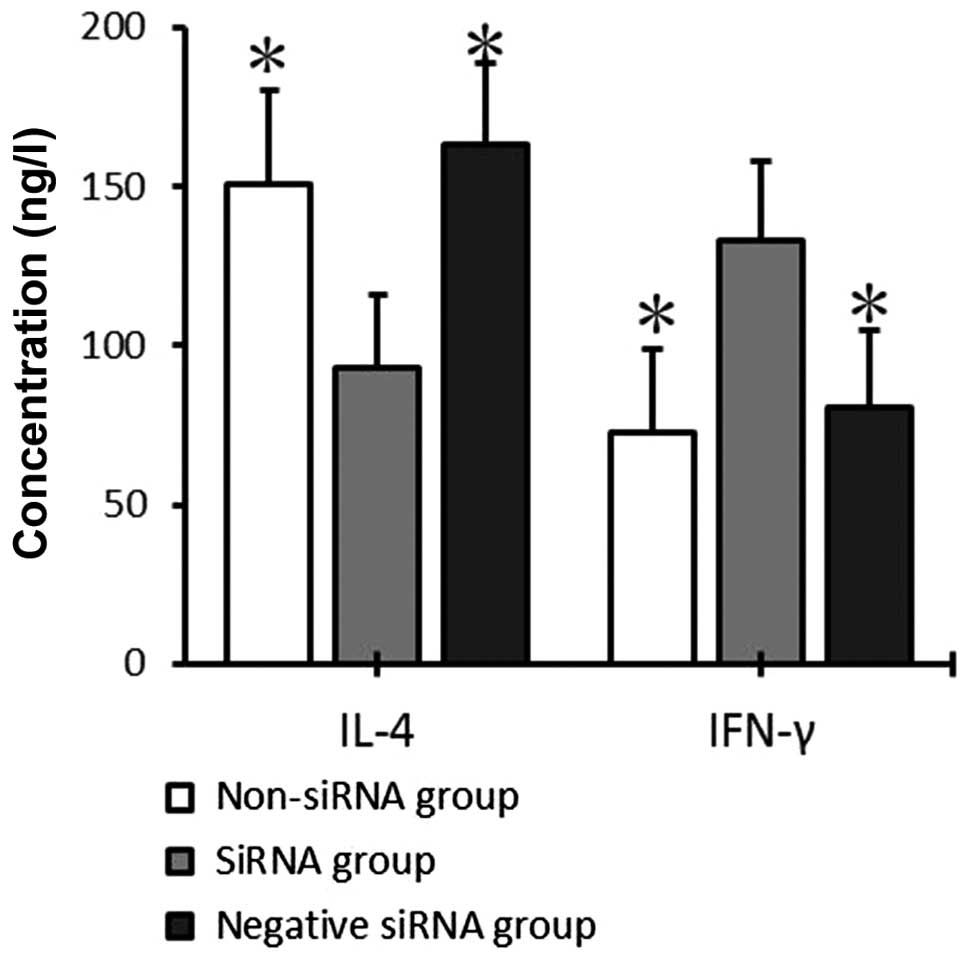
IFN-γ and IL-4 secretion by T cells
co-cultured with mDCs
After 72 h of co-culture, IFN-γ and IL-4 levels in
the supernatant of the mDC and T-cell co-culture system were
detected by ELISA. IFN-γ expression was significantly increased in
the siRNA group (132.73±25.04 pg/ml), as compared with the
non-siRNA and negative siRNA groups (72.56±26.30 and 80.21±24.42
pg/ml, respectively; P<0.05), whereas no significant differences
were detected between the non-siRNA and negative siRNA groups
(P>0.05). IL-4 expression levels were significantly decreased in
the siRNA group (93.04±23.13 pg/ml), as compared with the non-siRNA
and negative siRNA groups (150.69±29.50 and 163.19±25.36 pg/ml,
respectively; P<0.05), whereas no significant differences were
detected between the non-siRNA and negative siRNA groups
(P>0.05; Fig. 6).
Discussion
Bronchial asthma is a chronic inflammatory airway
disease involving a variety of inflammatory cells, including mast
cells, eosinophils, lymphocytes and other cell components (14–17).
Th1/Th2 imbalance is a key factor contributing to asthma severity
(18). APCs, including DCs,
macrophages and B cells (19), play
a crucial role in the stimulation of T cells (3). Among those, the DC is the most powerful
APC, contributing to primary and secondary immune responses,
including allergic immunity. Mature DCs (mDCs) with high levels of
expression of the co-stimulatory molecules CD80 and CD86 can
activate T cells, while immature dendritic cells (iDCs) with a low
level of expression of CD80 and CD86 suppress the T-cell response
and induce immune tolerance (20,21). DCs
in patients with asthma have been demonstrated to be hyperactive
(22).
CD80 and CD86 are two types of protein that are
expressed on the APC surface, and which work in tandem to provide
co-stimulatory signals necessary for T-cell activation and
survival. Previous studies have shown that the expression levels of
the co-stimulatory molecules CD80 and CD86 on the mDC surface are
closely associated with Th2-cell reaction and airway inflammation
(23,24). In asthmatic patients, mDCs that
highly express CD80 and CD86 can stimulate naïve CD4+
helper T-cell activation to differentiate toward Th2 cells,
resulting in a Th1/Th2 imbalance. Following that, the inadequate
secretion of Th1 cytokines such as IFN-γ, along with the increased
secretion of Th2 cytokines such as IL-4 and IL-5, causes
eosinophilic inflammation and allergic airway inflammation
(10,24,25). Two
signals are required for the promotion of Th2-cell activation
(26–28). The first signal is the formation of
antigen-MHC complexes on the mDC surface that bind specifically
with the T-cell receptor-CD3 receptor complex on T-cell surfaces,
and the second signal is co-stimulatory molecule expression and
functional activation on the mDC surfaces that specifically bind to
receptors on naïve T cells; the two signals form a co-stimulatory
pathway (29). It has also been
suggested that there is a third signal (30), as certain cellular molecules produced
by DCs, such as thymic stromal lymphopoietin (TSLP), affect the
direction of Th-cell differentiation. However, among all the
aforementioned signals, the CD80/CD86-CD28 co-stimulatory pathway
is the most classic and important. Previous research has indicated
that the CD80/CD86-CD28 co-stimulatory pathway may be an effective
target for asthma treatment by demonstrating that blocking the
CD80/CD86 co-stimulatory pathway by monoclonal antibody approaches
can inhibit inflammation in asthmatic mice (25). In addition, suppressing the CD80/CD86
co-stimulatory pathway using antisense oligonucleotides can
suppress airway hyperactivity (31).
RNAi is a gene-silencing phenomenon whereby
endogenous- or exogenous-specific double-stranded RNAs trigger the
degradation of homologous mRNA and induce the loss of corresponding
functional phenotypes. Since the technique was first discovered in
1998 by Fire et al (32), the
technique has undergone further development to attain a high degree
of specificity and efficiency. The therapeutic application of RNAi
technology is a topic that has been attracting high levels of
interest in basic medical and clinical research in recent years.
The ability of RNAi to inhibit virulent gene expression has been
widely used to treat a variety of diseases (33–35).
Also, a number of studies have reported that RNAi can be used in
DCs to diagnose and treat bronchial asthma (36–39).
Darcan-Nicolaisen et al (40)
discovered that the two major signs of allergic asthma in the
OVA-induced asthma mouse model, which are airway inflammation and
hyperactivity, were significantly ameliorated by signal transducer
and activator of transcription 6 (STAT6) silencing in airway
epithelial cells using the administration of siRNA nose drops.
Moriwaki et al (41)
demonstrated that siRNA-mediated suppressor of cytokine signaling 3
(SOCS3) gene silencing could suppress the airway reactivity and
eosinophilic infiltration that was induced by allergenic
stimulation in asthmatic mice. Zheng et al (42) found that the application of siRNA was
able to inhibit tyrosine protein kinase (TPK) gene expression in
the DCs of asthmatic mice, repressing the functional capability of
DCs as antigen-presenting cells, thereby inhibiting T-cell
activation and differentiation. However, studies concerning the
effects of siRNA-mediated CD80 and CD86 knockdown in DCs on T-cell
differentiation in asthmatic mice are lacking. In the present
study, CD80 and CD86 mRNA and protein expression in murine bone
marrow-derived DCs was successfully decreased with CD80- and
CD86-targeting siRNA, which verified the efficiency of RNAi.
In the present study, an asthmatic mouse model was
used that was established according to previously reported methods
(43). The results indicated that
the mDCs obtained from the asthmatic group exhibited increased CD80
and CD86 expression levels, which implies that the CD80/CD86
capacity may be heightened in asthmatic patients. Following
transfection of the mDCs with CD80- and CD86-targeted siRNA, the
mRNA expression levels and protein positive expression rates of
CD80 and CD86 were significantly decreased, confirming the
inhibitory effect that the siRNA approach had on the co-stimulatory
molecules CD80 and CD86 at the transcriptional and translational
levels. In the supernatant from the co-culture of mDCs and T cells,
RNAi induced an increase in IFN-γ expression and a reduction of
IL-4 levels, indicating that decreasing the expression of the
co-stimulatory molecules CD80 and CD86 in mDCs weakened the
CD80/CD86-CD28 co-stimulatory pathway in asthmatic mice. RNAi also
affected the expression of Th1/Th2 cytokines, indicating that the
original Th1/Th2 imbalance was changed and, consequently, immune
tolerance was induced. These findings indicate that CD80 and CD86
may be potential targets for RNAi application in asthma treatment,
and provide a new avenue for the gene therapy of asthma.
Acknowledgements
This study was supported by an Open Projects Grants
from the State Key Laboratory of Respiratory Disease (Guangzhou,
China) (grant no. 2007DA80154F1107).
References
|
1
|
Locksley RM: Asthma and allergic
inflammation. Cell. 140:777–783. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Holgate ST: Innate and adaptive immune
responses in asthma. Nat Med. 18:673–683. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Larché M, Robinson DS and Kay AB: The role
of T lymphocytes in the pathogenesis of asthma. J Allergy Clin
Immunol. 111:450–464. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lambrecht BN and Hammad H: The role of
dendritic and epithelial cells as master regulators of allergic
airway inflammation. Lancet. 376:835–843. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Holt PG and Upham JW: The role of
dendritic cells in asthma. Curr Opin Allergy Clin Immunol. 4:39–44.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lim TS, Goh JK, Mortellaro A, Lim CT,
Hämmerling GJ and Ricciardi-Castagnoli P: CD80 and CD86
differentially regulate mechanical interactions of T-cells with
antigen-presenting dendritic cells and B-cells. PLoS One.
7:e451852012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wong CK, Lun SW, Ko FW, Ip WK, Hui DS and
Lam CW: Increased expression of plasma and cell surface
co-stimulatory molecules CTLA-4, CD28 and CD86 in adult patients
with allergic asthma. Clin Exp Immunol. 141:122–129. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shi HZ, Xie ZF, Deng JM, Chen YQ and Xiao
CQ: Soluble CD86 protein in serum samples of patients with asthma.
Thorax. 59:870–875. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Van Rijt LS and Lambrecht BN: Dendritic
cells in asthma: a function beyond sensitization. Clin Exp Allergy.
35:1125–34. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Van Rijt LS, Vos N, Willart M, Kleinjan A,
Coyle AJ, Hoogsteden HC and Lambrecht BN: Essential role of
dendritic cell CD80/CD86 costimulation in the induction, but not
reactivation, of TH2 effector responses in a mouse model of asthma.
J Allergy Clin Immunol. 114:166–173. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu SG, Wang GL, Li LY and Ji J: Effects of
microRNA-21 on the interleukin 12/signal transducer and activator
of transcription 4 signaling pathway in asthmatic mice. Cent Eur J
Immunol. 39:40–45. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gu X, Xiang J, Yao Y and Chen Z: Effects
of RNA interference on CD80 and CD86 expression in bone
marrow-derived murine dendritic cells. Scand J Immunol. 64:588–594.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−∆∆Ct method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fanta CH: Asthma. N Engl J Med.
360:1002–1014. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lambrecht BN and Hammad H: Lung dendritic
cells in respiratory viral infection and asthma: From protection to
immunopathology. Annu Rev Immunol. 30:243–270. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wenzel SE: Asthma phenotypes: The
evolution from clinical to molecular approaches. Nat Med.
18:716–725. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hansel TT, Johnston SL and Openshaw PJ:
Microbes and mucosal immune responses in asthma. Lancet.
381:861–873. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Compalati E, Braido F and Canonica GW: An
update on allergen immunotherapy and asthma. Curr Opin Pulm Med.
20:109–117. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Goyvaerts C, Dingemans J, De Groeve K,
Heirman C, Van Gulck E, Vanham G, De Baetselier P, Thielemans K,
Raes G and Breckpot K: Targeting of human antigen-presenting cell
subsets. J Virol. 87:11304–11308. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Reise Sousa C: Dendritic cells in a mature
age. Nat Rev Immunol. 6:476–483. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gill MA: The role of dendritic cells in
asthma. J Allergy Clin Immunol. 129:889–901. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shi JH, LI YG and LI TS: The roles of
dendritic cells in antigen presentation and the pathogenesis of
asthma. Zhonghua Jie He He Hu Xi Za Zhi. 28:22–27. 2005.(In
Chinese). PubMed/NCBI
|
|
23
|
Wong CK, Lun SW, Ko FW, Ip WK, Hui DS and
Lam CW: Increased expression of plasma and cell surface
co-stimulatory molecules CTLA-4, CD28 and CD86 in adult patients
with allergic asthma. Clin Exp Immunol. 141:122–129. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bellou A and Finn PW: Costimulation:
Critical pathways in the immunologic regulation of asthma. Curr
Allergy Asthma Rep. 5:149–154. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen YQ and Shi HZ: CD28/CTLA-4-CD80/CD86
and ICOS-B7RP-1 costimulatory pathway in bronchial asthma. Allergy.
61:15–26. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bieber T, Novak N, Herrmann N and Koch S:
Role of dendritic cells in atopic dermatitis: An update. Clin Rev
Allergy Immunol. 41:254–258. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hespel C and Moser M: Role of inflammatory
dendritic cells in innate and adaptive immunity. Eur J Immunol.
42:2535–2543. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Novak N: An update on the role of human
dendritic cells in patients with atopic dermatitis. J Allergy Clin
Immunol. 129:879–886. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lombardi V, Singh AK and Akbari O: The
role of costimulatory molecules in allergic disease and asthma. Int
Arch Allergy Immunol. 151:179–189. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang Y, Zhou X and Zhou B: DC-derived
TSLP promotes Th2 polarization in LPS-primed allergic airway
inflammation. Eur J Immunol. 42:1735–1743. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Crosby JR, Guha M, Tung D, Miller DA,
Bender B, Condon TP, York-DeFalco C, Geary RS, Monia BP, Karras JG
and Gregory SA: Inhaled CD86 antisense oligonucleotide suppresses
pulmonary inflammation and airway hyper-responsiveness in allergic
mice. J Pharmacol Exp Ther. 321:938–946. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fire A, Xu S, Montgomery MK, Kostas SA,
Driver SE and Mello CC: Potent and specific genetic interference by
double-stranded RNA in Caenorhabditis elegans. Nature.
391:806–811. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ghafouri-Fard S: siRNA and cancer
immunotherapy. Immunotherapy. 4:907–917. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gavrilov K and Saltzman WM: Therapeutic
siRNA: Principles, challenges and strategies. Yale J Biol Med.
85:187–200. 2012.PubMed/NCBI
|
|
35
|
Yan WJ, Xu SJ, Xie MM and Wu BL: Effects
of exogenous cyclic dimeric guanosinemonophosphate on the gene
expression of Streptococcus mutans. Zhongguo Zu Zhi Gong
Cheng Yan Jiu. 16:1451–1454. 2012.(In Chinese).
|
|
36
|
Lee CC, Huang HY and Chiang BL:
Lentiviral-mediated interleukin-4 and interleukin-13 RNA
interference decrease airway inflammation and hyperresponsiveness.
Hum Gene Ther. 22:577–686. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li Y, Sun M, Cheng H, Li S, Liu L, Qiao H,
Hua S and Lu J: Silencing IL-23 expression by a small hairpin RNA
protects against asthma in mice. Exp Mol Med. 43:197–204. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Woska JJ and Gillespie ME:
Small-interfering RNA-mediated identification and regulation of the
ternary SNARE complex mediating RBL-2H3 mast cell degranulation.
Scand J Immunol. 73:8–17. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lu XX, McCoy KS, Xu JL, Hu WK and Chen HB:
Small interfering RNA targeting T-cell Ig mucin-3 decreases
allergic airway inflammation and hyperresponsiveness. Inflammation.
36:582–591. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Darcan-Nicolaisen Y, Meinicke H, Fels G,
Hegend O, Haberland A, Kühl A, Loddenkemper C, Witzenrath M, Kube
S, Henke W and Hamelmann E: Small interfering RNA against
transcription factor STAT6 inhibits allergic airway inflammation
and hyperreactivity in mice. J Immunol. 182:7501–7508. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Moriwaki A, Inoue H, Nakano T, Matsunaga
Y, Matsuno Y, Matsumoto T, Fukuyama S, Kan-O K, Matsumoto K,
Tsuda-Eguchi M, et al: T cell treatment with small interfering RNA
for suppressor of cytokine signaling 3 modulates allergic airway
responses in a murine model of asthma. Am J Respir Cell Mol Biol.
44:448–455. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zheng YH, Lu JJ, Guo ZL, Ren T and Liang
YJ: Small interfering RNAs specific for spleen tyrosine kinase
inhibit maturation of dendritic cells of asthmatic mice in vitro.
Zhongguo Hu Xi Yu Wei Zhong Jian Hu Za Zhi. 8:487–491. 2009.(In
Chinese).
|
|
43
|
Li JG, Zhuansun YX, Ran PX, Zhang W, Mo
XN, Wu H and Li YQ: Effects of bone marrow mesenchymal stem cells
on CD4+CD25+ regulatory T cells and airway
inflammation in asthmatic mice. Zhongguo Zu Zhi Gong Cheng Yan Jiu.
12:9302–9305. 2008.(In Chinese).
|