Introduction
Esophageal carcinoma is a common type of malignant
tumor that forms in the upper digestive tract. Based on worldwide
incidence rates, esophageal carcinoma is the ranks eighth most
common type of malignant tumor, and has the sixth highest mortality
rate of all types of cancer (1).
Esophageal carcinoma is insidious at onset and early diagnosis is
difficult. Furthermore, due to its ability to rapidly metastasize
to the lymph nodes, the prognosis for esophageal carcinoma is poor
(2). Esophageal carcinomas are
subdivided into esophageal squamous cell carcinoma (ESCC) and
adenocarcinoma, according to the tissue type. ESCC is more common
in the developing world. In China, >90% of all esophageal
carcinomas are ESCC. With the application and development of
surgery, chemotherapy, and immunotherapy, the prognosis for
patients with ESCC has markedly improved. However, five-year
survival rates for patients with ESCC remain low at ~10%, due to
the invasive and metastatic potential of ESCC (3,4). Earlier
diagnosis of ESCC may reduce the probability of tumor cell
metastasis and markedly improve the prognosis of patients (5,6).
Therefore, the identification of novel molecular markers for ESCC
has potential clinical application for the early diagnosis of
patients with ESCC.
MicroRNA (miRNA), which are small endogenic
non-coding RNA molecules (18–22 nt), regulate gene expression by
binding to 3′-untranslated regions of target mRNA (7,8). The
application of high-throughput sequencing and microarray technology
has produced notable alterations in the miRNA expression profiles
of tumor tissue, as compared with normal tissue (9,10).
Variations in miRNA expression levels between tumor tissues and
normal tissues may lead to the dysfunction of downstream signaling
pathways, which are associated with proliferation, invasion,
angiogenesis and the metastasis of tumors (11,12,13).
Furthermore, miRNA molecules are highly stable, which facilitates
their survival in tissue fluid, blood and urine (14). Therefore, miRNA has been demonstrated
to be a suitable biomarker for the diagnosis of tumors.
MicroRNA-202 (miR-202) is a recently identified
tumor miRNA molecule with various biological functions (15). The expression levels of miR-202 are
associated with the proliferation, invasion, metastasis, apoptosis
and angiogenesis of numerous tumor tissue types, including gastric,
liver and pancreatic cancer (16–19).
Analysis of the expression levels of genes in ESCC tissue, using
the database of Gene Expression Omnibus (GSE20347), indicated that
1,755 genes were differentially expressed in ESCC tissues,
including miR-202 (20). However,
the expression levels and biological function of miR-202 in
peripheral blood remains unclear. In the present study, the
expression levels of miR-202 in the peripheral blood of patients
with ESCC were analyzed using reverse transcription-quantitative
polymerase chain reaction (RT-qPCR). In addition, the effects of
miR-202 on cellular proliferation, migration and invasiveness were
evaluated.
Materials and methods
Reagents
TRIzol® used for RNA extraction was purchased from
Invitrogen (Thermo Fisher Scientific Inc., Waltham, MA, USA).
Takara PrimeScript™ RT Reagent kit and SYBR® PrimeScript™ RT-PCR
Kit II (Perfect Real Time) were purchased from Takara Biotechnology
Co., Ltd. (Tokyo, Japan). Transwell® chambers were purchased from
Corning Life Sciences (Lowell, MA, USA).
Patients
In the present study, ESCC tissues and peripheral
blood samples were harvested from 60 patients with ESCC (38 men and
22 women) recruited from the Laiwu People's Hospital (Laiwu,
China). Patients had not received chemotherapy or anti-tumor
therapy and were aged between 38.5 and 67 years (average age, 52.6
years). A total of 29 cases of well differentiated, 22 cases of
moderately differentiated and 9 cases of poorly differentiated
cells were included in the ESCC tissue samples. Lymph node
metastases were categorized as follows: N0, no evidence of lymph
node metastasis; and N1, metastasis to lymph node. In the present
study, patients were separated into N1 lymph node metastases (n=36)
and N0 lymph node metastasis (n=24) groups. For a control group, 30
healthy individuals were enrolled. For patients with ESCC,
peripheral blood (5 ml) was collected from the cubital vein one
week prior to the surgical procedure. For healthy controls,
peripheral blood (5 ml) was collected from the cubital vein when
the patients underwent physical examination. Peripheral blood
samples were stored at −80°C for further analysis. ESCC tissue
samples were collected from the 60 patients with ESCC during
surgery and maintained at −80°C for further analysis. Prior written
and informed consent was obtained from every patient. The present
study was approved by the Ethics Review Board of Laiwu People's
Hospital.
Cell culture and transfection
EC9706 ESCC cell lines were cultured in RPMI-1640
medium supplemented with Gibco 10% fetal bovine serum (FBS; Thermo
Fisher Scientific Inc.), 100 U/ml penicillin and 100 µg/ml
streptomycin (both obtained from Beyotime Institute of
Biotechnology, Shanghai, China) in a humidified atmosphere
containing 5% CO2 at 37°C. Cells in the logarithmic
growth phase were used in the present study. Transfection of
miR-202 mimics (Guangzhou RiboBio Co., Ltd., Guangzhou, China) was
performed in 70% confluent cells using Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific Inc.), according to the
manufacturer's protocol. MiR-202 independent sequence was used as
the negative control.
RT-qPCR analysis
Total RNA was extracted from peripheral blood
samples using TRIzol® reagent. RNA quality was assessed using RNA
electrophoresis and a spectrophotometer at optical density (OD)
260/280. RNA was reverse transcribed into cDNA using the
PrimeScript™ RT Reagent kits. RT-qPCR was conducted using SYBR®
PrimeScript™ RT-PCR kit II (Perfect Real Time), according to the
manufacturer's protocol. U6 was used as an internal control for
normalization. PCR reactions were repeated ≥3 times for each
sample. Oligonucleotide primers of miR-202 and U6 were synthesized
by Guangzhou RiboBio Co., Ltd. (Guangzhou, China), as follows:
miR-202, forward 5′-CCTCCCAGGCTCACGAGGCT-3′ and reverse
5′-GGTGCAGGTGCACTGGTGCA-3′; and U6, forward
5′-CAAAGTCAGTGCAGGTAGGCTTA-3′ and reverse
5′-AACGCTTCACGAATTTGCGT-3′. The RT-qPCR reaction system included 10
µl qRT-PCR-Mix, 0.5 µl forward primer, 0.5 µl reverse primer, 2 µl
cDNA and 7 µl ddH2O. The RT-qPCR was performed using an
ABI 7300 thermal cycler (Applied Biosystems Inc., Foster City, CA,
USA). The reaction procedure was as follows: Pre-denaturation at
95°C for 10 min and 40 cycles of 95°C for 1 min and 60°C for 30
sec. The 2−ΔΔCQ method (21) was used to analyze the relative
expression levels of miR-202.
Methylthiazolyl-tetrazolium bromide
(MTT) proliferation assay
EC9706 cells in the logarithmic phase were routinely
digested for 12 min at 37°C using 0.2% trypsin/EDTA (Beyotime
Institute of Biotechnology) and prepared as single-cell suspensions
containing 5×105 cells/ml. Each well of a 96-well plate
was seeded with 200 µl cell suspension (2×103
cells/well). All 96-well plates were incubated at 37°C in an
atmosphere containing 5% CO2 for 0, 24, 48 or 72 h,
after which the culture medium was discarded and replaced with 180
µl RPMI-1640 medium and 20 µl MTT solution (5 g/l; Beyotime
Institute of Biotechnology). Following incubation at 37°C in an
atmosphere containing 5% CO2 for a further 4 h, the
supernatant was discarded, each well was supplemented with 150
µl/well dimethyl sulphoxide (Sigma-Aldrich, St. Louis, MO, USA) and
the plates were shaken for 10 min using a shaking table
concentrator (Molecular Devices LLC, Sunnyvale, CA, USA). The OD of
each well was determined using a FlexStation® 3 Multi-Mode
Microplate Reader (Molecular Devices LLC) at 490 nm. Tests were
repeated in triplicate for each group.
In vitro scratch assay
Each well of a 24-well plate was seeded with 800 µl
EC9706 cell suspension (2×103 cells/well) and incubated
for 24 h at 37°C in an atmosphere containing 5% CO2.
Once a confluent monolayer was formed, cells were serum-starved for
24 h and the cell monolayers were subsequently scratched using a
1,000-µl pipette tip. Scratched cells were cultured in RPMI-1640
medium supplemented with 10% FBS for 24 h and observed under an
inverted microscope (Olympus BX50; Olympus Corporation, Tokyo,
Japan; magnification, ×10). The migratory ability of the cells was
assessed by comparing the respective repair distances.
Transwell® chamber assay
Metastasis and invasive ability of the EC9706 cells
was examined using a Transwell® chamber assay. Matrigel (BD
Biosciences, Franklin Lakes, NJ, USA) was hydrated with serum-free
medium (dilution, 1:2) and added to the upper chambers for ≥1 h at
37°C in a CO2 incubator. Following rehydration of the
Matrigel, the lower chamber was filled with 500 µl RPMI-1640 medium
supplemented with 10% FBS and the upper chamber was filled with
cell suspension diluted to 1×105 cells and 200 µl
serum-free RPMI-1640 medium. The invasion chamber plate was
incubated for 24 h. Cells invading through the Matrigel were fixed
with 4% formaldehyde and subjected to Giemsa staining (Beyotime
Institute of Biotechnology). These processes were all performed at
4°C. Five fields were randomly selected from each section and the
number of cells that invaded the Matrigel layer were subsequently
counted using an inverted microscope (magnification, ×10).
Statistical analysis
Results are expressed as the mean ± standard
deviation. Statistical analyses were performed using SPSS 11.0 for
Windows (SPSS Inc., Chicago, IL, USA). Paired t-test was used to
analyze comparisons between the groups and paired data. P<0.05
was considered to indicate a significantly significant
difference.
Results
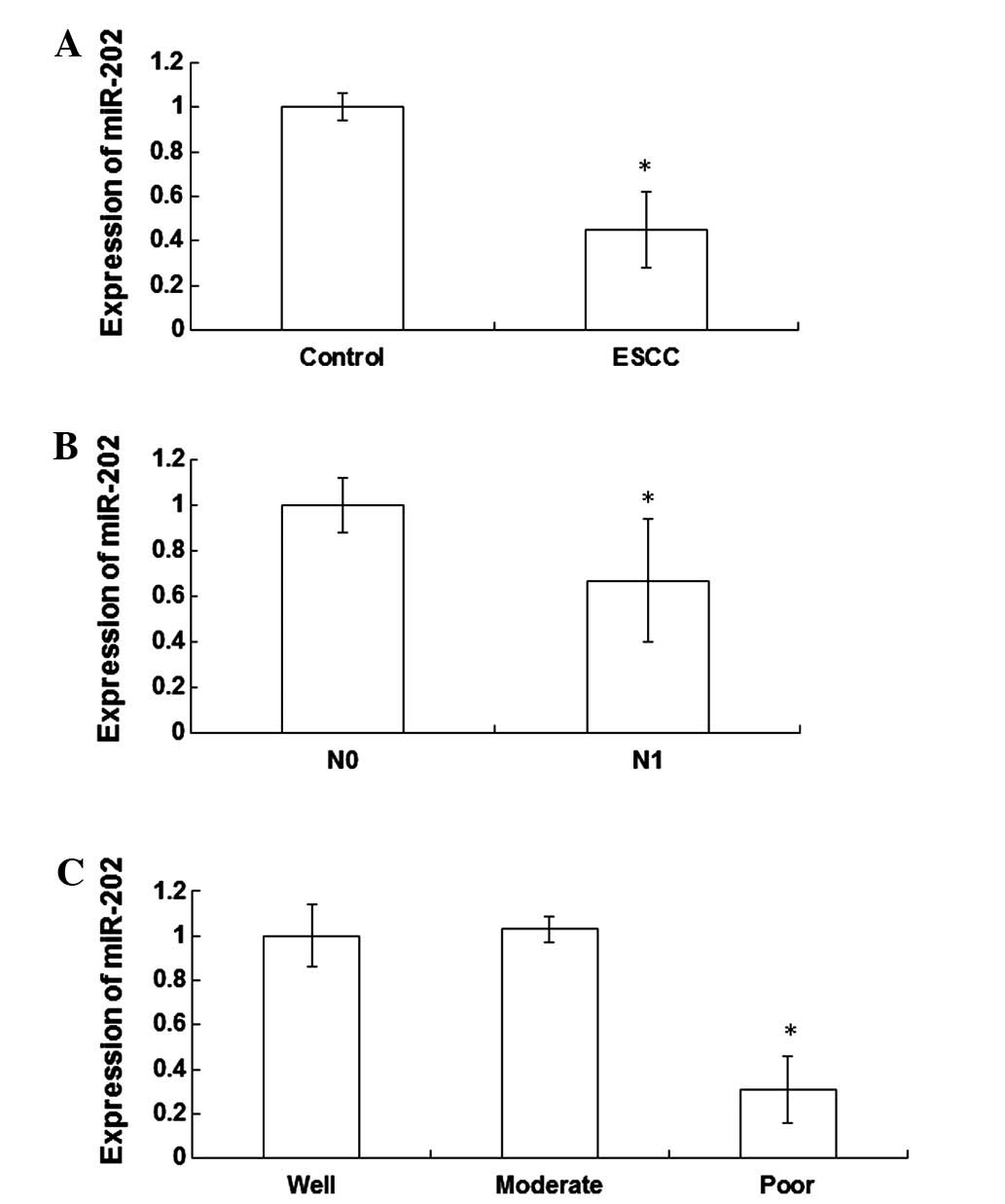
Expression levels of miR-202 are
significantly decreased in the peripheral blood of patients with
ESCC
In order to detect the expression levels of miR-202
in the peripheral blood of patients with ESCC and healthy
individuals, RT-qPCR was performed. The expression levels of
miR-202 were significantly reduced (0.45±0.17 fold; P<0.05) in
the peripheral blood cells of patients with ESCC, as compared with
the healthy individuals (Fig. 1A).
Furthermore, the expression levels of miR-202 were significantly
reduced (0.67±0.27 fold; P<0.05) in the N1 lymph node metastasis
group, as compared with the N0 lymph node metastasis group
(Fig. 1B). Notably, the expression
levels of miR-202 were significantly decreased in patients with
poorly differentiated ESCC tissues, as compared with patients with
well and moderately differentiated ESCC tissues (P<0.05).
Furthermore, miR-202 expression levels were significantly reduced
(0.31±0.15 fold; P<0.05) in patients with poorly differentiated
ESCC tissues, as compared with patients with well differentiated
ESCC tissues (Fig. 1C). These
results indicate that the expression levels of miR-202 are
significantly decreased in the peripheral blood of patients with
ESCC, which may be associated with the degree of differentiation
and lymph node metastasis.
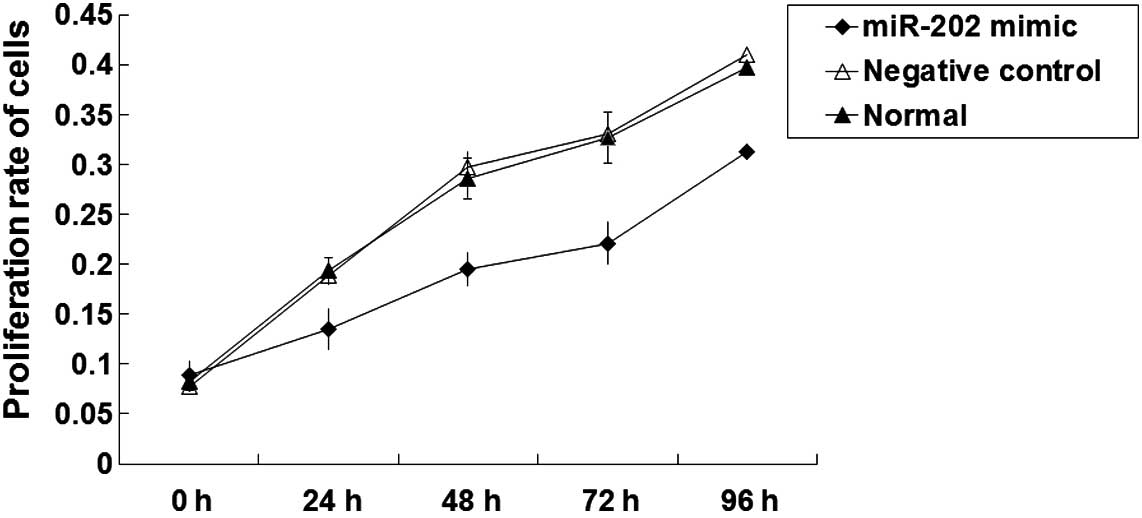
Proliferation of EC9706 cell lines is
inhibited by the expression of miR-202
In order to investigate the effects of miR-202
expression levels on cell proliferation, an MTT proliferation assay
was performed. As outlined in Fig.
2, the proliferation rates of the negative control and normal
EC9706 cells were normal. An 8-fold increase in the number of
EC9706 cells was about detected following 96 h culture (P<0.05).
Following transfection with miR-202 mimics, the proliferation of
EC9706 cells gradually increased. As compared with pre-transfection
cells, the number of EC9706 cells significantly increased by
<2-fold at 96 h following transfection (P<0.05). These
results suggest that the proliferation of EC9706 ESCC cell lines
may be inhibited by the expression of miR-202.
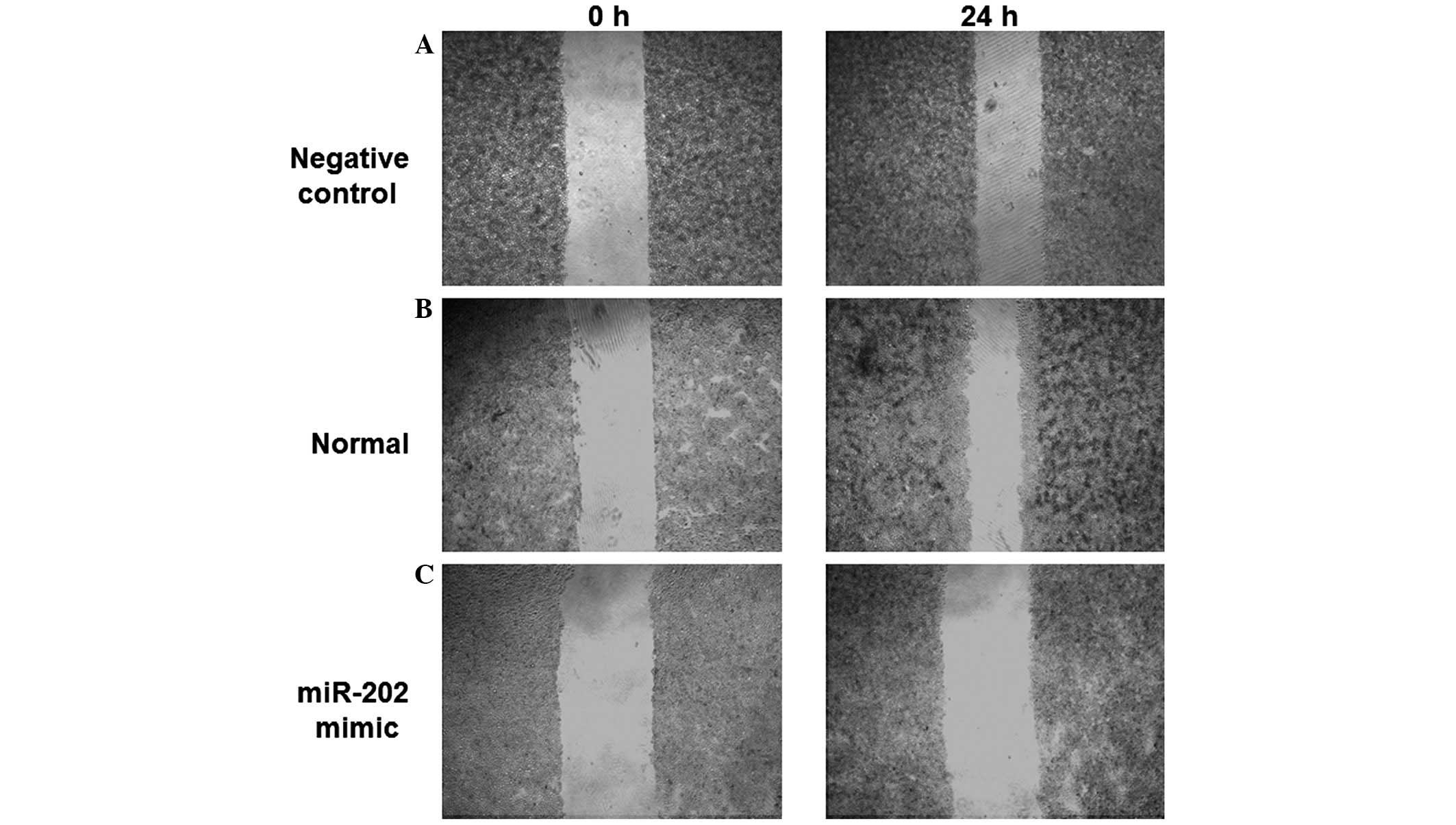
Migratory ability of EC9706 ESCC cell
lines is inhibited by the expression of miR-202
In order to determine the effects of miR-202
expression levels on the migratory ability of ESCC cells, an in
vitro scratch assay was performed. The scratched areas of the
negative control and normal EC9706 cells were decreased 24 h
post-scratch, as compared with 0 h (Fig.
3A and B). However, no significant difference in the scratched
area of EC9706 cells transfected with miR-202 mimics was detected
following 24 h, as compared with 0 h (Fig. 3C). These results demonstrated that
the migratory ability of EC9706 cells was reduced following
transfection with miR-202 mimics, indicating that the migratory
ability of ESCC cells may be inhibited by the expression of
miR-202.
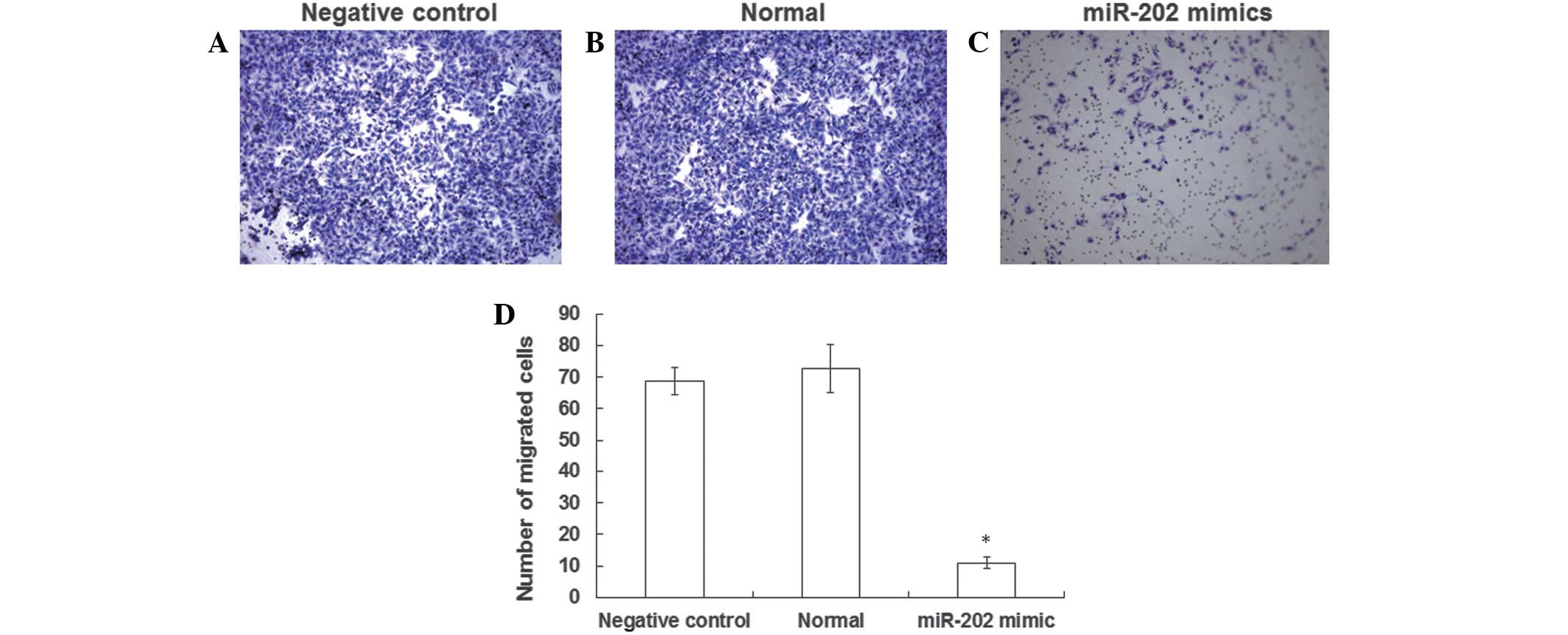
Invasive ability of EC9706 ESCC cell
lines is inhibited by the expression of miR-202
In order to determine the effects of miR-202
expression levels on the invasive ability of EC9706 cells, a
Transwell® chamber assay was performed. The number of negative
control and normal EC9706 cells (Fig. 4A
and B) that successfully invaded the Matrigel layer was
increased, as when compared with EC9706 cells transfected with
miR-202 mimics (Fig. 4C). In
particular, the number of cells that invaded the Matrigel layer was
7 fold higher in the negative control group, as compared with the
EC9706 cells transfected with miR-202 mimics, a result that was
significantly different (Fig. 4D).
The results of the Transwell® chamber assay suggest that the
invasive ability of ESCC cell lines may be inhibited by the
expression of miR-202.
Discussion
Numerous genes are associated with the development,
invasion and metastasis of ESCC. Previous studies have demonstrated
that oncogenes, such as p53, c-Myc, matrix metallopeptidase 9 and
human sterile alpha motif domain-containing 9, are abnormally
expressed in ESCC tissues (22–24).
Furthermore, increased expression levels of transforming growth
factor-β may stimulate the PTEN/PI3K pathway and promote
epithelial-mesenchymal transition in the human esophageal
epithelium (25).
Epithelial-mesenchymal transition is a risk factor for ESCC, as
cell junctions are disrupted and the invasive ability of ESCC is
enhanced (26).
It has previously been demonstrated that there are
evident alterations in the miRNA expression profiles of ESCC
tissues and peripheral blood (27).
The expression levels of miR-145, miR-143 and miR-21, which were
associated with the differentiation, invasion and metastasis of
ESCC, have demonstrated clinical significance in the diagnosis and
prognosis of patients with ESCC (28). In the present study, RT-qPCR was
performed in order to determine the expression levels of miR-202 in
the peripheral blood of patients with ESCC. Furthermore, in
vitro proliferation, migration and invasion assays were
performed to investigate the biological function of miR-202 in the
progression of ESCC, which may be useful in the early diagnosis and
prognosis of patients with ESCC.
The results of the present study demonstrated that
the expression levels of miR-202 were significantly decreased in
the peripheral blood of patients with ESCC, which may be associated
with the degree of differentiation and lymph node metastasis.
Furthermore, the proliferative, migratory and invasive abilities of
ESCC cells were inhibited by the expression of miR-202. These
results suggested that low miR-202 expression levels in peripheral
blood may increase the risk of metastasis in patients with
ESCC.
In conclusion, miR-202 expression levels are reduced
in the peripheral blood of patients with ESCC. As miR-202 is
capable of inhibiting the proliferation, migration and invasion of
ESCC cells, assessing miR-202 expression levels may facilitate
early diagnosis of ESCC and thus improve the prognosis of patients
with ESCC.
Acknowledgements
The authors thank Professor Lijuan Liu at the
Department of Clinical Laboratory (Laiwu People's Hospital, Laiwu,
China).
References
|
1
|
Xing SZ and Zhang Y: Efficacy and safety
of transdermal fentanyl for the treatment of oral mucositis pain
caused by chemoradiotherapy in patients with esophageal squamous
cell carcinoma. Support Care Cancer. 23:753–759. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Guo XF, Mao T, Gu ZT, Ji CY, Fang WT and
Chen WH: Clinical study on postoperative recurrence in patients
with pN0 esophageal squamous cell carcinoma. J Cardiothorac Surg.
9:1502014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jing W, Zhu H, Guo H, Zhang Y, Shi F, Han
A, Li M, Kong L and Yu J: Feasibility of elective nodal irradiation
(ENI) and involved field irradiation (IFI) in radiotherapy for the
elderly patients (aged ≥ 70 years) with esophageal squamous cell
cancer: A retrospective analysis from a single institute. PLoS One.
10:e01430072015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu X, Song N, Liu Y, Liu Y, Li J, Ding J
and Tong Z: Efficient induction of anti-tumor immune response in
esophageal squamous cell carcinoma via dendritic cells expressing
MAGE-A3 and CALR antigens. Cell Immunol. 295:77–82. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Song B, Cui H, Li Y, Cheng C, Yang B, Wang
F, Kong P, Li H, Zhang L, Jia Z, et al: Mutually exclusive
mutations in NOTCH1 and PIK3CA associated with clinical prognosis
and chemotherapy responses of esophageal squamous cell carcinoma in
China. Oncotarget. doi: 10.18632/oncotarget.6120 [Epub ahead of
print]. 2015.
|
|
6
|
Arigami T, Okumura H, Matsumoto M,
Uchikado Y, Uenosono Y, Kita Y, Owaki T, Mori S, Kurahara H, Kijima
Y, et al: Analysis of the fibrinogen and neutrophil-lymphocyte
ratio in esophageal squamous cell carcinoma: A promising blood
marker of tumor progression and prognosis. Medicine (Baltimore).
94:e17022015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhong Y, Xuan P, Han K, Zhang W and Li J:
Improved pre-miRNA classification by reducing the effect of class
imbalance. Biomed Res Int. doi: 10.1155/2015/960108, Epub 2015 Nov
10.
|
|
8
|
Lu Y, Gao W, Zhang C, Wen S, Huangfu H,
Kang J and Wang B: Hsa-miR-301a-3p acts as an oncogene in laryngeal
squamous cell carcinoma via target regulation of Smad4. J Cancer.
6:1260–1275. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Taccioli C, Garofalo M, Chen H, Jiang Y,
Tagliazucchi GM, Di Leva G, Alder H, Fadda P, Middleton J, Smalley
KJ, et al: Repression of esophageal neoplasia and inflammatory
signaling by anti-miR-31 delivery in vivo. J Natl Cancer Inst.
107:pii: djv2202015. View Article : Google Scholar
|
|
10
|
Meng XR, Lu P, Mei JZ, Liu GJ and Fan QX:
Expression analysis of miRNA and target mRNAs in esophageal cancer.
Braz J Med Biol Res. 47:811–817. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang M, Liu R, Li X, Liao J, Pu Y, Pan E,
Yin L and Wang Y: miRNA-183 suppresses apoptosis and promotes
proliferation in esophageal cancer by targeting PDCD4. Mol Cells.
37:873–880. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Okumura T, Shimada Y, Moriyama M, Takei Y,
Omura T, Sekine S, Nagata T, Shimizu K and Tsukada K: MicroRNA-203
inhibits the progression of esophageal squamous cell carcinoma with
restored epithelial tissue architecture in vivo. Int J Oncol.
44:1923–1932. 2014.PubMed/NCBI
|
|
13
|
Yu X, Jiang X, Li H, Guo L, Jiang W and Lu
SH: miR-203 inhibits the proliferation and self-renewal of
esophageal cancer stem-like cells by suppressing stem renewal
factor Bmi-1. Stem Cells Dev. 23:576–585. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sakai NS, Samia-Aly E, Barbera M and
Fitzgerald RC: A review of the current understanding and clinical
utility of miRNAs in esophageal cancer. Semin Cancer Biol.
23:512–521. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gadducci A, Sergiampietri C, Lanfredini N
and Guiggi I: Micro-RNAs and ovarian cancer: The state of art and
perspectives of clinical research. Gynecol Endocrinol. 30:266–271.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rawlings-Goss RA, Campbell MC and Tishkoff
SA: Global population-specific variation in miRNA associated with
cancer risk and clinical biomarkers. BMC Med Genomics. 7:532014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sun Z, Zhang T, Hong H, Liu Q and Zhang H:
miR-202 suppresses proliferation and induces apoptosis of
osteosarcoma cells by downregulating Gli2. Mol Cell Biochem.
397:277–283. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang Y, Zheng D, Xiong Y, Xue C, Chen G,
Yan B and Ye Q: miR-202 suppresses cell proliferation in human
hepatocellular carcinoma by downregulating LRP6
post-transcriptionally. FEBS Lett. 588:1913–1920. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu JJ, Shen XJ, Wang XD and Ju SQ: Effect
of miR-202 on the growth of multiple myeloma cells via regulating B
cell-activating factor and the underlying mechanism. Zhonghua Zhong
Liu Za Zhi. 35:886–891. 2013.(In Chinese). PubMed/NCBI
|
|
20
|
Hu N, Clifford RJ, Yang HH, Wang C,
Goldstein AM, Ding T, Taylor PR and Lee MP: Genome wide analysis of
DNA copy number neutral loss of heterozygosity (CNNLOH) and its
relation to gene expression in esophageal squamous cell carcinoma.
BMC Genomics. 11:5762010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yin X, Zhang R, Feng C, Zhang J, Liu D, Xu
K, Wang X, Zhang S, Li Z, Liu X and Ma H: Diallyl disulfide induces
G2/M arrest and promotes apoptosis through the p53/p21 and MEK-ERK
pathways in human esophageal squamous cell carcinoma. Oncol Rep.
32:1748–1756. 2014.PubMed/NCBI
|
|
23
|
Zhong C, Fan L, Yao F, Shi J, Fang W and
Zhao H: HMGCR is necessary for the tumorigenecity of esophageal
squamous cell carcinoma and is regulated by Myc. Tumour Biol.
35:4123–4129. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tang S, Zheng X, He L, Qiao L, Jing D, Ni
Z, Zeng W and Jian M: Significance of SAMD9 expression in
esophageal squamous cell carcinoma. Xi Bao Yu Fen Zi Mian Yi Xue Za
Zhi. 30:411–413. 2014.(In Chinese). PubMed/NCBI
|
|
25
|
Zhang HY, Wang ZQ, Li YY, Wang F, Zeng QR,
Gao Y, Xuan XY and Li SS: Transforming growth factor-β1-induced
epithelial-mesenchymal transition in human esophageal squamous cell
carcinoma via the PTEN/PI3K signaling pathway. Oncol Rep.
32:2134–2142. 2014.PubMed/NCBI
|
|
26
|
Niwa Y, Yamada S, Koike M, Kanda M, Fujii
T, Nakayama G, Sugimoto H, Nomoto S, Fujiwara M and Kodera Y:
Epithelial to mesenchymal transition correlates with tumor budding
and predicts prognosis in esophageal squamous cell carcinoma. J
Surg Oncol. 110:764–769. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wen J, Luo K, Liu H, Liu S, Lin G, Hu Y,
Zhang X, Wang G, Chen Y, Chen Z, et al: MiRNA expression analysis
of pretreatment biopsies predicts the pathological response of
esophageal squamous cell carcinomas to neoadjuvant
chemoradiotherapy. Ann Surg 2015 Oct 1 [Epub ahead of print].
|
|
28
|
Akagi I, Miyashita M, Ishibashi O, Mishima
T, Kikuchi K, Makino H, Nomura T, Hagiwara N, Uchida E and Takizaw
T: Relationship between altered expression levels of MIR21, MIR143,
MIR145, and MIR205 and clinicopathologic features of esophageal
squamous cell carcinoma. Dis Esophagus. 24:523–530. 2011.
View Article : Google Scholar : PubMed/NCBI
|