Introduction
Resveratrol (RSV; 3,5,4-trihydroxystilbene) is a
natural polyphenol found in grapes, red wine, peanuts and berries
(1,2). RSV possesses anti-oxidative and
anti-inflammatory properties and is able to modulate lipid
metabolism (3–7). Therefore, a diet rich in RSV may
attenuate diabetes, and cardiovascular and stress-related diseases
(8–11). The liver is the major organ
responsible for metabolizing nutrients in the body and is easily
damaged by an imbalance in redox status (12). However, the underlying mechanism by
which RSV exerts its beneficial effects on hepatic damage remains
unclear.
Alcoholic drinks are widely consumed throughout the
world and have been implicated in many diseases such as liver
cirrhosis and liver cancer, which globally account for an equal
number of cases of mortality and disability as tobacco (13). The liver is the primary organ
responsible for alcohol metabolism in the human body, and alcohol
is primarily metabolized in hepatocytes. Reactive oxygen species
(ROS) are known to cause ethanol-induced liver damage (14,15), and
a manifestation of alcoholic liver injury is lipid accumulation
(16). Imbalanced lipid homeostasis,
in addition to the generation of ROS, promotes the hepatic symptoms
of steatosis, fibrosis, cirrhosis and hepatitis (17,18). The
antioxidative properties of RSV suggest that it may be a promising
protective agent against alcoholic liver disease. In order to
delineate the protective effect of RSV against oxidative damage
in vivo, hepatic malondialdehyde (MDA, a lipid peroxidation
product) levels and histopathology of the livers of ethanol plus
RSV-treated mice were examined in the present study.
Endogenous antioxidative enzymes such as superoxide
dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPx)
are able to protect the liver against oxidative damage (19). SOD scavenges the superoxide anion
radical (O2•−) to oxygen and hydrogen
peroxide (H2O2). H2O2
may then be converted into water by CAT. By contrast, GPx is able
to protect the cell against oxidative stress via the reduction of
H2O2 and lipid peroxides, using glutathione
as an electron donor (20). The
present study provides an insight into the role of RSV in the
regulation of antioxidative enzyme activity and gene expression,
which contribute towards the defense mechanism against ROS in
hepatocytes under oxidative stress.
Peroxisome proliferator-activated receptors (PPARs)
are nuclear receptors that contribute towards nutrient-gene
interactions, which are involved in the maintenance of metabolic
homeostasis (21,22). PPARs bind to retinoid-X receptors to
form heterodimers and regulate the expression of their target
genes, which have PPAR-response elements (PPREs) in their promoter
regions (23). PPARs consist of 3
isoforms, namely PPARα, PPARβ/δ and PPARγ (23). Among these PPARs, PPARα is expressed
at a relatively high concentration in the liver, where it oxidizes
fatty acids and metabolizes lipids (24,25).
PPARβ/δ is involved in cell differentiation and epidermal wound
healing (26–28). However, PPARγ regulates antioxidative
enzyme genes, including SOD (29)
and CAT (30). It has been suggested
that PPARγ may serve an essential role in protecting organs against
oxidative stress (31). The present
study hypothesizes that RSV may display a modulatory role on PPARs
in maintaining hepatic lipid homeostasis and in executing its
antioxidative properties.
In the current study, the ability of RSV to modulate
the activity of antioxidative enzymes in the mouse liver and in
HepG2 cells was investigated. Furthermore, the effects of RSV on
the expression of PPARs and the transcriptional activities of the
reporter genes were evaluated using various PPREs in the
5′-upstream of the luciferase gene.
Materials and methods
Experimental animals
Thirty-two male C57BL/6J mice (6–8 weeks old,
18.5–20.3 g) were obtained from the Laboratory Animal Center of the
National Yang Ming University (Taipei, Taiwan), and randomly
assigned to one of four groups (n=8 per group). Two animals were
housed in each cage and maintained at 25°C in under a 12-h
light/dark cycle. All experimental procedures were approved by the
Animal Care and Use Committee of the Chang Gung Medical Foundation
(Chiayi, Taiwan) and complied with the NIH Guide for Care and Use
of Laboratory Animals (National Research Council, 1996) (32). Mice in the four groups were
administered daily, via oral gavage for 28 consecutive days, with
the following: Ethanol (200 mg/kg); RSV (200 mg/kg; Sigma-Aldrich,
St. Louis, MA, USA); ethanol + RSV (100 mg/kg each); or distilled
water (control group). RSV was diluted in sterile water. Food and
tap water were available ad libitum and body weight was
recorded weekly throughout the experiment.
Following treatment, mice were anesthetized with
CO2 and blood samples were extracted following a 12-h
overnight fast. Plasma was isolated by centrifugation at 1,500 × g
for 15 min and was stored at −35°C for subsequent biochemical
analyses. Plasma concentrations of total cholesterol and
triglycerol were determined spectrophotometrically using commercial
kits (Merck Millipore, Darmstadt, Germany) according to
manufacturer's instructions. The liver was rapidly removed, blotted
dry, weighed, frozen in liquid nitrogen and stored at −80°C until
use. To analyze antioxidative enzyme activity and expression, the
liver tissue was homogenized in 10 volumes of 50 mM phosphate
buffer (pH 7.4; Sigma-Aldrich) on ice using a Polytron homogenizer
(model 099C-K54; Glas-Col LLC, Terre Haute, IN, USA). The
homogenate was transferred into centrifuge tubes and centrifuged at
9,000 × g at 4°C for 20 min. The supernatant was separated for use
in the subsequent measurement of antioxidative enzyme activity.
Measurement of MDA and hepatic lipid
accumulation
Plasma levels of MDA, an oxidative stress marker,
were monitored by quantifying thiobarbituric acid (TBA)-reactive
substances as previously described (33). Briefly, 1 g liver tissue was
homogenized in 10 ml 1.15% KCl buffer (Sigma-Aldrich). The
homogenate was mixed with 1% H3PO4
(Sigma-Aldrich) and 0.6% TBA (Sigma-Aldrich), and heated at 100°C
for 45 min. The samples were cooled to room temperature and
combined with n-butanol (Merck Millipore). Following vigorous
vortexing, the butanolic phase was centrifuged at 4,000 × g for 10
min. 1,1,3,3-Tetraethoxypropane (Merck Millipore) was used as the
standard. The histology of hepatic microvesicular steatosis was
assessed using the Oil Red O (Sigma-Aldrich) staining method as
previously described (34).
Cell culture, cell viability and ROS
assays
HepG2 is a well-differentiated human hepatocarcinoma
cell line commonly used in hepatic studies. HepG2 cells were
provided by Prof. An-Na Chiang and cultured in Dulbecco's Modified
Eagle's Medium (HyClone; GE Healthcare, Logan, UT, USA) containing
10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml
streptomycin (GE Healthcare). After 24 h of growth at 37°C in 5%
CO2, cells were treated with ethanol or RSV (50, 100,
200 or 400 mM) for 24 h. The
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay (Sigma-Aldrich) was used to evaluate the cytotoxic effects of
ethanol and RSV (35). The ROS
levels in HepG2 cells were measured using the dye,
2′,7′-dichlorodihydrofluorescein diacetate (Molecular Probes;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), as described
previously (36). This reduced dye
was added to cells at a final concentration of 10 µM. The
fluorescence of the oxidized dichlorofluorescein was recorded, with
an excitation wavelength of 488 nm and an emission wavelength of
525 nm, using flow cytometry (model FC500; Beckman Coulter, Inc.,
Brea, CA, USA). The results were expressed as the relative
fluorescence intensity. Measurements of ROS levels without ethanol
or RSV treatment were used as the control.
Measurement of antioxidative enzyme
activity
SOD activity in the liver extracts of C57BL/6 mice
or HepG2 cells was assayed using the hydroxylamine reduction method
(37). The hypoxanthine/xanthine
oxidase system (38) was used to
measure the reduction of hydroxylamine by
O2•−, which was monitored at 550 nm. One unit
of SOD activity was recorded as the quantity of enzyme required to
decrease the reduction of hydroxylamine by 50%. Mouse liver or
HepG2 cell CAT activity in the extract was assayed using the method
described by Aebi (39).
Decomposition of H2O2 resulting from CAT
activity was assayed by monitoring H2O2 and
the reduction in absorbance at 240 nm. One unit of CAT activity was
recorded as the quantity of enzyme catalyzing 1 µmol
H2O2 per min at 25°C. GPx activity was
quantified according to a coupled enzyme (GPx and glutathione
reductase) procedure (40), which
measures the decrease in absorbance at 340 nm as NADPH is converted
to NADP. One unit of GPx activity was recorded as the quantity of
enzyme oxidizing 1 µmol NADPH per min. The specific activity of
SOD, CAT and GPx are expressed as U/mg protein. The protein content
of the liver or cell extract was determined using the Bradford
method (Bio-Rad Laboratories, Inc., Hercules, CA, USA) (41).
Western blot analysis
Protein levels of antioxidative enzymes and PPARs
were determined using western blot analysis in HepG2 cells
supplemented with ethanol and/or RSV for 24 h. Total cell protein
was extracted using lysis buffer containing 1% Triton X-100, 50 mM
HEPES, 6 mM EDTA, and 150 mM NaCl supplemented with complete
protease inhibitor cocktail (Roche Diagnostics GmbH, Mannheim,
Germany). Cell lysates were centrifuged, and the supernatants were
collected. The protein concentration was determined using the
Bradford method (Bio-Rad Laboratories, Inc.) using bovine serum
albumin as a standard, and equal quantities of protein (30 µg) were
analyzed by 10% sodium dodecyl sulfate polyacrylamide gel
electrophoresis and transferred to nitrocellulose membranes (Pall
Corporation, Port Washington, NY, USA). The immunoblots were
blocked with 5% non-fat milk and then incubated at 4°C for 16–32 h
with the following primary antibodies: Rabbit anti-human SOD
polyclonal antibody (1:5,000; cat. no. ab13533; Abcam, Cambridge,
UK); rabbit anti-human CAT polyclonal antibody (1:2,000; cat. no.
ab16731; Abcam); goat anti-human GPx IgG (1:1,000; cat. no.
sc-22145; Santa Cruz Biotechnology, Inc., Dallas, TX, USA); rabbit
anti-human PPARα IgG (1:1,000; cat. no. sc-9000; Santa Cruz
Biotechnology, Inc.); rabbit anti-human PPARβ/δ IgG (1:1,000; cat.
no. sc-7197; Santa Cruz Biotechnology, Inc.); goat anti-human PPARγ
IgG (1:1,000; cat. no. sc-1981; Santa Cruz Biotechnology, Inc.).
Following the washing stage, the blots were incubated with
horseradish peroxidase-conjugated secondary antibodies
(Sigma-Aldrich) for 1 h at 4°C, and the target protein bands were
visualized using Western Lightning Plus-Enhanced Chemiluminescence
reagents (PerkinElmer, Inc., Waltham, MA, USA). The blots were then
stripped for further probing, using β-actin (rabbit anti-human
β-actin polyclonal antibody; cat. no. ab189073; Abcam) as an
internal control. Relative intensities of protein bands were
quantified by densitometry using ImageQuant software version 5.0
(Molecular Dynamics, Sunnyvale, CA, USA).
RNA extraction and quantitative
polymerase chain reaction (qPCR) analysis
Total RNA was extracted from HepG2 cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Total RNA (2 µg) was
reverse transcribed using Moloney murine leukemia virus reverse
transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.). The
resulting cDNA (1 µg) was used as a template for qPCR analysis
using SYBR Green Master Mixture (QuantiTect SYBR Green PCR kit 200;
cat. no. 204143; Qiagen, Inc., Valencia, CA, USA) in a Roche
LightCycler system (Roche Diagnostics GmbH). The specific primers
(50 µM in 0.08 µl) for SOD, CAT and GPx genes were used as
previously described (42). The
following primer sequences were used: Cu/Zn-SOD forward,
5′-CAGGTCCTCACTTCAATCC-3′ and reverse, 5′-CCAAACGACTTCCAGCAT-3′;
CAT forward, 5′-CGAAGGCGAAGGTGTTTG-3′ and reverse,
5′-AGTGTGCGATCCATATCC-3′; GPx forward, 5′-CACAACGGTGCGGGACTA-3′ and
reverse, 5′-CATTGCGACACACTGGAGAC-3′; and glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) forward, 5′-GAAGGTGAAGGTCGGAGTC-3′ and
reverse, 5′-GAAGATGGTGATGGGATTTC-3′. Comparative assessment of
target gene mRNA expression was performed using GAPDH mRNA as an
internal control. Reaction conditions were 95°C for 10 min,
followed by 40 cycles of 95°C for 20 sec, 60°C for 20 sec and 72°C
for 20 sec. Relative quantification of antioxidative enzyme mRNA
was calculated using the Cq method (ratio = 2 − (Cq
(antioxidant enzyme) - Cq (GAPDH)) as described previously
(43).
Transient transfection assays
HepG2 cells were transiently transfected with 0.2 µg
tk-PPREx-Luc reporter plasmid and 0.2 µg PPARα (pGShPPARα), PPARβ
(pCMX-hPPARβ/δ) or PPARγ (pCMX-hPPARγ) expression vectors, which
were constructed by Prof. An-Na Chiang's laboratory. For each
transfection, the internal control vector, pCMV-β-gal, was also
co-transfected. The luciferase assay was measured using a
luminometer (EG&G Berthold; Berthold Technologies USA LLC, Oak
Ridge, TN, USA) and normalized against the activity of
β-galactosidase (44).
Statistical analysis
Data are presented as the mean ± standard error.
Each experiment was repeated at least three times. Statistical
analysis between two groups was performed using the Student's
t-test. One-way analysis of variance combined with Tukey's
multiple-comparison test was used to evaluate the statistical
significance of differences among the four groups. Statistical
analysis was performed using SPSS software version 17.0 (SPSS,
Inc., Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
Effect of ethanol and RSV on plasma
lipid profile and hepatic antioxidative enzyme activity
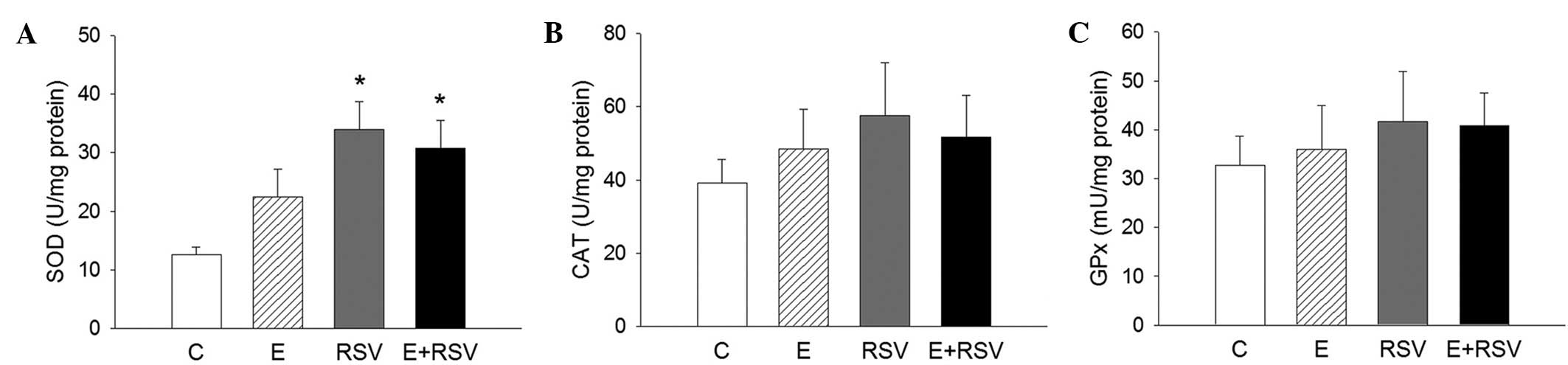
Compared with the control mice, no significant
differences were detected in the body weight and concentrations of
plasma cholesterol levels among the groups (Table I). However, plasma triglyceride and
MDA levels were 75.3 and 56.1% higher, respectively, in the
ethanol-treated mice in comparison with the control mice (Table I). After 28 days of treatment, mice
in the RSV group presented a 79.5% increase in SOD activity;
however, no change was observed in the ethanol and ethanol + RSV
groups. Moreover, ethanol and RSV exerted no significant effect on
CAT and GPx activity compared with the control group in the livers
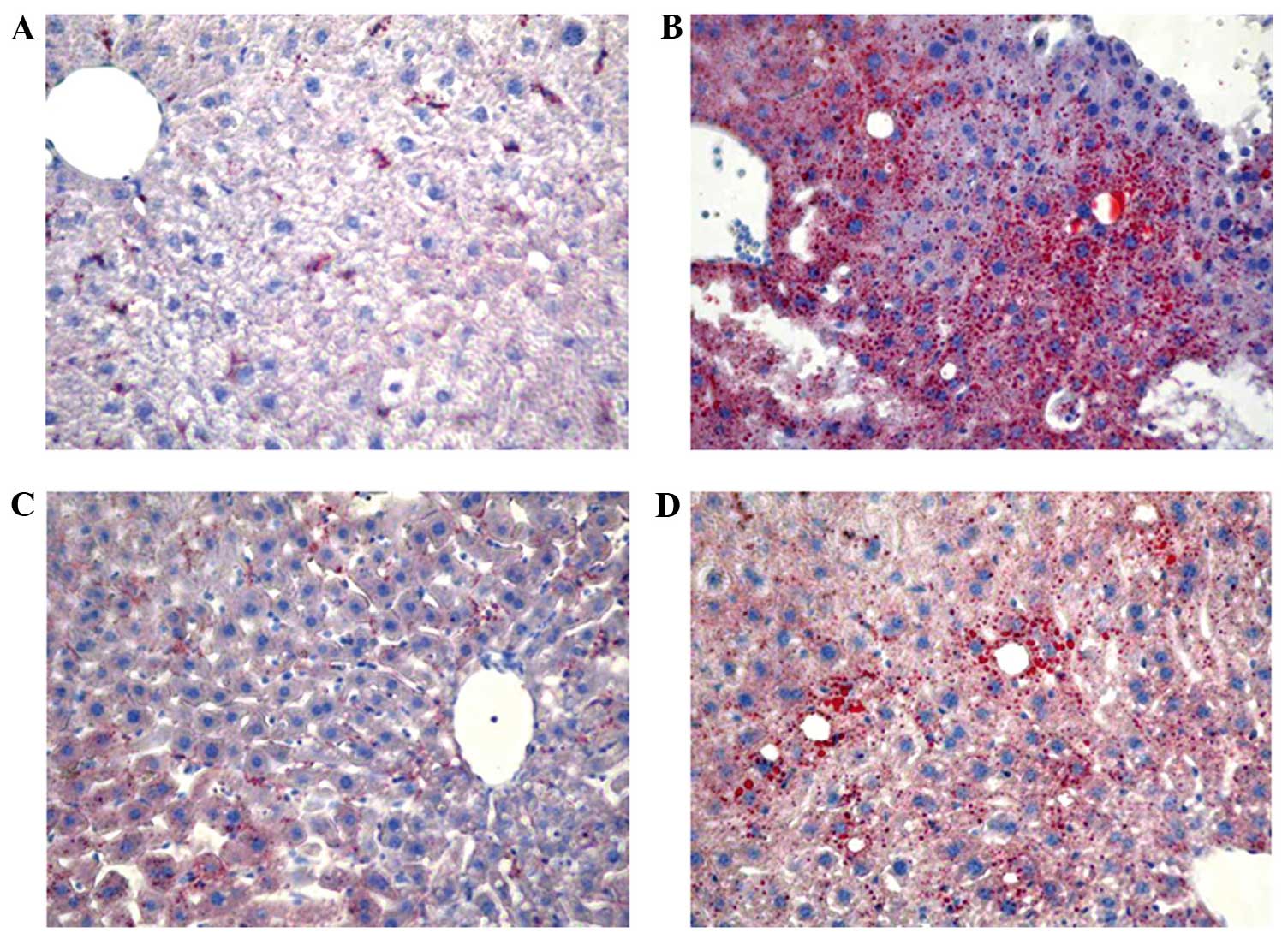
of C57BL/6 mice. Furthermore, lipid accumulation was markedly
increased in the liver of mice treated with ethanol, moderately
increased in mice treated with ethanol + RSV and remained unchanged
in mice treated with RSV (Fig.
1).
 | Table I.Effect of ethanol and resveratrol on
plasma lipid profile and hepatic antioxidative enzyme activity
in vivo. |
Table I.
Effect of ethanol and resveratrol on
plasma lipid profile and hepatic antioxidative enzyme activity
in vivo.
| Parameter | Control | Ethanol | Resveratrol | Ethanol +
resveratrol |
|---|
| Body weight, g | 25.5±0.68 | 26.1±0.58 | 26.3±0.83 | 25.9±0.66 |
| Plasma lipid |
|
|
|
|
| Total
cholesterol, mg/dl | 96.4±10.3 | 130±14.6 | 121±12.8 | 129±14.9 |
|
Triglyceride, mg/dl | 81.0±10.8 |
142±13.4a | 103±12.7 | 122±15.4 |
|
Malondialdehyde, nmol/l | 254±30.4 |
396±37.5a | 297±27.2 | 333±41.2 |
| Hepatic
antioxidative enzyme |
|
|
|
|
|
Superoxide dismutase,
U/mg | 15.1±1.5 | 18.7±2.2 |
27.1±3.1a | 22.6±2.8 |
|
Catalase, U/mg | 42.1±4.8 | 45.4±5.0 | 47.8±4.7 | 46.5±3.9 |
|
Glutathione peroxidase,
mU/mg | 34.3±4.3 | 39.5±4.1 | 37.1±4.0 | 38.9±4.4 |
Effect of ethanol and RSV on cell
viability and ROS production
No significant alteration of cell viability was
detected by the MTT assay in the HepG2 cells exposed to ethanol,
RSV or ethanol + RSV (Fig. 2A). By
contrast, ethanol treatment appeared to enhance ROS production in a
dose-dependent manner (Fig. 2B).
However, no significant change in ROS production was observed in
the ethanol + RSV group. This indicates that RSV is able to
scavenge ROS from ethanol-induced ROS generation.
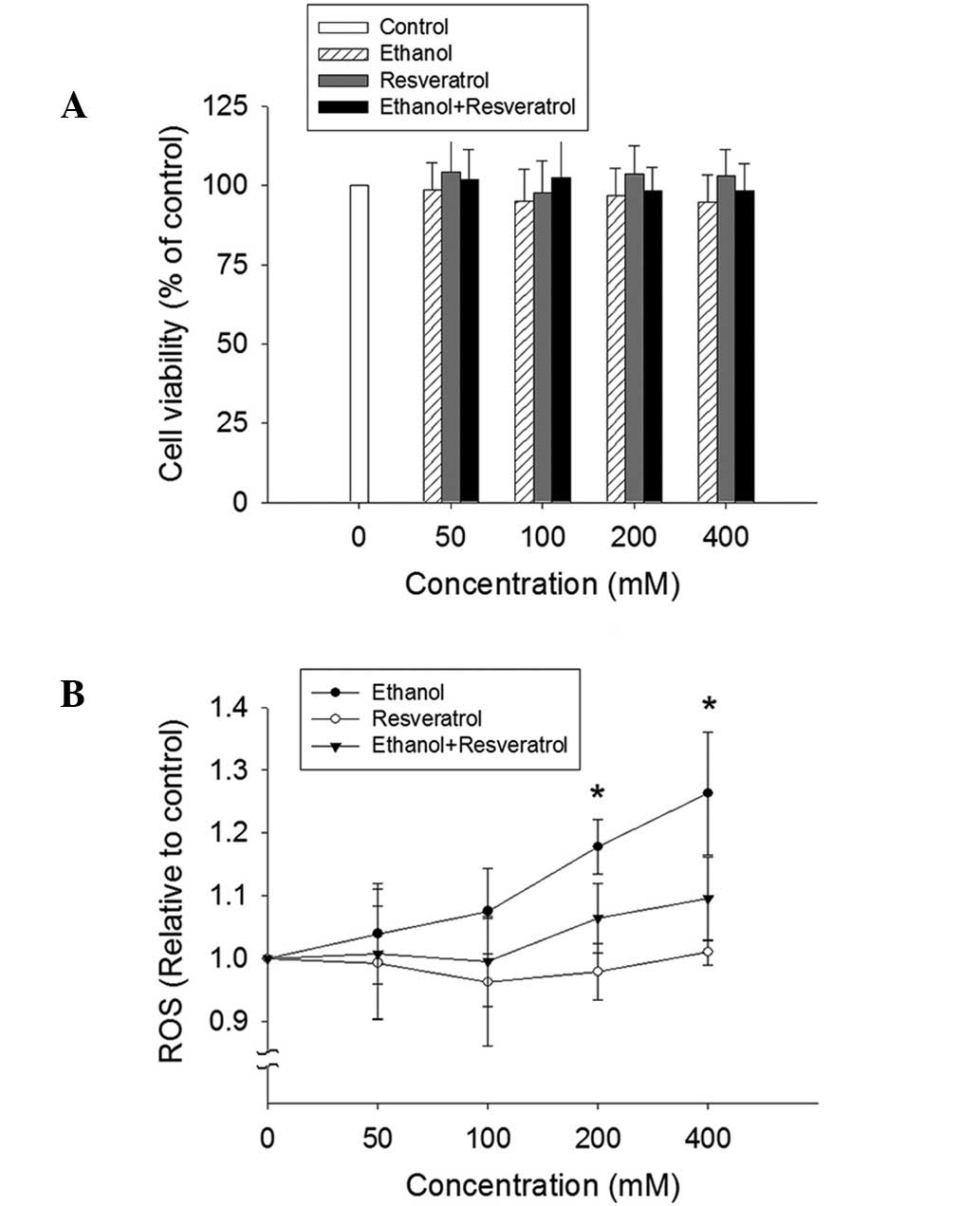 | Figure 2.Effect of ethanol and resveratrol on
cell viability and ROS production. (A) HepG2 cells were treated
with 50, 100, 200 and 400 mM ethanol, resveratrol, and ethanol plus
resveratrol for 24 h. The impact of resveratrol on cell viability
was assessed using an MTT assay. All values are expressed as a
percentage of the control group, which is set as 100%. (B) ROS
production was determined by the conversion of
2′,7′-dichlorodihydrofluorescein diacetate to
2′,7′-dichlorodihydrofluorescein. The results were expressed as the
relative fluorescence intensity compared to the control, which is
set as 1. Data are presented as the mean ± standard error (n=3-5)
*P<0.05, vs. control. ROS, reactive oxygen species. |
Effect of ethanol and RSV on the
activity and expression of antioxidative enzymes in HepG2
cells
To assess whether antioxidative enzymes are involved
in RSV-mediated protection against oxidative stress, the activity
of SOD, CAT and GPx was determined in HepG2 cells. As presented in
Fig. 3, SOD activity significantly
increased in cells exposed to RSV in comparison with the control
(P<0.05), whereas the activity of CAT and GPx was not affected
by ethanol or RSV treatment. The expression levels of SOD, CAT, and
GPx protein (Fig. 4A–C) and mRNA
(Fig. 4D–F) in HepG2 cells were
determined by western blot and qPCR analysis. In comparison with
the control cells, the expression levels of SOD protein and mRNA
were 1.9- and 1.95-fold higher in the RSV group, respectively.
There was no change in the expression of CAT and GPx in the three
experimental groups in comparison with the control group.
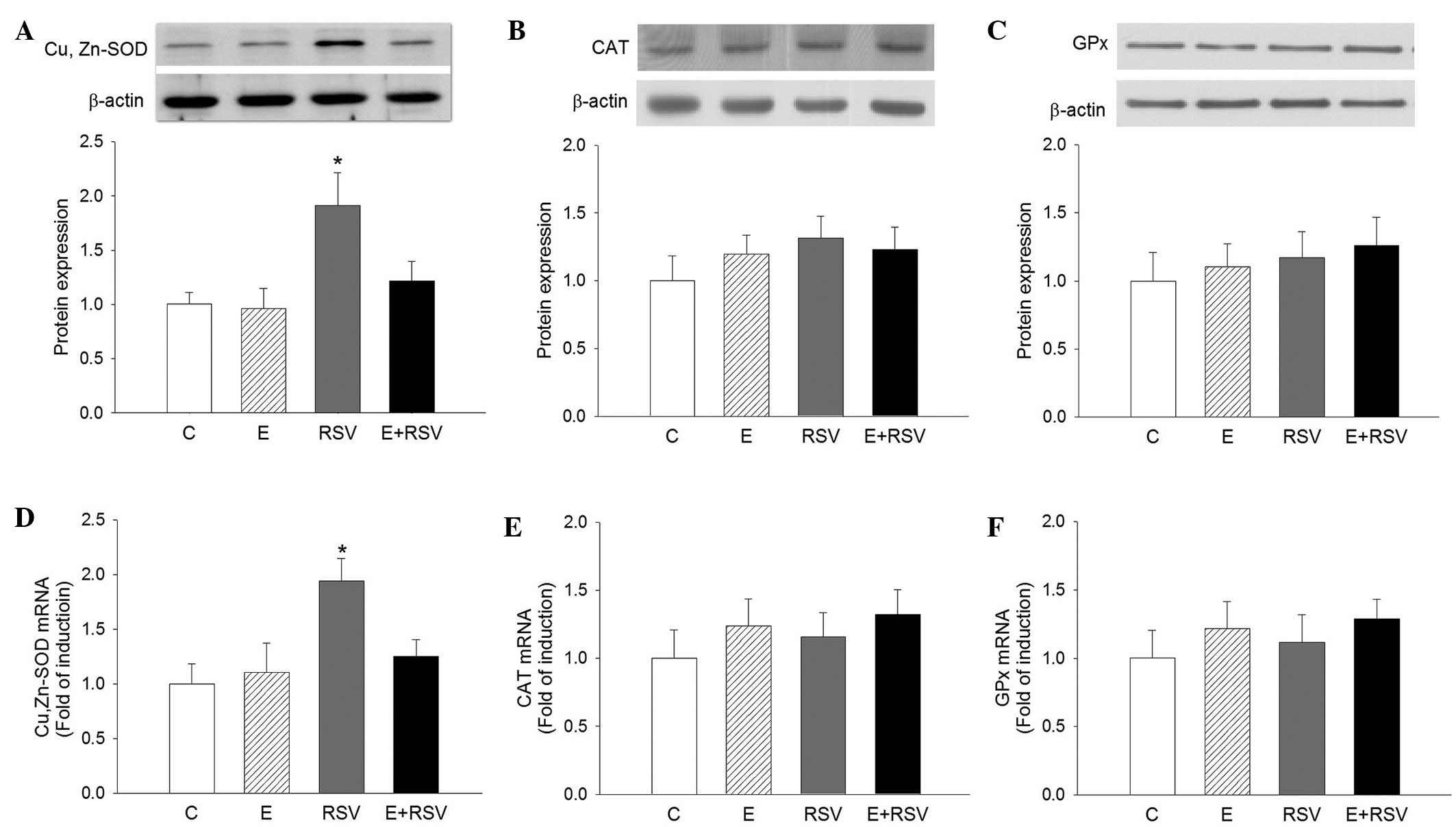 | Figure 4.Effects of E and RSV on antioxidative
enzyme mRNA and protein expression. Western blot and quantitative
protein expression levels of (A) SOD, (B) CAT, and (C) GPx, and
mRNA levels of (D) SOD, (E) CAT and (F) GPx were analyzed. The
results are presented as the mean ± standard error from four
experiments. *P<0.05 vs. control group. C, control; E, ethanol;
RSV, resveratrol; SOD, superoxide dismutase; CAT, catalase; GPx,
glutathione peroxidase. |
Effect of ethanol and RSV on the
protein transactivation activity of PPARs
To determine the mechanism underlying the induction
of SOD gene expression by RSV, the expression and transactivation
activity of PPARs in HepG2 cells was determined. PPARα, PPARβ and
PPARγ protein expression was not affected in cells treated with
ethanol and/or RSV (Fig. 5A and B).
However, the transfection of RSV-treated HepG2 cells with the
tk-PPREγ-Luc reporter construct and pCMX-hPPARγ resulted in a
2.1-fold increase in promoter activity in comparison with the
untreated control cells (Fig. 5C).
The treatment of HepG2 cells with 200 mM ethanol alone or 100 mM
ethanol + 100 mM RSV did not exert any significant effect on PPAR
transactivation.
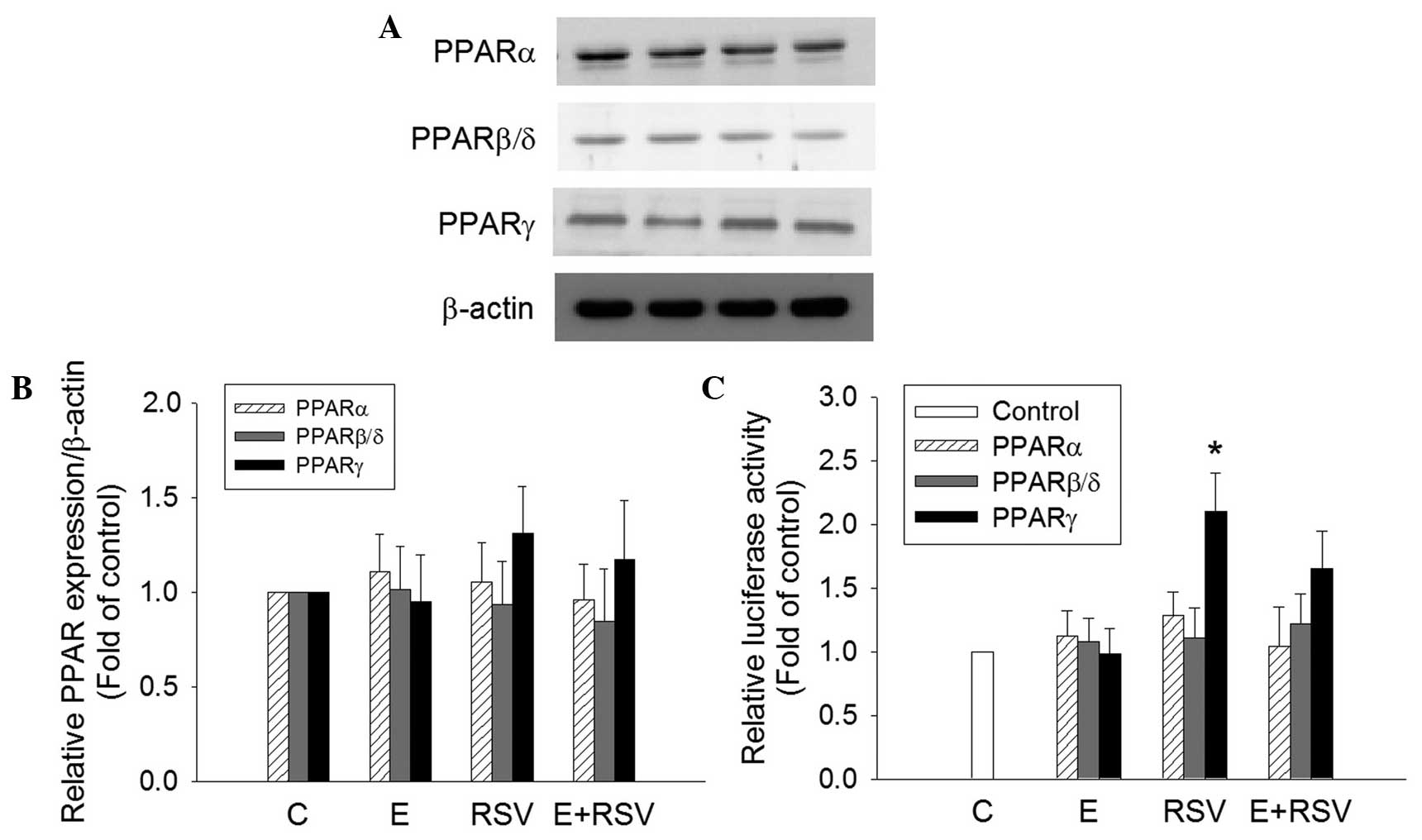 | Figure 5.Effect of E and RSV on PPAR
expression and activity. (A) Relative protein expression levels of
PPARα, PPARβ/δ, and PPARγ were measured using western blot analysis
of nuclear extracts from HepG2 cells supplemented with ethanol
and/or RSV for 24 h. (B) The fold inductions of PPAR protein levels
were expressed relative to the control, which is set as 1. Data are
presented as the mean ± standard error (n=4). (C) Transcriptional
activity of PPARα, PPARβ/δ, and PPARγ were evaluated by
transfection assays using HepG2 cells with PPRE-luc together with
pGSH PPARα, pCMX-hPPARβ/δ or pCMX-hPPARγ plasmids, respectively.
Results are presented as the relative luciferase activity obtained
by dividing normalized luciferase activity from the reporter vector
PPRE-luc. *P<0.05 vs. control cells (n=4). PPAR,
proliferator-activated receptor; C, control; E, ethanol; RSV,
resveratrol. |
Discussion
Alcoholic drinks typically range between ~4.5%
ethanol (900 mM) in beer and ~12% ethanol (2 M) in wine (45). A large quantity of ingested ethanol
circulates to the liver via the portal vein and creates a state of
oxidative stress and lipid accumulation in hepatocytes (14,46). In
the present study, plasma triglyceride levels and MDA generation
were increased in ethanol-treated mice, indicating that ethanol
causes lipid metabolism disorder and increases lipid peroxidation
in vivo. Oil Red O staining identified RSV-attenuated
ethanol-induced lipid accumulation in the liver, which may be the
cause or effect of ethanol on liver damage. The ethanol + RSV group
had a lower accumulation of lipids in the liver compared with the
ethanol treated group.
An imbalance of redox status may result in a wide
range of health complications and various pathophysiological
syndromes (47). Ethanol-induced
oxidative stress causes liver damage in alcoholism (47). RSV, a widely used nutritional
component, possesses various biological functions, including
antioxidative effects (10). A
previous study demonstrated that RSV inhibits foam cell formation
via the suppression of ROS generation in macrophages (48). The present study aimed to determine
whether RSV is able to prevent the ethanol-induced oxidative stress
in hepatocytes.
An endogenous antioxidant system composed of
antioxidative enzymes such as SOD, CAT and GPx, is responsible for
the protection against free radical damage in vivo (49). The results of the present study
suggest that the protective effect of RSV against ethanol-induced
liver damage results from its ability to induce the activity of
hepatic SOD. This was previously indicated in a study by
Kasdallah-Grissa et al (50)
who reported that the hepatic activity of SOD, CAT and GPx was
suppressed in ethanol-treated rats, which was reversed by treatment
with RSV. In the current study, the mechanisms underlying the
protective action of RSV against oxidative stress, using HepG2
cells as the hepatic cellular model, were investigated. The results
demonstrated that ethanol enhances ROS in a dose-dependent manner,
and RSV protects cells against oxidative damage by suppressing ROS
generation. Moreover, in the present study, RSV administration was
demonstrated to enhance the activity and expression of SOD in
comparison with the control group.
Previously, RSV has been observed to selectively
activate PPARs in neuronal cells and adipocytes (51,52). The
SOD gene has known to be a PPARγ target gene (29). The present study hypothesized that
RSV activates hepatic SOD gene expression through PPAR activation.
The effect of RSV on PPAR expression and its transcriptional
activation was investigated using western blot analysis and a
luciferase assay. It was observed that RSV significantly
upregulated PPARγ activity; however, its fundamental mechanism
still remains to be elucidated.
In summary, the present study demonstrates that RSV
serves an essential role in mitigating ethanol-induced hepatic
oxidative stress in vivo and in vitro. This
protective effect may result from the suppression of lipid
peroxidation and accumulation, and the activation of SOD gene
expression. Furthermore, it may be suggested that PPARγ activation
is involved in RSV-enhanced SOD gene regulation. The results in the
current study identify novel mechanisms underlying the protective
actions of RSV and provide a novel insight into the prevention of
ethanol-induced hepatic damage and liver disease.
Acknowledgements
The present study was supported by the Chang Gung
Medical Foundation of Taiwan (grant no. CMRP:G6A0311). The authors
thank Prof. An-Na Chiang (Institute of Biochemistry and Molecular
Biology, National Yang-Ming University, Taipei, Taiwan) for
providing HepG2 cells and technical assistance.
References
|
1
|
Pirola L and Fröjdö S: Resveratrol: One
molecule, many targets. IUBMB Life. 60:323–332. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shakibaei M, Harikumar KB and Aggarwal BB:
Resveratrol addiction: To die or not to die. Mol Nutr Food Res.
53:115–128. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Leonard SS, Xia C, Jiang BH, Stinefelt B,
Klandorf H, Harris GK and Shi X: Resveratrol scavenges reactive
oxygen species and effects radical-induced cellular responses.
Biochem Biophys Res Commun. 309:1017–1026. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lasa A, Schweiger M, Kotzbeck P, Churruca
I, Simón E, Zechner R and Portillo MP: Resveratrol regulates
lipolysis via adipose triglyceride lipase. J Nutr Biochem.
23:379–384. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mikuła-Pietrasik J, Kuczmarska A, Rubiś B,
Filas V, Murias M, Zieliński P, Piwocka K and Książek K:
Resveratrol delays replicative senescence of human mesothelial
cells via mobilization of antioxidative and DNA repair mechanisms.
Free Radic Biol Med. 52:2234–2245. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Park DW, Kim JS, Chin BR and Baek SH:
Resveratrol inhibits inflammation induced by heat-killed
Listeria monocytogenes. J Med Food. 15:788–794. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Voloshyna I, Hussaini SM and Reiss AB:
Resveratrol in cholesterol metabolism and atherosclerosis. J Med
Food. 15:763–773. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bhatt JK, Thomas S and Nanjan MJ:
Resveratrol supplementation improves glycemic control in type 2
diabetes mellitus. Nutr Res. 32:537–541. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chang CC, Chang CY, Huang JP and Hung LM:
Effect of resveratrol on oxidative and inflammatory stress in liver
and spleen of streptozotocin-induced type 1 diabetic rats. Chin J
Physiol. 55:192–201. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sadruddin S and Arora R: Resveratrol:
Biologic and therapeutic implications. J Cardiometab Syndr.
4:102–106. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tomé-Carneiro J, Gonzálvez M, Larrosa M,
García-Almagro FJ, Avilés-Plaza F, Parra S, Yáñez-Gascón MJ,
Ruiz-Ros JA, García-Conesa MT, Tomás-Barberán FA and Espín JC:
Consumption of a grape extract supplement containing resveratrol
decreases oxidized LDL and ApoB in patients undergoing primary
prevention of cardiovascular disease: A triple-blind, 6-month
follow-up, placebo-controlled, randomized trial. Mol Nutr Food Res.
56:810–821. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cesaratto L, Vascotto C, Calligaris S and
Tell G: The importance of redox state in liver damage. Ann Hepatol.
3:86–92. 2004.PubMed/NCBI
|
|
13
|
Room R, Babor T and Rehm J: Alcohol and
public health. Lancet. 365:519–530. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cahill A, Cunningham CC, Adachi M, Ishii
H, Bailey SM, Fromenty B and Davies A: Effects of alcohol and
oxidative stress on liver pathology: The role of the mitochondrion.
Alcohol Clin Exp Res. 26:907–915. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hoek JB and Pastorino JG: Cellular
signaling mechanisms in alcohol-induced liver damage. Semin Liver
Dis. 24:257–272. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu KC, Liu J and Klaassen CD: Role of Nrf2
in preventing ethanol-induced oxidative stress and lipid
accumulation. Toxicol Appl Pharmacol. 262:321–329. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
de Alwis NM and Day CP: Non-alcoholic
fatty liver disease: The mist gradually clears. J Hepatol. 48(Suppl
1): S104–S112. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mann RE, Smart RG and Govoni R: The
epidemiology of alcoholic liver disease. Alcohol Res Health.
27:209–219. 2003.PubMed/NCBI
|
|
19
|
Kumar M, Sharma VL, Sehgal A and Jain M:
Protective effects of green and white tea against benzo(a)pyrene
induced oxidative stress and DNA damage in murine model. Nutr
Cancer. 64:300–306. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pigeolet E, Corbisier P, Houbion A,
Lambert D, Michiels C, Raes M, Zachary MD and Remacle J:
Glutathione peroxidase, superoxide dismutase, and catalase
inactivation by peroxides and oxygen derived free radicals. Mech
Ageing Dev. 3:283–297. 1990. View Article : Google Scholar
|
|
21
|
Jonker JW, Suh JM, Atkins AR, Ahmadian M,
Li P, Whyte J, He M, Juguilon H, Yin YQ, Phillips CT, et al: A
PPARγ-FGF1 axis is required for adaptive adipose remodelling and
metabolic homeostasis. Nature. 485:391–394. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kidani Y and Bensinger SJ: Liver X
receptor and peroxisome proliferator-activated receptor as
integrators of lipid homeostasis and immunity. Immunol Rev.
249:72–83. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Berger J and Moller DE: The mechanisms of
action of PPARs. Annu Rev Med. 53:409–435. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang J, Jia Y, Fu T, Viswakarma N, Bai L,
Rao MS, Zhu Y, Borensztajn J and Reddy JK: Sustained activation of
PPARα by endogenous ligands increases hepatic fatty acid oxidation
and prevents obesity in ob/ob mice. FASEB J. 26:628–638. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Takahashi N, Senda M, Lin S, Goto T, Yano
M, Sasaki T, Murakami S and Kawada T: Auraptene regulates gene
expression involved in lipid metabolism through PPARα activation in
diabetic obese mice. Mol Nutr Food Res. 55:1791–1797. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chong HC, Tan MJ, Philippe V, Tan SH, Tan
CK, Ku CW, Goh YY, Wahli W, Michalik L and Tan NS: Regulation of
epithelial-mesenchymal IL-1 signaling by PPARbeta/delta is
essential for skin homeostasis and wound healing. J Cell Biol.
184:817–831. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Man MQ, Barish GD, Schmuth M, Crumrine D,
Barak Y, Chang S, Jiang Y, Evans RM, Elias PM and Feingold KR:
Deficiency of PPARbeta/delta in the epidermis results in defective
cutaneous permeability barrier homeostasis and increased
inflammation. J Invest Dermatol. 128:370–377. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Müller R, Rieck M and Müller-Brüsselbach
S: Regulation of cell proliferation and differentiation by PPARβ/δ.
PPAR Res. 2008:6148522008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yoo HY, Chang MS and Rho HM: Induction of
the rat Cu/Zn superoxide dismutase gene through the peroxisome
proliferator-responsive element by arachidonic acid. Gene.
234:87–91. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Girnun GD, Domann FE, Moore SA and Robbins
ME: Identification of a functional peroxisome
proliferator-activated receptor response element in the rat
catalase promoter. Mol Endocrinol. 16:2793–2801. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jung KH, Chu K, Lee ST, Kim SJ, Song EC,
Kim EH, Park DK, Sinn DI, Kim JM, Kim M and Roh JK: Blockade of AT1
receptor reduces apoptosis, inflammation, and oxidative stress in
normotensive rats with intracerebral hemorrhage. J Pharmacol Exp
Ther. 322:1051–1058. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
National Research Council (US) Institute
for Laboratory Animal Research: Guide for the Care and Use of
Laboratory Animals. Washington (DC): National Academies Press (US).
1996.
|
|
33
|
Lin KY, Chen YL, Shih CC, Pan JP, Chan WE
and Chiang AN: Contribution of HDL-apolipoproteins to the
inhibition of low density lipoprotein oxidation and lipid
accumulation in macrophages. J Cell Biochem. 86:258–267. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cui W, Chen SL and Hu KQ: Quantification
and mechanisms of oleic acid-induced steatosis in HepG2 cells. Am J
Transl Res. 2:95–104. 2010.PubMed/NCBI
|
|
35
|
Carmichael J, DeGraff WG, Gazdar AF, Minna
JD and Mitchell JB: Evaluation of a tetrazolium-based semiautomated
colorimetric assay: Assessment of chemosensitivity testing. Cancer
Res. 47:936–942. 1987.PubMed/NCBI
|
|
36
|
LeBel CP, Ischiropoulos H and Bondy SC:
Evaluation of the probe 2′,7′-dichlorofluorescin as an indicator of
reactive oxygen species formation and oxidative stress. Chem Res
Toxicol. 5:227–231. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Oyanagui Y: Reevaluation of assay methods
and establishment of kit for superoxide dismutase activity. Anal
Biochem. 142:290–296. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen J, Rogers SC and Kavdia M: Analysis
of kinetics of dihydroethidium fluorescence with superoxide using
xanthine oxidase and hypoxanthine assay. Ann Biomed Eng.
41:327–337. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Aebi H: Catalase in vitro. Methods
Enzymol. 105:121–126. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Flohé L and Günzler WA: Assays of
glutathione peroxidase. Methods Enzymol. 105:114–121. 1984.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bradford MM: A Rapid and sensitive method
for the quantitation of microgram quantities of protein utilizing
the principle of protein-dye binding. Anal Biochem. 72:248–254.
1976. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xu CL, Wang YZ, Guo J, Liu JX and Feng J:
Comparison of age-related differences in expression of antioxidant
enzyme mRNA and activity in various tissues of pigs. Comp Biochem
Physiol B Biochem Mol Biol. 147:445–451. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Patel DN, King CA, Bailey SR, Holt JW,
Venkatachalam K, Agrawal A, Valente AJ and Chandrasekar B:
Interleukin-17 stimulates C-reactive protein expression in
hepatocytes and smooth muscle cells via p38 MAPK and
ERK1/2-dependent NF-kappaB and C/EBPbeta activation. J Biol Chem.
282:27229–27238. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Young DC, Kingsley SD, Ryan KA and Dutko
FJ: Selective inactivation of eukaryotic β-galactosidase in assays
for inhibitors of HIV-1 TAT using bacterial β-galactosidase as a
reporter enzyme. Anal Biochem. 215:24–30. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pochareddy S and Edenberg HJ: Chronic
alcohol exposure alters gene expression in HepG2 cells. Alcohol
Clin Exp Res. 36:1021–1033. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tsukamoto H and Lu SC: Current concepts in
the pathogenesis of alcoholic liver injury. FASEB J. 15:1335–1349.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Comporti M, Signorini C, Leoncini S, Gardi
C, Ciccoli L, Giardini A, Vecchio D and Arezzini B: Ethanol-induced
oxidative stress: Basic knowledge. Genes Nutr. 2:101–109. 2010.
View Article : Google Scholar
|
|
48
|
Park DW, Baek K, Kim JR, Lee JJ, Ryu SH,
Chin BR and Baek SH: Resveratrol inhibits foam cell formation via
NADPH oxidase 1-mediated reactive oxygen species and monocyte
chemotactic protein-1. Exp Mol Med. 41:171–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Matés JM, Pérez-Gómez C and de Núñez
Castro I: Antioxidant enzymes and human diseases. Clin Biochem.
32:595–603. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kasdallah-Grissa A, Mornagui B, Aouani E,
Hammami M, El May M, Gharbi N, Kamoun A and El-Fazaâ S:
Resveratrol, a red wine polyphenol, attenuates ethanol-induced
oxidative stress in rat liver. Life Sci. 80:1033–1039. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cheng G, Zhang X, Gao D, Jiang X and Dong
W: Resveratrol inhibits MMP-9 expression by up-regulating PPAR
alpha expression in an oxygen glucose deprivation-exposed neuron
model. Neurosci Lett. 451:105–108. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kennedy A, Overman A, Lapoint K, Hopkins
R, West T, Chuang CC, Martinez K, Bell D and McIntosh M: Conjugated
linoleic acid-mediated inflammation and insulin resistance in human
adipocytes are attenuated by resveratrol. J Lipid Res. 50:225–232.
2009. View Article : Google Scholar : PubMed/NCBI
|