Introduction
Esophageal cancer (EC) is among the most common
types of cancer associated with the digestive system. Due to a lack
of efficient early diagnostic measurements, the majority of
patients with EC are in an advanced stage when treatment is
initiated, which is associated with a poor prognosis. Mortality
rates of patients with EC are high (>300,000 patients per year),
and the incidence of EC among Kazakh people in the Xinjiang Uyghur
Autonomous Region of northwest China is 155.9/100,000, which is
higher than any other ethnic group in China (1–3).
Conventional methods of pathological diagnosis
provide crucial information regarding tumor differentiation and the
morphological characteristics (4,5) in
patients with EC. However, clinical diagnosis and decisions on
treatment are difficult due to various limitations, including
tissue disfigurement following extrusion, inadequate biopsy depth
and discrepancies between pathological diagnosis and actual
diagnosis. Therefore, a more objective and quantitative method is
required to improve the accuracy of EC diagnosis (6).
Previous studies have suggested that the incidence
and evolution of EC is associated with various chromosomal
anomalies (7,8). During carcinogenesis, cells undergo
molecular cytogenetic changes prior to any alterations in
morphology. Nuclear chromosome abnormality, which is observed in
cancer cells, is an early event during the process of
tumorigenesis, and it has become the objective index for
determining cancerous cells (9,10).
Nuclear aneuploidy is a prevalent feature of various types of
cancer, including EC (9,10); therefore, the detection of
aneuploidy, which is usually found in aneusomic nuclei, may be used
to detect cancerous cells. Fluorescence in situ
hybridization (FISH) technology is a rapid and sensitive method
used for the detection of aneusomy on a specific chromosome
(11). FISH has been utilized for
the diagnosis of various types of cancer with high sensitivity and
specificity, including hematological malignancies and lung, breast
and kidney cancer (12,13). The advantage of FISH is that it is an
objective and quantitative method for identifying cancerous cells.
Previous studies have demonstrated that the priority of FISH in
cancer detection is higher than conventional cytology and,
therefore, may be advantageous to the early diagnosis of various
types of cancer (14,15).
In the present study, chromosomal loci centromere of
chromosome (CEP) 3 and 17 were selected to fabricate the DNA probe
as they have an increased aberration rate in esophageal and lung
cancer (16,17). In order to analyze the clinical
application of FISH and any correlations between the prognosis of
patients in the diagnosis of ESCC, 40 Kazakh patients with
esophageal squamous cell carcinoma (ESCC) underwent FISH
examination and conventional pathological diagnosis using surgical
samples.
Patients and methods
Patients
Between June 2011 and September 2012, 40 Kazakh
patients with ESCC were admitted to the Department of Thoracic
Surgery at The First Affiliated Hospital of Xinjiang Medical
University (Urumqi, China) and underwent surgical resection. Among
the 40 patients enrolled in the present study, 34 were male and 6
were female, with an average age of 57.4 years (Table I). None of the patients had
previously received any preoperative radiotherapy, chemotherapy or
other treatment. Pathology was graded according to the 7th American
Joint Committee on Cancer staging manual (18). Final post-operative pathological
diagnosis confirmed ESCC in all 40 cases, including well
differentiated (n=13), moderately differentiated (n=16) and poorly
differentiated tumors (n=11). Lymph node metastasis was detected in
55% (22/40) of cases; therefore, 45% (18/40) of the cases enrolled
in the present study were non-metastatic. A total of 10 esophageal
tissue samples (>5 cm distant from the tumor) were harvested as
normal controls from healthy individuals. Informed consent was
obtained from all patients.
 | Table I.Clinical characteristics of the
patients with esophageal squamous cell carcinoma. |
Table I.
Clinical characteristics of the
patients with esophageal squamous cell carcinoma.
| Case | Age (years) | Gender | Pathology |
|---|
| 1 | 71 | M | T1bN0M0, IA |
| 2 | 75 | M | T2N0M0, IB |
| 3 | 71 | M | T3N0M0, IIA |
| 4 | 55 | M | T2N0M0, IB |
| 5 | 58 | M | T3N1M0, IIIA |
| 6 | 54 | F | T3N0M0, IIB |
| 7 | 69 | M | T2N0M0, IB |
| 8 | 55 | M | T3N2M0, IIIB |
| 9 | 62 | M | T2N0M0, IB |
| 10 | 45 | F | T3N1M0, IIIA |
| 11 | 54 | M | T2N0M0, IB |
| 12 | 60 | M | T2N0M0, IIA |
| 13 | 57 | M | T1N0M0, IA |
| 14 | 70 | M | T3N2M0, IIIB |
| 15 | 58 | F | T2N0M0, IIA |
| 16 | 56 | M | T3N1M0, IIIA |
| 17 | 73 | M | T2N1M0, IIB |
| 18 | 68 | M | T3N1M0, IIIA |
| 19 | 40 | M | T1bN0M0, IA |
| 20 | 43 | F | T2N1M0, IIB |
| 21 | 54 | M | T3N2M0, IIIB |
| 22 | 47 | M | T3N1M0, IIIA |
| 23 | 44 | M | T3N1M0, IIIA |
| 24 | 56 | M | T2N1M0, IIB |
| 25 | 42 | M | T3N1M0, IIIA |
| 26 | 45 | M | T3N0M0, IIB |
| 27 | 50 | M | T3N1M0, IIIA |
| 28 | 56 | F | T2N0M0, IB |
| 29 | 72 | M | T3N1M0, IIIA |
| 30 | 65 | M | T2N1M0, IIB |
| 31 | 67 | F | T3N1M0, IIIA |
| 32 | 52 | M | T1aN0M0, IA |
| 33 | 50 | M | T3N1M0, IIIA |
| 34 | 48 | M | T3N1M0, IIIA |
| 35 | 62 | M | T2N1M0, IIB |
| 36 | 50 | M | T2N0M0, IB |
| 37 | 63 | M | T3N0M0, IIB |
| 38 | 55 | M | T1bN0M0, IA |
| 39 | 72 | M | T3N1M0, IIIA |
| 40 | 50 | M | T2N1M0, IIB |
FISH method
Touch preparations of cells were performed on glass
slides from fresh specimens and air-dried for 24 h at room
temperature and were subsequently stored at −80°C in preparation
for FISH. The same specimens were stained with hematoxylin and
eosin for pathological evaluation (Beijing Zhongshan Jinqiao
Biotechnology Co., Ltd., Beijing, China).
Cells were denatured with 70% formaldehyde at 74°C
in a water bath and subsequently washed twice with standard saline
citrate (SSC; Abbott Molecular, Inc., Des Plaines, IL, USA) at room
temperature. Slides were dehydrated through a graded ethanol series
(70, 85 and 100%) prior to the application of 10 µl hybridization
solution, containing 1 µl orange fluorescently-labeled CEP 3 probe,
1 µl green fluorescently-labeled CEP 17 probe, 7 µl hybridization
buffer and 1 µl double-distilled water (all Abbott Molecular,
Inc.). Slides were covered with a cover slip and sealed with rubber
cement (Fixogum; Marabu GmbH & Co. KG, Tamm, Germany).
Following incubation for 16 h at 42°C in a humidity-controlled
chamber, the slides were washed with SSC at 74°C and at room
temperature for 2 min. Subsequently, 5 µl diamidinophenylindole
(DAPI II; Abbott Molecular, Inc.) was applied to each spot and
covered with a cover slip.
Two fluorescently-labeled probes were used, Cy3 and
FITC (Thermo Fisher Scientific, Inc., Waltham, MA, USA), and each
of the protocols included at least one normal esophageal tissue as
a control. FISH signal analysis was performed according to the kit
instructions (Vysis CEP 3 (D3Z1) SpectrumOrange Probe Kit and CEP
17 Probe Kit; Abbott Molecular, Inc.) A fluorescence microscope and
an image acquisition and analysis system (DM6000; Leica
Microsystems GmbH, Wetzlar, Germany) was used to determine the
signal count. Results were analyzed by two individual
statisticians. All cells were evaluated, with the exception of
damaged cells or those with overlapping nuclei. A total of 100
nuclei were counted from each patient, and the total number of
centromeric signals was recorded. Cells were deemed positive for
aneuploidy when the percentage of hyperdisomic nuclei with >3
copies of at least one nucleus was >10%.
Statistical analysis
SPSS 17.0 statistical software (SPSS, Inc., Chicago,
IL, USA) was used to perform statistical analyses in the present
study. The Rank sum test and t-test were used to analyze the data
from each group. Throughout the study, P<0.05 was considered to
indicate a statistically significant difference.
Results
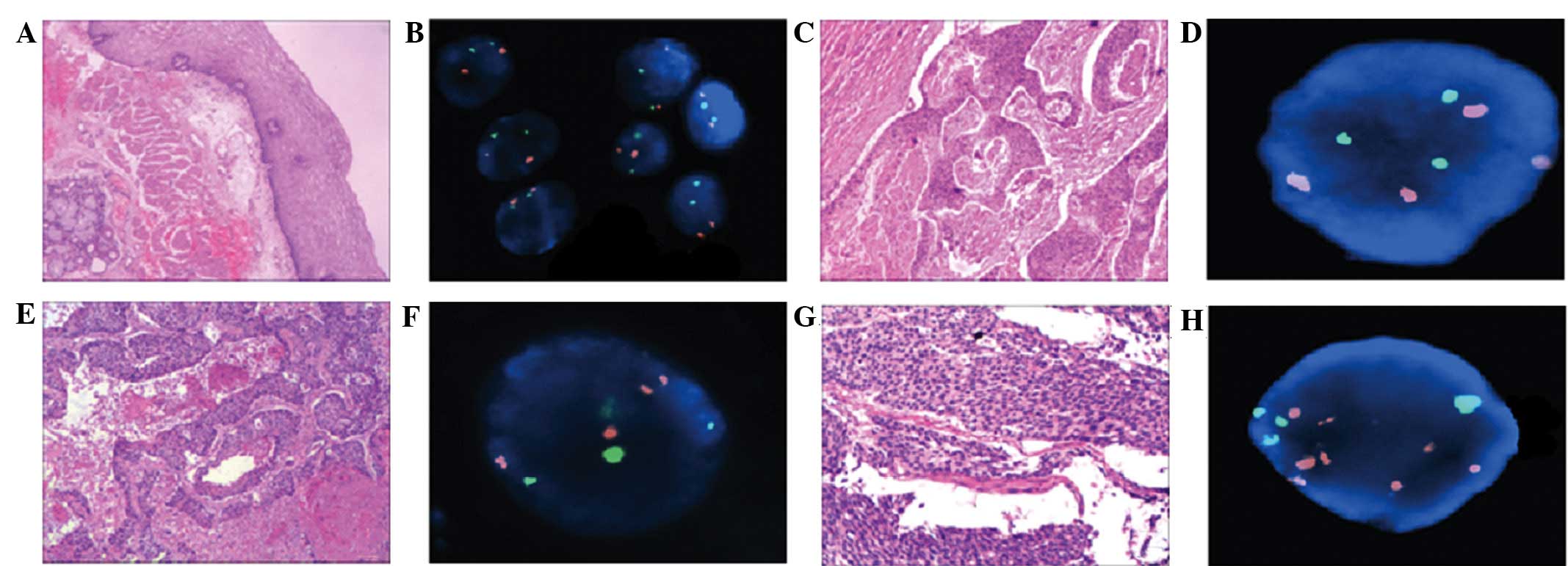
Polyploidy rates of the centromeres of
chromosomes 3 and 17 were detected using CEPs and FISH
analysis
The percentage of diploid CEP3 and 17 was >93% in
the control group (Table II).
Abnormal polyploidy copy numbers of CEP3 and 17 were detected in
all 40 ESCC specimens (Fig. 1).
 | Table II.Results of fluorescence in
situ hybridization in normal esophageal tissue. |
Table II.
Results of fluorescence in
situ hybridization in normal esophageal tissue.
|
| Aneuploidy (CEP
3/CEP 17) |
|
|---|
|
|
|
|
|---|
| Case | 2 copies | 3 copies | 4 copies | Multiple
copies |
|---|
| 1 | 96/96 | 1/2 | 3/2 | 4/4 |
| 2 | 97/95 | 2/3 | 1/2 | 3/5 |
| 3 | 97/97 | 2/2 | 1/1 | 3/3 |
| 4 | 98/98 | 2/2 | 0 | 2/2 |
| 5 | 96/97 | 3/2 | 1/1 | 4/3 |
| 6 | 96/95 | 4/2 | 0/3 | 4/5 |
| 7 | 95/95 | 2/2 | 3/3 | 5/5 |
| 8 | 95/96 | 3/2 | 2/2 | 5/4 |
| 9 | 93/94 | 3/3 | 4/3 | 7/6 |
| 10 | 97/95 | 2/3 | 1/2 | 3/5 |
CEP3 and CEP17 aberrations correlated
with lymph node metastasis
Average mutation rates of CEP 3 and 17 were 61.2%
and 50.95%, respectively (Table
III). The sensitivity and specificity of the FISH results were
100% with no false positives or negatives, calculated according to
the final pathological diagnosis. Lymph node metastasis was
detected in 55% of cases, therefore, 45% of the cases enrolled in
the present study were non-metastatic. Furthermore, polyploidy
rates were significantly increased in the metastatic lymph node
group, as compared with the non-metastatic group (CEP 3, P=0.0001;
CEP 17, P=0.012) (Table IV).
 | Table III.Results of fluorescence in
situ hybridization in esophageal cancer tissue. |
Table III.
Results of fluorescence in
situ hybridization in esophageal cancer tissue.
|
|
| Aneuploidy (CEP
3/CEP 17) |
|
|---|
|
|
|
|
|
|---|
| Case | Pathology | 2 copies | 3 copies | 4 copies | ≥5 copies | Multiple
copies |
|---|
| 1 | WDSCC | 45/39 | 25/31 | 20/22 | 10/8 | 55/61 |
| 2 | WDSCC | 71/73 | 19/15 | 10/12 | 0/0 | 29/27 |
| 3 | WDSCC | 73/77 | 17/12 | 9/11 | 1/0 | 27/23 |
| 4 | WDSCC | 65/63 | 21/16 | 11/12 | 3/9 | 35/37 |
| 5 | WDSCC | 60/72 | 28/12 | 5/8 | 7/8 | 40/28 |
| 6 | WDSCC | 82/80 | 17/15 | 0/5 | 1/0 | 18/20 |
| 7 | WDSCC | 76/74 | 13/11 | 6/7 | 5/8 | 24/26 |
| 8 | WDSCC | 72/68 | 13/15 | 7/9 | 8/8 | 28/32 |
| 9 | WDSCC | 66/72 | 20/17 | 10/9 | 4/2 | 34/28 |
| 10 | WDSCC | 59/68 | 26/21 | 6/7 | 9/4 | 41/32 |
| 11 | WDSCC | 60/65 | 21/19 | 11/8 | 8/8 | 40/35 |
| 12 | WDSCC | 50/57 | 20/17 | 25/20 | 5/6 | 50/43 |
| 13 | WDSCC | 71/74 | 11/11 | 10/10 | 8/5 | 29/26 |
| 14 | MDSCC | 41/59 | 11/12 | 36/20 | 12/9 | 59/41 |
| 15 | MDSCC | 38/56 | 25/17 | 28/20 | 9/7 | 62/44 |
| 16 | MDSCC | 43/59 | 28/18 | 13/12 | 16/11 | 57/41 |
| 17 | MDSCC | 64/65 | 18/13 | 12/15 | 6/7 | 36/35 |
| 18 | MDSCC | 38/57 | 26/29 | 28/11 | 8/3 | 62/43 |
| 19 | MDSCC | 31/56 | 36/20 | 19/12 | 16/12 | 69/44 |
| 20 | MDSCC | 52/78 | 32/17 | 13/4 | 3/1 | 48/22 |
| 21 | MDSCC | 60/70 | 9/11 | 12/12 | 9/7 | 40/30 |
| 22 | MDSCC | 45/55 | 26/21 | 12/9 | 17/15 | 55/45 |
| 23 | MDSCC | 39/46 | 45/39 | 9/13 | 7/2 | 61/54 |
| 24 | MDSCC | 48/57 | 25/20 | 19/16 | 8/7 | 52/43 |
| 25 | MDSCC | 44/54 | 31/26 | 23/15 | 3/5 | 56/46 |
| 26 | MDSCC | 45/52 | 25/25 | 12/8 | 18/15 | 55/48 |
| 27 | MDSCC | 24/24 | 55/36 | 9/30 | 12/10 | 76/76 |
| 28 | MDSCC | 15/26 | 35/44 | 20/20 | 30/10 | 85/74 |
| 29 | MDSCC | 80/75 | 19/15 | 1/10 | 0/0 | 20/25 |
| 30 | PDSCC | 30/38 | 27/21 | 24/26 | 19/15 | 70/62 |
| 31 | PDSCC | 25/28 | 40/40 | 25/23 | 10/9 | 75/72 |
| 32 | PDSCC | 21/27 | 30/23 | 39/40 | 10/10 | 79/73 |
| 33 | PDSCC | 13/9 | 55/46 | 17/30 | 15/15 | 87/91 |
| 34 | PDSCC | 14/30 | 41/29 | 23/30 | 22/11 | 86/70 |
| 35 | PDSCC | 14/6 | 40/41 | 20/24 | 26/31 | 86/94 |
| 36 | PDSCC | 28/40 | 28/25 | 27/23 | 17/12 | 72/60 |
| 37 | PDSCC | 32/40 | 39/26 | 25/24 | 4/10 | 68/60 |
| 38 | PDSCC | 12/25 | 58/52 | 12/12 | 18/11 | 88/75 |
| 39 | PDSCC | 32/24 | 47/55 | 10/12 | 11/9 | 68/76 |
| 40 | PDSCC | 40/42 | 20/17 | 27/26 | 13/15 | 60/58 |
 | Table IV.CEP3 and CEP17 aberrations correlated
with lymph node metastasis. |
Table IV.
CEP3 and CEP17 aberrations correlated
with lymph node metastasis.
| Lymph node
metastasis | CEP3 | CEP17 |
|---|
| Negative | 44.38±3.820 | 40.57±3.486 |
| Positive | 66.32±3.946 | 56.21±4.898 |
| T-value | −3.990 | −2.639 |
| P-value | 0.000 | 0.012 |
Results of differentiation degree of
CEP3 and CEP17 in three groups
The polyploidy rates for CEP3 and 17 were 35.38 and
30.92% in well-differentiated ESCC, 55.81 and 44.43% in moderately
differentiated ESCC, and 76.27 and 71.90% in poorly differentiated
ESCC, respectively. Significant differences were detected between
the poorly differentiated, moderately differentiated and
well-differentiated ESCC specimens (CEP 3, P<0.05; CEP 17,
P<0.05) (Table V).
 | Table V.Differentiation degree of CEP3 and
CEP17 in the three groups. |
Table V.
Differentiation degree of CEP3 and
CEP17 in the three groups.
|
Differentiation | CEP3 | CEP17 |
|---|
| WD |
35.3846±12.2647 |
30.9231±10.9199 |
| MD |
55.8125±15.3849 |
44.4375±14.5692 |
| PD | 76.2727±9.5403 |
71.9091±12.0785 |
| F-value | 8.616 | 7.975 |
| P-value | <0.05 | <0.05 |
Discussion
The incidence of EC is high in the Xinjiang Uyghur
Autonomous Region of northwestern China, and the Kazakh ethnic
group in particular suffers from an increased incidence of EC, as
compared with other ethnic groups (2). Therefore, improved diagnosis and
treatment of EC is required to enhance the survival rate and
quality of life of patients with EC.
Previous studies have demonstrated that various
genetic mutation occur during carcinogenesis (10,15).
FISH is a sensitive and specific diagnostic technique that is
capable of detecting various types of cytogenetic alterations,
including aneusomy, amplification and deletion. This technique has
become a widely used diagnostic method in cytogenetic studies
(11,19); however, to the best of our knowledge
there are no previous studies investigating the application of FISH
in the diagnosis of Kazakh esophageal cancer.
Human chromosome 3 carries hyperdense genes, which
are associated with numerous types of cancer, including raf-1 and
erb-A; whereas chromosome 17 carries certain oncogenes and tumor
suppressor genes, including p53, c-erbB2, BRCA1 and nm23 (9,14,20).
Previous studies have demonstrated elevated aberration rates in CEP
3 and 17 in patients with EC, which indicates that EC may be
correlated with constitutional cytogenetic alterations (12,20).
In the present study, CEP 3 and 17 DNA probes were
selected, and the cut-off value for the percentage of hyperdisomic
cells was set at 10%. Normal cells often have <6% hyperdisomic
cells and this discrepancy is likely to be due to sister chromatids
being counted as copies (6,8).
In a previous study conducted by Fiegl et al
(21) FISH was applied in the
diagnosis of lung, breast, liver and stomach cancer, and the
confirmed diagnostic rate was increased. Furthermore, Fritcher
et al (22) analyzed
esophageal adenocarcinoma using the FISH method with c-Myc, P16,
HER2 and 20q13 centromeric region probes, and demonstrated that the
sensitivity of cytology was only 45% for the detection of
esophageal adenocarcinoma; however, a detection rate of 100% was
achieved using FISH.
In the present study, FISH was applied to Kazakh
patients with ESCC to elucidate the diagnostic value of FISH in the
detection of cancer cells and as a prognostic indicator. Polyploidy
of CEP 3 and 17 was detected in all 40 ESCC specimens and
significant differences in the rates of polyploidy were detected
between the poorly differentiated, moderately differentiated and
well-differentiated ESCC specimens for both CEPs (CEP 3, P<0.05;
CEP 17, P<0.05). Furthermore, polyploidy was significantly
increased in the metastatic lymph node group, as compared with the
non-metastatic group (CEP 3, P=0.0001; CEP 17, P=0.012). The
average mutation rates of CEP 3 and 17 were 61.2 and 50.95%,
respectively.
The results of the present study demonstrated that
the aberration rates of CEP 3 and 17 were correlated with the level
of ESCC differentiation. This may due to the eating habits of the
Kazakh population, in particular the over consumption of smoked
meat, fermented food, heavy smoking or drinking, and the reduced
consumption of fresh fruits and vegetables (2,23,24).
In conclusion, the present study successfully used
CEP 3 and 17 probes to detect cancerous cells in Kazakh patients
with ESCC. In particular, aneuploidy was significantly higher in
poorly differentiated squamous cells and the metastatic lymph node
group. Therefore, DNA probes may be used as predictive biological
markers for the prognosis of patients with ESCC. Furthermore, as an
objective and qualitative method, FISH technology is capable of
detecting CEP 3 and 17 variations in the diagnosis of Kazakh
patients with ESCC, which may be used to genetically diagnose EC in
the future. Further studies are required.
Acknowledgements
This study was supported by the Returned Overseas
Students to Science and Technology Activities Fund (no. 2012-111)
and National Natural Science Foundation of China (no. 81160279).
The authors thank the Department of Hematology at the First
Affiliated Hospital of Xinjiang Medical University for technical
support.
Glossary
Abbreviations
Abbreviations:
|
FISH
|
fluorescence in situ
hybridization
|
|
EC
|
esophageal cancer
|
|
ESCC
|
esophageal squamous cell carcinoma
|
|
CEP
|
centromere of chromosome
|
|
SSC
|
standard saline citrate
|
References
|
1
|
Parkin DM: Global cancer statistics in the
year 2000. Lancet Oncol. 2:533–543. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lu XM, Zhang YM, Lin RY, Arzi G, Wang X,
Zhang YL, Zhang Y, Wang Y and Wen H: Relationship between genetic
polymorphisms of metabolizing enzymes CYP2E1, GSTM1 and Kazakh's
Esophageal squamous cell cancer (ESCC) in Xinjiang, China. World J
Gastroenterol. 11:3651–3654. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Krasna MJ: Multimodality therapy for
esophageal cancer. Oncology (Williston Park). 24:1134–1138.
2010.PubMed/NCBI
|
|
4
|
Odze RD: Barrett esophagus: Histology and
pathology for the clinician. Nat Rev Gastroenterol Hepatol.
6:478–490. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yerian L: Histology of metaplasia and
dysplasia in Barrett's esophagus. Surg Oncol Clin N Am. 18:411–422.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Papanicolaou GN: Criteria of malignancy.
Atlas of Exfoliative Cytology. Harvard University Press.
(Cambridge, MA). 13–21. 1954.
|
|
7
|
Awut I, Niyaz M, Huizhong X, Biekemitoufu
H, Yan ZH, Zhu Z, Sheyhedin I, Changmin Z, Wei Z and Hao W: Genetic
diagnosis of patients with esophageal cancer using FISH. Oncol
Lett. 1:809–814. 2010.PubMed/NCBI
|
|
8
|
Niyaz M, Turghun A, Ping ZH, Zhu Z,
Sheyhedin I, Ren C and Awut I: TP53 gene deletion in esophageal
cancer tissues of patients and its clinical significance. Mol Med
Rep. 7:122–126. 2013.PubMed/NCBI
|
|
9
|
Rajagopalan H and Lengauer C: Aneuploidy
and cancer. Nature. 432:338–341. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chao WR, Lee MY, Lin WL, Koo CL, Sheu GT
and Han CP: Assessing the HER2 status in mucinous epithelial
ovarian cancer on the basis of the 2013 ASCO/CAP guideline update.
Am J Surg Pathol. 38:1227–1234. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Halling KC and Kipp BR: Fluorescence in
situ hybridization in diagnostic cytology. Hum Pathol.
38:1137–1144. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tafe LJ, Allen SF, Steinmetz HB, Dokus BA,
Cook LJ, Marotti JD and Tsongalis GJ: Automated processing of
fluorescence in-situ hybridization slides for HER2 testing in
breast and gastro-esophageal carcinomas. Exp Mol Pathol.
97:116–119. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Prins MJ, Ruurda JP, van Diest PJ, van
Hillegersberg R and ten Kate FJ: Evaluation of the HER2
amplification status in oesophageal adenocarcinoma by conventional
and automated FISH: A tissue microarray study. J Clin Pathol.
67:26–32. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gordon MA, Gundacker HM, Benedetti J,
Macdonald JS, Baranda JC, Levin WJ, Blanke CD, Elatre W, Weng P,
Zhou JY, et al: Assessment of HER2 gene amplification in
adenocarcinomas of the stomach or gastroesophageal junction in the
INT-0116/SWOG9008 clinical trial. Ann Oncol. 24:1754–1761. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yoshizawa A, Sumiyoshi S, Sonobe M,
Kobayashi M, Uehara T, Fujimoto M, Tsuruyama T, Date H and Haga H:
HER2 status in lung adenocarcinoma: A comparison of
immunohistochemistry, fluorescence in situ hybridization (FISH),
dual-ISH, and gene mutations. Lung Cancer. 85:373–378. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Awut I, Niyaz M, Biekemitoufu H, Zhang Z,
Sheyhedin I and Hao W: Molecular pathological diagnosis for early
esophageal cancer in Kazakh patients. Oncol Lett. 3:549–553.
2012.PubMed/NCBI
|
|
17
|
Nakamura H, Saji H, Idiris A, Kawasaki N,
Hosaka M, Ogata A, Saijo T and Kato H: Chromosomal instability
detected by fluorescence in situ hybridization in surgical
specimens of non-small cell lung cancer is associated with poor
survival. Clin Cancer Res. 9:2294–2299. 2003.PubMed/NCBI
|
|
18
|
Rice TW, Blackstone EH and Rusch VW: 7th
edition of the AJCC Cancer Staging Manual: esophagus and
esophagogastric junction. Ann Surg Oncol. 17:1721–1724. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li C, Ba J, Hao X, Zhang S, Hu Y, Zhang X,
Yuan W, Hu L, Cheng T, Zetterberg A, et al: Multi-gene fluorescence
in situ hybridization to detect cell cycle gene copy number
aberrations in young breast cancer patients. Cell Cycle.
13:1299–1305. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nakamura H, Idiris A, Kawasaki N, Taguchi
M, Ohira T and Kato H: Quantitative detection of lung cancer cells
by fluorescence in situ hybridization: Comparison with conventional
cytology. Chest. 128:906–911. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fiegl M, Massoner A, Haun M, Sturm W,
Kaufmann H, Hack R, Krugmann J, Fritzer-Szekeres M, Grünewald K and
Gastl G: Sensitive detection of tumour cells in effusions by
combining cytology and fluorescence in situ hybridisation (FISH).
Br J Cancer. 91:558–563. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fritcher EG, Brankley SM, Kipp BR, Voss
JS, Campion MB, Morrison LE, Legator MS, Lutzke LS, Wang KK, Sebo
TJ and Halling KC: A comparison of conventional cytology, DNA
ploidy analysis, and fluorescence in situ hybridization for the
detection of dysplasia and adenocarcinoma in patients with
Barrett's esophagus. Hum Pathol. 39:1128–1135. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bahmanyar S and Ye W: Dietary patterns and
risk of squamous-cell carcinoma and adenocarcinoma of the esophagus
and adenocarcinoma of the gastric cardia: A population-based
case-control study in Sweden. Nutr Cancer. 54:171–178. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Morita M, Kumashiro R, Kubo N, Nakashima
Y, Yoshida R, Yoshinaga K, Saeki H, Emi Y, Kakeji Y, Sakaguchi Y,
et al: Alcohol drinking, cigarette smoking, and the development of
squamous cell carcinoma of the esophegus: Epidemiology, clinical
findings, and prevention. Int J Clin Oncol. 15:126–134. 2010.
View Article : Google Scholar : PubMed/NCBI
|