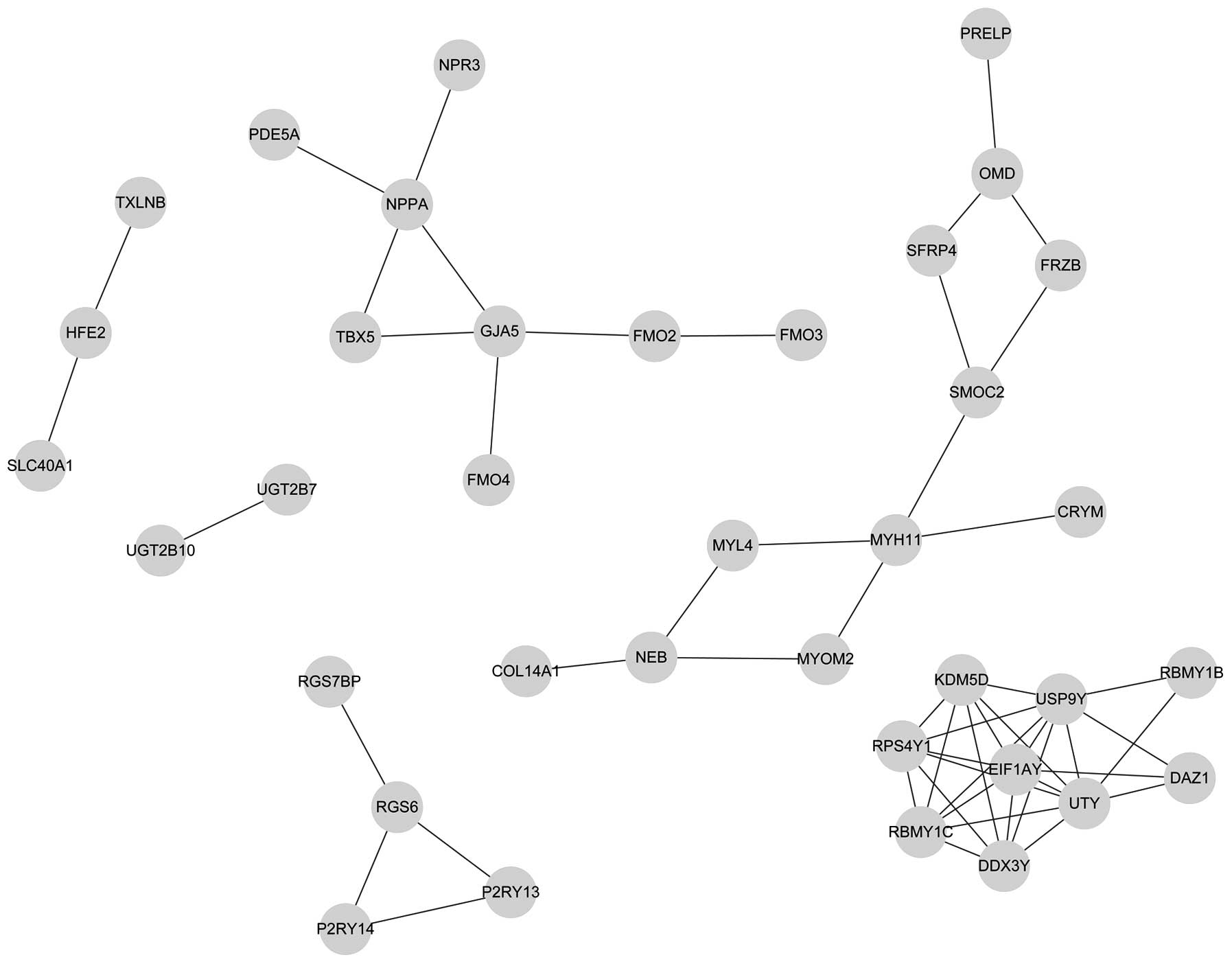
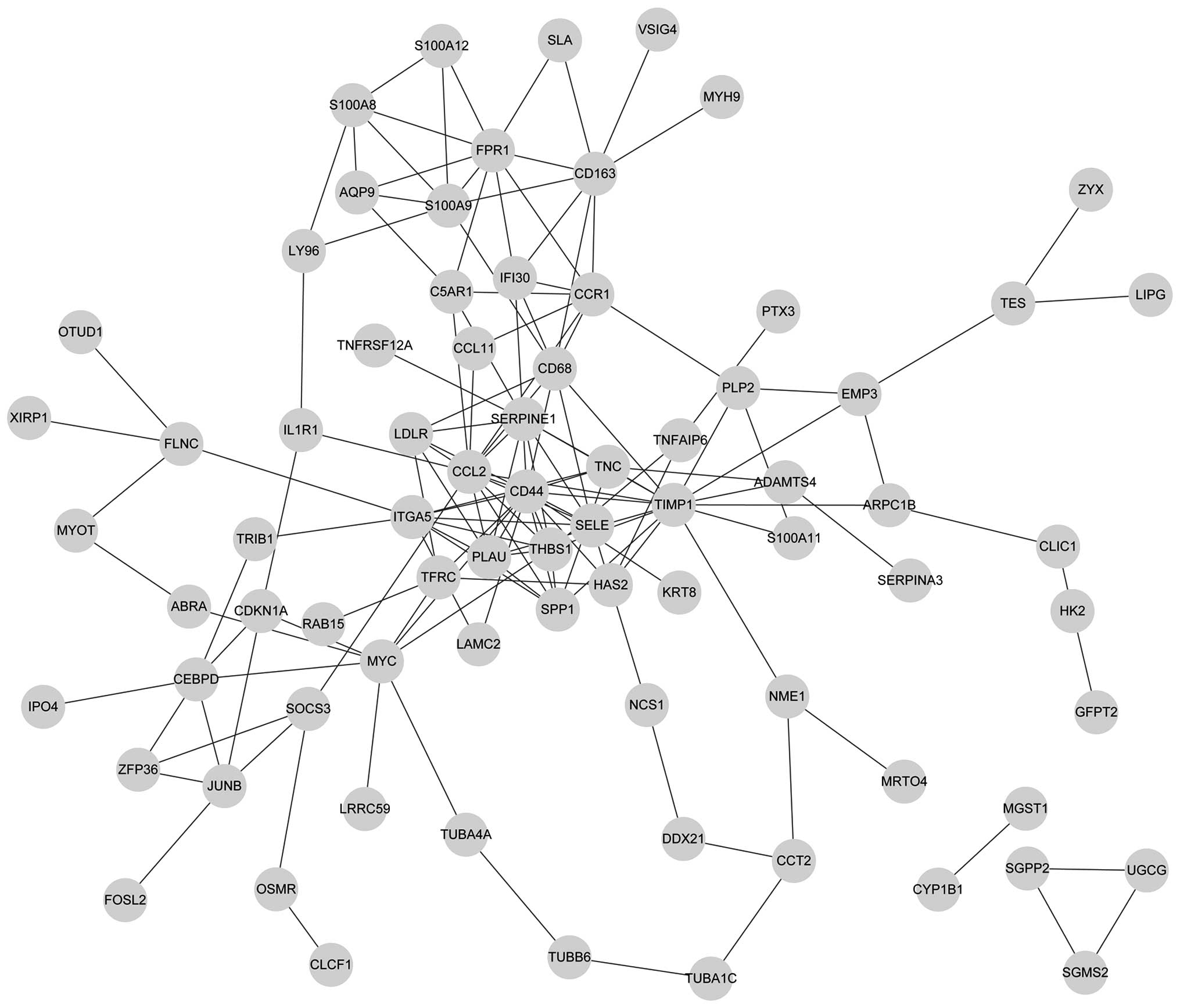
|
1
|
Van Diepen S, Hellkamp AS, Patel MR,
Becker RC, Breithardt G, Hacke W, Halperin JL, Hankey GJ, Nessel
CC, Singer DE, et al: Efficacy and safety of rivaroxaban in
patients with heart failure and nonvalvular atrial fibrillation:
Insights from ROCKET AF. Circ Heart Fail. 6:740–747. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Carr HJ, McDermott A, Tadbiri H, Uebbing
A-M and Londrigan M: The effectiveness of computer-based learning
in hospitalized adults with heart failure on knowledge,
re-admission, self-care, quality of life and patient satisfaction:
A systematic review protocol. The JBI Database of Systematic
Reviews and Implementation Reports. 11:129–145. 2013. View Article : Google Scholar
|
|
3
|
Suskin N, McKelvie RS, Burns RJ, Latini R,
Pericak D, Probstfield J, Rouleau JL, Sigouin C, Solymoss CB,
Tsuyuki R, et al: Glucose and insulin abnormalities relate to
functional capacity in patients with congestive heart failure. Eur
Heart J. 21:1368–1375. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Diercks G, Van Boven A, Hillege H, Janssen
W, Kors J, De Jong P, Grobbee D, Crijns H and Van Gilst W:
Microalbuminuria is independently associated with ischaemic
electrocardiographic abnormalities in a large non-diabetic
population. The PREVEND (Prevention of REnal and Vascular ENdstage
Disease) study. Eur Heart J. 21:1922–1927. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Klamann A, Sarfert P, Launhardt V, Schulte
G, Schmiegel WH and Nauck MA: Myocardial infarction in diabetic vs
non-diabetic subjects. Survival and infarct size following therapy
with sulfonylureas (glibenclamide). Eur Heart J. 21:220–229. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Anker SD, Egerer KR, Volk H-D, Kox WJ,
Poole-Wilson PA and Coats AJ: Elevated soluble CD14 receptors and
altered cytokines in chronic heart failure. Am J Cardiol.
79:1426–1430. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Krishnan E: Hyperuricemia and incident
heart failure. Circ Heart Fail. 2:556–562. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Harutyunyan M, Christiansen M, Johansen
JS, Køber L, Torp-Petersen C and Kastrup J: The inflammatory
biomarker YKL-40 as a new prognostic marker for all-cause mortality
in patients with heart failure. Immunobiology. 217:652–656. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kusumoto A, Miyata M, Kubozono T, Ikeda Y,
Shinsato T, Kuwahata S, Fujita S, Takasaki K, Yuasa T and Hamasaki
S: Highly sensitive cardiac troponin T in heart failure: Comparison
with echocardiographic parameters and natriuretic peptides. J
Cardiol. 59:202–208. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Troughton RW, Frampton CM, Yandle TG,
Espiner EA, Nicholls MG and Richards AM: Treatment of heart failure
guided by plasma aminoterminal brain natriuretic peptide (N-BNP)
concentrations. The Lancet. 355:1126–1130. 2000. View Article : Google Scholar
|
|
11
|
Greco S, Fasanaro P, Castelvecchio S,
D'Alessandra Y, Arcelli D, Di Donato M, Malavazos A, Capogrossi MC,
Menicanti L and Martelli F: MicroRNA dysregulation in diabetic
ischemic heart failure patients. Diabetes. 61:1633–1641. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kapoun AM, Liang F, O'Young G, Damm DL,
Quon D, White RT, Munson K, Lam A, Schreiner GF and Protter AA:
B-type natriuretic peptide exerts broad functional opposition to
transforming growth factor-beta in primary human cardiac
fibroblasts: Fibrosis, myofibroblast conversion, proliferation and
inflammation. Circ Res. 94:453–461. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. Journal of the Royal Statistical Society. Series
B (Methodological). 289–300. 1995.
|
|
15
|
Von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
He X and Zhang J: Why do hubs tend to be
essential in protein networks? PLoS Genet. 2:e882006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Alvord WG, Roayaei J, Stephens R, Baseler
MW, Lane HC, Lempicki RA, Huang DW, Sherman BT, Tan Q and Collins
JR: The DAVID Gene Functional Classification Tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bottomley PA, Panjrath GS, Lai S, Hirsch
GA, Wu K, Najjar SS, Steinberg A, Gerstenblith G and Weiss RG:
Metabolic rates of ATP transfer through creatine kinase (CK Flux)
predict clinical heart failure events and death. Sci Transl Med.
5:215re32013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Burchell AE, Sobotka PA, Hart EC,
Nightingale AK and Dunlap ME: Chemohypersensitivity and autonomic
modulation of venous capacitance in the pathophysiology of acute
decompensated heart failure. Curr Heart Fail Rep. 10:139–146. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tulassay T, Seri I and Rascher W: Atrial
natriuretic peptide and extracellular volume contraction after
birth. Acta Paediatr Scand. 76:444–446. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Samson WK: Atrial natriuretic factor
inhibits dehydration and hemorrhage-induced vasopressin release.
Neuroendocrinology. 40:277–279. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ren X, Xu C, Zhan C, Yang Y, Shi L, Wang
F, Wang C, Xia Y, Yang B, Wu G, et al: Identification of NPPA
variants associated with atrial fibrillation in a Chinese GeneID
population. Clin Chim Acta. 411:481–485. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Maeda K, Tsutamoto T, Wada A, Hisanaga T
and Kinoshita M: Plasma brain natriuretic peptide as a biochemical
marker of high left ventricular end-diastolic pressure in patients
with symptomatic left ventricular dysfunction. Am Heart J.
135:825–832. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Seronde MF, Gayat E, Logeart D, Lassus J,
Laribi S, Boukef R, Sibellas F, Launay JM, Manivet P, Sadoune M, et
al: Comparison of the diagnostic and prognostic values of B-type
and atrial-type natriuretic peptides in acute heart failure. Int J
cardiol. 168:3404–3411. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Potocki M, Breidthardt T, Reichlin T,
Hartwiger S, Morgenthaler NG, Bergmann A, Noveanu M, Freidank H,
Taegtmeyer AB, Wetzel K, et al: Comparison of midregional
pro-atrial natriuretic peptide with N-terminal pro-B-type
natriuretic peptide in the diagnosis of heart failure. J Intern
Med. 267:119–129. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lip GY and Gibbs CR: Does heart failure
confer a hypercoagulable state? Virchow's triad revisited. J Am
Coll Cardiol. 33:1424–1426. 1999.PubMed/NCBI
|
|
30
|
Rengo G, Pagano G, Squizzato A, Moja L,
Femminella GD, de Lucia C, Komici K, Parisi V, Savarese G, Ferrara
N, et al: Oral anticoagulation therapy in heart failure patients in
sinus rhythm: A systematic review and meta-analysis. PLoS One.
8:e529522013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Camp NP, Jones SD, Liebeschuetz JW, et al:
Serine protease inhibitors. Journal. 2005.
|
|
32
|
Askari AT, Brennan ML, Zhou X, Drinko J,
Morehead A, Thomas JD, Topol EJ, Hazen SL and Penn MS:
Myeloperoxidase and plasminogen activator inhibitor 1 play a
central role in ventricular remodeling after myocardial infarction.
J Exp Med. 197:615–624. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xu Z, Castellino FJ and Ploplis VA:
Plasminogen activator inhibitor-1 (PAI-1) is cardioprotective in
mice by maintaining microvascular integrity and cardiac
architecture. Blood. 115:2038–2047. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Spiroski I, Kedev S, Antov S, Trajkov D,
Petlichkovski A, Dzhekova-Stojkova S, Kostovska S and Spiroski M:
Investigation of SERPINE1 genetic polymorphism in Macedonian
patients with occlusive artery disease and deep vein thrombosis.
Kardiol Pol. 67:1088–1094. 2009.PubMed/NCBI
|
|
35
|
Iwanaga Y, Aoyama T, Kihara Y, Onozawa Y,
Yoneda T and Sasayama S: Excessive activation of matrix
metalloproteinases coincides with left ventricular remodeling
during transition from hypertrophy to heart failure in hypertensive
rats. J Am Coll Cardiol. 39:1384–1391. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Janicki JS and Brower GL: The role of
myocardial fibrillar collagen in ventricular remodeling and
function. J Card Fail. 8(Suppl 6): S319–S325. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zheng J, Chen Y, Pat B, Dell'italia LA,
Tillson M, Dillon AR, Powell PC, Shi K, Shah N, Denney T, et al:
Microarray identifies extensive downregulation of noncollagen
extracellular matrix and profibrotic growth factor genes in chronic
isolated mitral regurgitation in the dog. Circulation.
119:2086–2095. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chatila K, Ren G, Xia Y, Huebener P, Bujak
M and Frangogiannis NG: The role of the thrombospondins in healing
myocardial infarcts. Cardiovasc Hematol Agents Med Chem. 5:21–27.
2007. View Article : Google Scholar : PubMed/NCBI
|