Introduction
Liver cancer is the fifth most common type of cancer
and the third leading cause of cancer-associated mortality in the
worldwide population (1). The
incidence and mortality rates of liver cancer are continuously
increasing, with ~700,000 people diagnosed with liver cancer
annually, among which hepatocellular carcinoma (HCC) constitutes
>80% of cases (2,3). The hepatitis B virus (HBV) is a major
risk factor for liver cancer (4).
HBV is an oncogenic virus that can cause HCC through direct and
indirect signaling pathways (5).
Continuous hepatocyte injury and regeneration in liver disease
result in increased liver cell turnover and accumulation of
critical mutations in the host genome, which can in turn cause gene
alterations, including the activation of oncogenes or inactivation
of tumour suppressor genes (5).
Since early diagnosis of liver cancer is challenging, the majority
of HCC patients suffer from poor prognosis. Although great progress
has been achieved in cancer treatment using methods such as
surgery, liver transplantation, radiofrequency ablation and
transcatheter arterial chemoembolization, the majority of HCC
patients succumb due to invasion or distant metastasis to other
organs (6,7). Therefore, research has focused on
examining the molecular mechanisms underlying invasion and
metastasis, as well as investigating the significant molecular
markers of HCC metastasis and identifying novel targets. One of the
most effective strategies for the successful management of HCC is
the prevention of metastasis of cancer cells in order to reduce the
mortality rate and improve the prognosis of liver cancer (8).
The underlying mechanism of cancer invasion and
metastasis is a complicated multistep process involving multiple
genetic and molecular alterations (9). Chromosome region maintenance 1 (CRM1),
also known as exportin 1, was initially identified in eukaryotic
cells, while human CRM1 was first attained by cloning in 1997 by
Kudo et al (10). It consists
of 1,071 amino acids, which can maintain the structure and function
of chromosomes in the process of mitosis (10). CRM1 is a major nuclear export protein
in mammals, which facilitates the transport of RNAs, proteins or
other macromolecules across the nuclear membrane to the cytoplasm
(11–14). In addition, CRM1 recognizes the
leucine-rich nuclear export signal (NES) sequence. Numerous cargo
proteins that are rich in NES sequences, including tumor suppressor
protein p53, p27, p21 and Forkhead box O, depend on CRM1 for
nuclear export function (15–19).
The transportation of macromolecules across the
nuclear membrane is critical for the proper functioning of living
cells. Accumulating evidence has suggested that cancer cells are
able to escape antineoplastic mechanisms and benefit from
prosurvival signals through the dysregulation of this system
(20). The protein CRM1 is the only
member of the karyopherin-β protein family that contributes to the
nucleocytoplasmic trafficking, and is considered to be an
anti-apoptotic oncogenic protein in transformed cells (21). In cancer cells, overexpression of
CRM1 results in alterations in nucleocytoplasmic trafficking and
deregulation of ribosomal biogenesis, as well as in aberrant
cytoplasmic localization of tumor suppressor proteins, cell cycle
regulators and pro-apoptotic proteins (20). A large number of studies (22–24) have
shown that imbalance of cell proliferation and cell death due to
disorder of the cell cycle is the major cause of malignancy.
Therefore, abnormally high expression of CRM1 is correlated with
poor patient prognosis in various malignancies. For instance,
Giovanni et al (25) have
reported that overexpression of CRM1 is associated with survival
difference of various tumors, including pancreatic, lung, ovarian
and cervical cancer, as well as in osteosarcoma and leukemia.
Anguinomycin and goniothalamin were found to be inhibitors of CRM1
(26,27), thus the development of CRM1-specific
small molecules as novel anti-cancer agents is considered.
Currently, therapeutic targeting of CRM1 has emerged as the
original specific inhibitor of CRM1, followed by the development of
several next-generation small molecules, such as KPT-330 which is a
selective inhibitor of nuclear export (20,28–29).
Since the role of CRM1 in oncogenesis has been
revealed by various studies and the anti-tumor mechanism of CRM1
inhibition is gradually elucidated (30–33), it
is crucial to focus on the association of CRM1 expression with
malignancy and clinical features. Therefore, the present study
aimed to detect the CRM1 protein expression in primary liver
carcinoma and adjacent cancer tissues in order to investigate its
association with clinical and pathological features using an
immunohistochemical assay. Furthermore, the study aimed to provide
new experimental evidence for the molecular mechanism of tumor
growth, invasion, metastasis and molecular therapy of primary liver
carcinoma.
Materials and methods
Human tissue samples
A total of 152 tumor tissues and adjacent normal
tissues (which were located <2 cm from the cancer tissue and
were used as the controls) were obtained between January 2009 and
June 2014. The liver samples were provided by the First Affiliated
Hospital of Dalian Medical University (Dalian, China). All liver
samples were collected during surgery and preserved in 10%
formaldehyde solution at room temperature. Approximately 1×1×0.3
cm3 liver samples were obtained and fixed with 10%
formalin for 24 h. The tissue samples were subsequently dehydrated
using a fully-automated tissue processor (Tissue-Tek®
VIP® 6) and embedded in paraffin using a
paraffin-embedding device (Leica EG1160). All enrolled patients
were diagnosed with primary cancer of the liver by pathological
examination, which was based on the World Health Organization
classification of tumors of the digestive system (34). Communication was conducted with
patients prior to surgery and all patients voluntarily participate
in the present study, providing written informed consents.
Information collected from the patient records included the gender,
age, cirrhosis status, hepatitis B surface antigen (HBsAg),
hepatitis B envelope antigen (HBeAg), α-fetoprotein (AFP),
carcinoembryonic antigen (CEA), differentiation degree (including
high, middle or low differentiation) (35) and American Joint Committee on Cancer
(AJCC) stage (36). HBsAg, HBeAg,
CEA and AFP levels were measured using an ELISA kits (Roche Cobas
e601 Analyzer; Roche Diagnostics GmbH, Mannheim, Germany). Detailed
clinical and pathological features of these patients are shown in
Table I.
 | Table I.Clinical and pathological features of
patients with primary cancer of the liver. |
Table I.
Clinical and pathological features of
patients with primary cancer of the liver.
| Variable | Percentage (%) |
|---|
| No. of
patients | 152 (100.0) |
| Gender |
|
|
Male | 122 (80.3) |
|
Female | 30 (19.7) |
| Age, years |
|
|
≤55 | 77 (50.7) |
|
>55 | 75 (49.3) |
| Diameter, cm |
|
| ≤5 | 86 (56.6) |
|
>5 | 66 (43.4) |
| No. of tumors |
|
| 1 | 136 (89.5) |
| ≥2 | 16 (10.5) |
| Cirrhosis |
|
| + | 102 (67.1) |
| − | 50 (32.9) |
| HBsAg |
|
| + | 139 (91.4) |
| − | 13 (8.6) |
| HBeAg |
|
| + | 31 (20.4) |
| − | 121 (79.6) |
| AFP, µg/l |
|
|
≤400 | 120 (79.0) |
|
>400 | 32 (21) |
| CEA, µg/l |
|
| ≤5 | 116 (76.3) |
|
>5 | 36 (23.7) |
| Differentiation
degree |
|
|
High | 32 (21.0) |
|
Middle | 53 (34.9) |
|
Low | 67 (44.1) |
| AJCC stage |
|
| I | 131 (86.2) |
| II | 9 (5.9) |
|
III | 10 (6.6) |
| IV | 2 (1.3) |
The expression levels of CRM1 in the tumor and
adjacent normal control tissues were then examined. Furthermore,
the clinical and pathological features of 152 liver cancer patients
and their association with CRM1 were also investigated. The present
study was approved by the Ethics Committee of the The First
Affiliated Hospital of Dalian Medical University.
Instruments and reagents
A Leica EG1160 paraffin-embedding device, a Leica
RM2245 microtome, a drying device and a Leica DM2500 microscope
were purchased from Leica Biosystems (Wetzlar, Germany). In
addition, a Tissue-Tek VIP6 fully-automated tissue processor was
purchased from Sakura Finetek (Tokyo, Japan). The primary antibody
used in the present study was a rabbit anti-human CRM1 antibody
(dilution, 1:100; ab191081; Abcam, Cambridge UK), and the secondary
antibody was a horseradish peroxidase-conjugated goat
anti-mouse/rabbit immunoglobulin G polymer (dilution, 1:1,000;
Fuzhou Maixin Biological Technology Development Company, Fujian,
China). Furthermore, phosphate-buffered saline (PBS),
3,3′-diaminobenzidine (DAB) chromogenic reagent and Tris-EDTA
buffer (pH 9.0) were purchased from Zhongshan Jinqiao Biotechnology
Co., Ltd. (Beijing, China). Other reagents, including xylene and
ethanol (70, 90 and 100%), were purchased from Shunda Chemicals Co.
Ltd. (Xi'an, China).
Immunohistochemical analysis
The expression and intracellular localization of
CRM1 in tumor and adjacent normal tissues were determined
immunohistochemically. The tissue samples were dehydrated using the
fully automated tissue processor (Tissue-Tek VIP6) and embedded in
paraffin using a paraffin-embedding device (Leica EG1160). The
tissue samples were then sectioned in 2-µm specimens with a
microtome (Leica RM2245) and dried at 60°C for 2 h using a drying
device. Subsequent to deparaffinization in xylene for 10 min and
rehydration in ascending grades of ethanol (70% for 2 min, 95% for
2 min and 100% for 2 min), high temperature antigen retrieval was
conducted for 3 min using 0.01 M Tris-EDTA buffer (pH 9.0) and a
standard pressure-cooker. All sections were subjected to endogenous
biotin blocking (3% H2O2)and incubated for 10
min at room temperature. Tissues were washed with PBS and then
incubated with the CRM1 primary antibody (ab191081) at 4°C
overnight. Subsequently, the tissues were washed with PBS and
incubated with secondary antibody for 1 h at room temperature.
After tissues were washed with PBS, a DAB chromagen substrate was
used, and the tissues were washed in PBS and stained with
hematoxylin (Shunda Chemical Co., Ltd., Xi'an, China). Following
dehydration in ascending grades of alcohol, clearing in xylene,
drying and sealing with resinene tablets (Ziyi Co., Shanghai,
China), the tissue sections were examined under a Leica DM2500
microscope.
Two experienced pathologist independently observed
the distribution characteristics of CRM1 expression using an
optical microscope. Brown staining indicated positive staining for
CRM1. Five high-power fields (magnification, ×200) of each section
were randomly selected and images were captured with the Leica
DM2500 microscope. Next, the area of interest was selected and the
value of optical density of each photo was detected by an image
analysis software (Image-Pro Plus version 6.0; Media Cybernetics,
Inc., Rockville, MD, USA). The mean value of the
immunohistochemical optical density was determined as the final
expression value of each tissue sample.
Statistical analysis
All statistical analyses were performed using SPSS
version 19.0 (IBM Corp., Armonk, NY, USA). The results are
expressed as the mean ± standard deviation. Comparisons between the
mean values were performed using Student's t test, analysis of
variance and Fisher's least significant difference test. A P-value
of <0.05 was considered as statistically significant.
Results
Immunohistochemical analysis
results
Since all patients did not exhibit CRM1 expression
in the adjacent normal tissue samples, only 101 cases with CRM1
expression in both the tumor tissue and adjacent normal tissue
samples were further analyzed. The results indicated that CRM1 was
highly expressed in both tumor and adjacent normal tissues, but the
expression was not statistically significant between the two groups
(P=0.106; Table II). Subsequently,
the association between CRM1 expression and individual clinical
features of the patients was further investigated. CRM1 expression
of adjacent normal tissues was found to be significantly higher
compared with that of the tumor tissues in the low differentiation
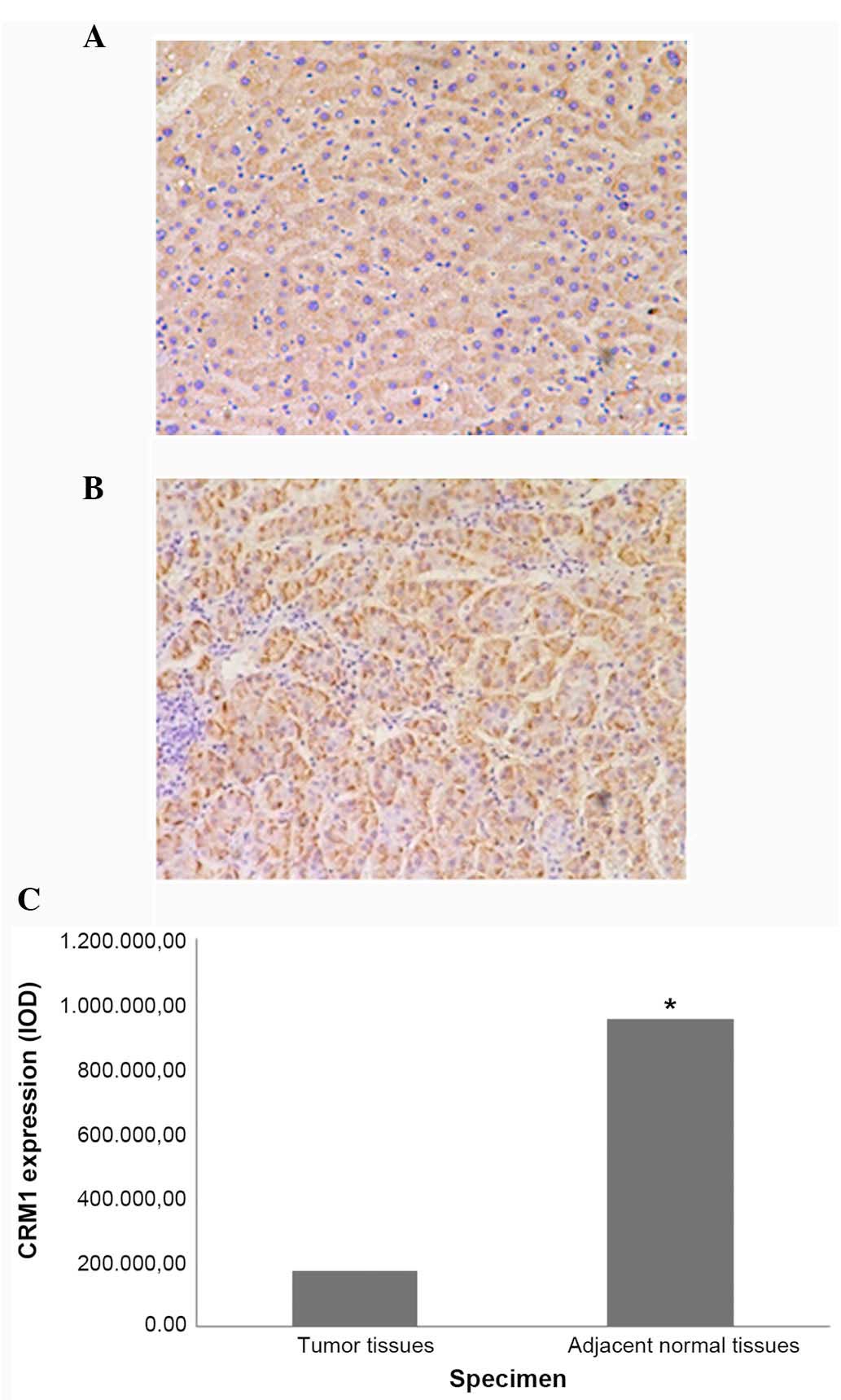
(P=0.004; Fig. 1) and negative HBeAg
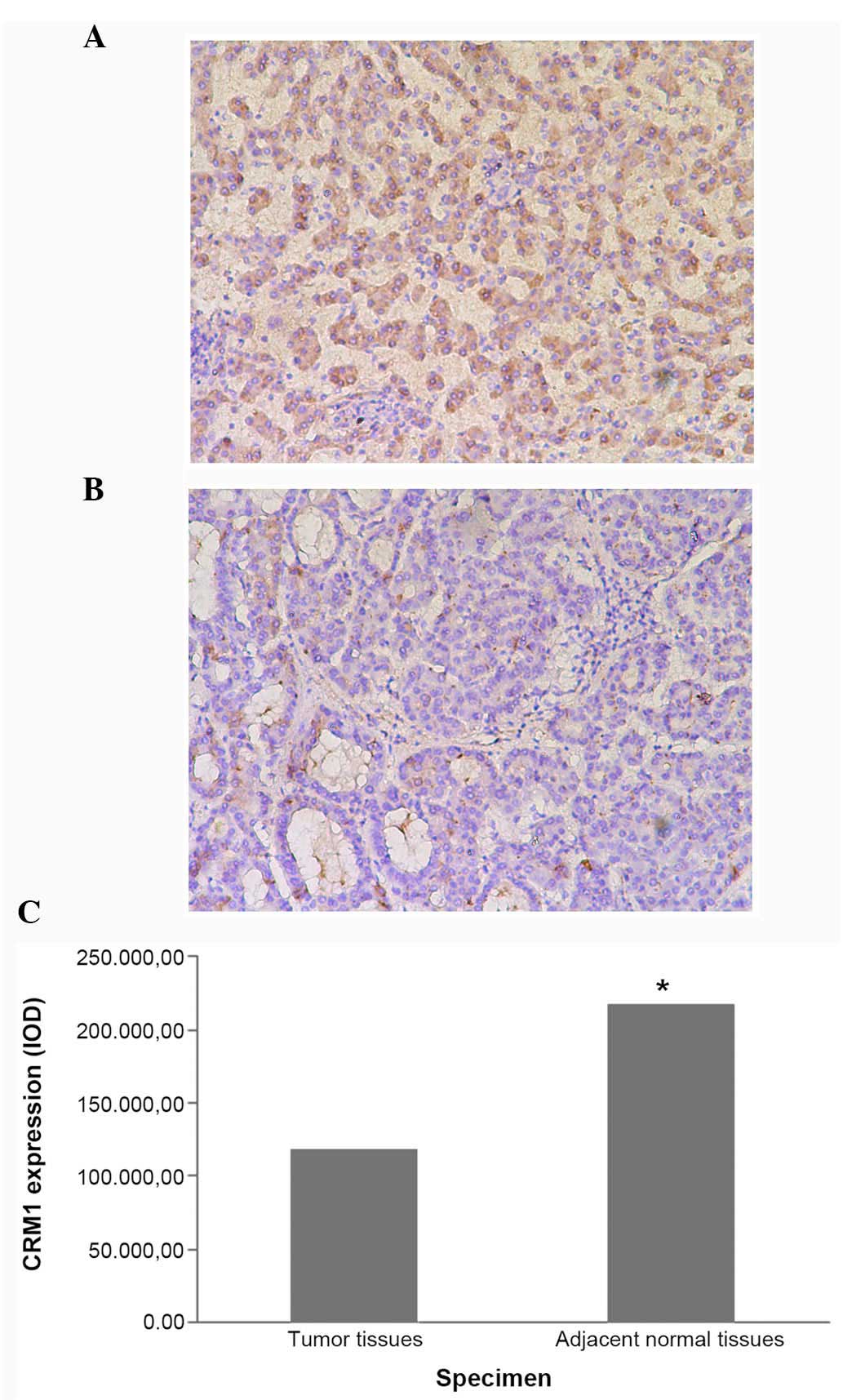
(P=0.035; Fig. 2) specimens.
However, as shown in Table II, the
CRM1 expression of tumor tissues was not significantly lower
compared with that in the adjacent normal tissues for other
parameters (all P>0.05), including gender (male, P=0.144;
female, P=0.508), age (patients ≤55 years, P=0.614; patients >55
years, P=0.069), cirrhosis status (positive, P=0.434; negative,
P=0.080), positive HBsAg (P=0.091), negative HBsAg (P=0.691),
positive HBeAg (P=0.474), AFP (≤400 µg/l, P=0.255; >400 µg/l,
P=0.133), CEA (≤5 µg/l, P=0.174; >5 µg/l, P=0.843), tumor
diameter (≤5 cm, P=0.097; >5 cm, P=0.680) and AJCC stage I
(P=0.084; Fig. 3). Since patients
with AJCC stage II, III or IV were few in numbers, further analysis
was not performed.
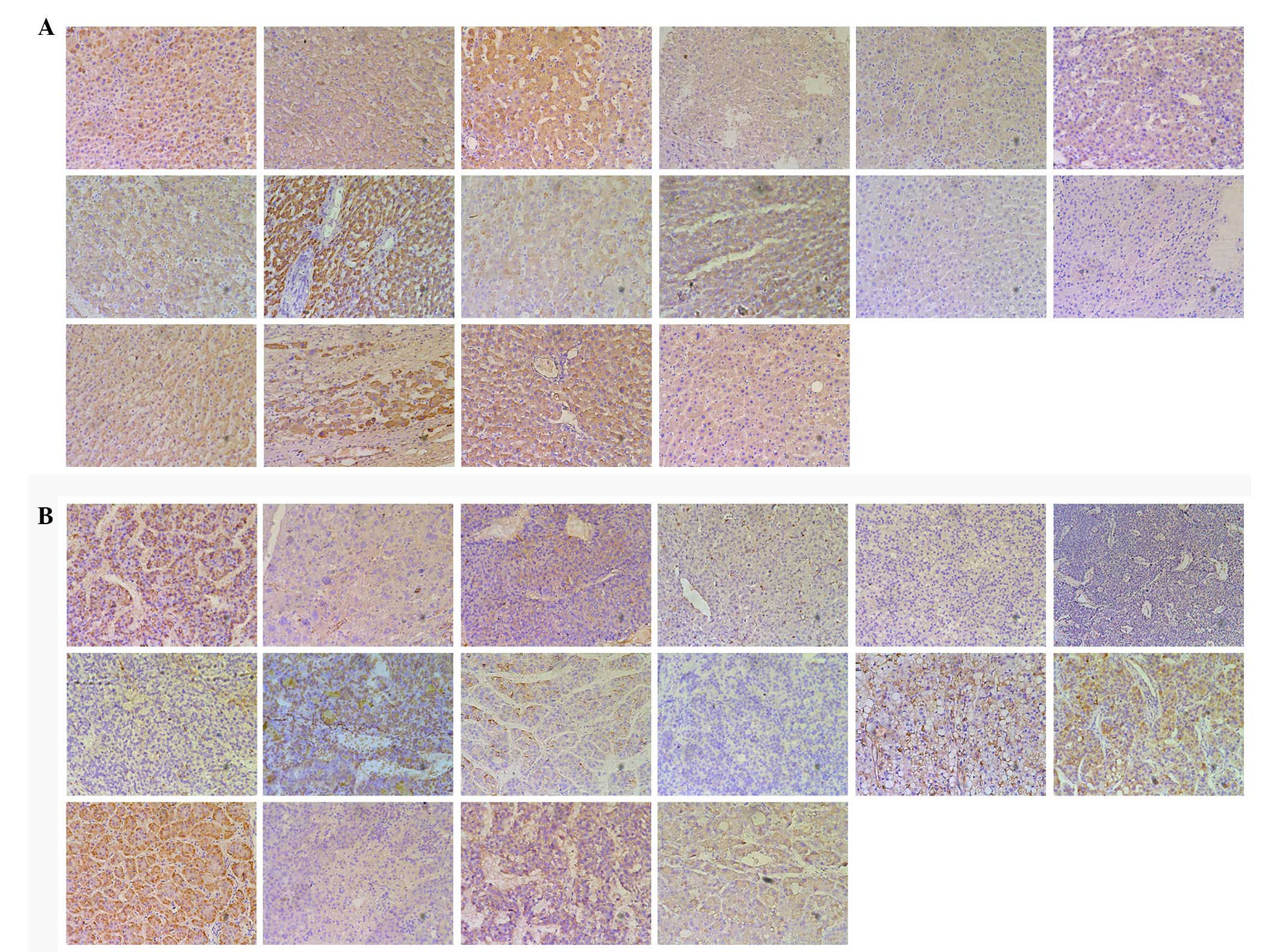 | Figure 3.Immunohistochemical analysis of CRM1
expression in (A) adjacent normal control and (B) tumor tissues,
including the following groups from the first column of the first
row to the fourth column of the third row: Male, female, age >55
years, age ≤55 years, cirrhosis, absence of cirrhosis, positive
HBsAg, negative HBsAg, positive HBeAg, AFP >400 µg/l, AFP ≤400
µg/l, CEA >5 µg/l, CEA ≤5 µg/l, tumor diameter >5 cm, tumor
diameter ≤5 cm and AJCC stage I. CRM1, chromosome region
maintenance 1; HBeAg, hepatitis B envelope antigen; AFP,
α-fetoprotein; CEA, carcinoembryonic antigen. |
 | Table II.Association of CRM1 expression of
tumor and adjacent normal tissues with clinical pathological
features of patients. |
Table II.
Association of CRM1 expression of
tumor and adjacent normal tissues with clinical pathological
features of patients.
|
|
| CRM1 (IOD) |
|
|
|---|
|
|
|
|
|
|
|---|
| Feature | No. | Tumor tissues | Adjacent normal
tissues | T-value | P-value |
|---|
| All patients | 101 |
134,737.84±144,343.88 |
165,603.86±141,841.21 | 1.632 | 0.106 |
| Gender |
|
|
Male | 83 |
142,175.00±199,338.96 |
172,651.86±137,014.50 | 1.477 | 0.144 |
|
Female | 18 |
100,444.32±116,058.22 |
133,104.73±162,584.74 | 0.676 | 0.508 |
| Age, years |
|
|
≤55 | 54 |
141,103.59±165,342.28 |
154,336.02±128,576.80 | 0.507 | 0.614 |
|
>55 | 47 |
127,424.01±117,018.72 |
178,549.88±156,105.03 | 1.861 | 0.069 |
| Cirrhosis |
|
| + | 65 |
149,593.95±128,705.42 |
169,193.31±140,717.66 | −0.787 | 0.434 |
| − | 36 |
107,914.32±167,596.64 |
159,122.90±145,626.71 | −1.806 | 0.080 |
| HBsAg |
|
| + | 94 |
132,439.61±146,800.11 |
166,820.07±144,761.11 | −1.710 | 0.091 |
| − | 7 |
165,599.83±109,678.66 |
149,271.89±100,829.23 | 0.418 | 0.691 |
| HBeAg |
|
| + | 82 |
138,606.82±155,005.00 |
153,497.11±131,531.72 | −0.719 | 0.474 |
| − | 19 |
118,040.13±85,257.80 |
217,854.03±174,268.98 | −2.285 | 0.035 |
| AFP, µg/l |
|
|
≤400 | 86 |
136,995.87±149,979.55 |
160,477.47±140,133.43 | −1.146 | 0.255 |
|
>400 | 15 |
121,691.05±113,876.58 |
204,903.30±153,643.40 | −1.603 | 0.133 |
| CEA, µg/l |
|
| ≤5 | 81 |
130,923.14±148,572.40 |
153,332.91±136,638.43 | 1.732 | 0.174 |
|
>5 | 20 |
192,052.42±145,804.10 |
201,119.63±175,908.90 | −0.214 | 0.843 |
| Diameter, cm |
|
| ≤5 | 65 |
131,421.49±153,865.43 |
172,529.67±154,822.13 | −1.683 | 0.097 |
|
>5 | 36 |
140,725.71±127,204.82 |
153,098.93±115,767.30 | −0.416 | 0.680 |
|
Differentiation |
|
|
Low | 44 |
168,498.02±137,486.15 |
952,982.20±103,493.34 | 3.017 | 0.004 |
|
Middle | 35 |
153,890.49±139,782.16 |
164,034.91±135,824.15 | −0.294 | 0.770 |
|
High | 22 |
172,440.15±201,919.78 |
178,450.43±158,258.18 | 0.141 | 0.889 |
| AJCC stage |
|
| I | 91 |
133,074.18±141,755.58 |
167,146.51±144,340.17 | −1.750 | 0.084 |
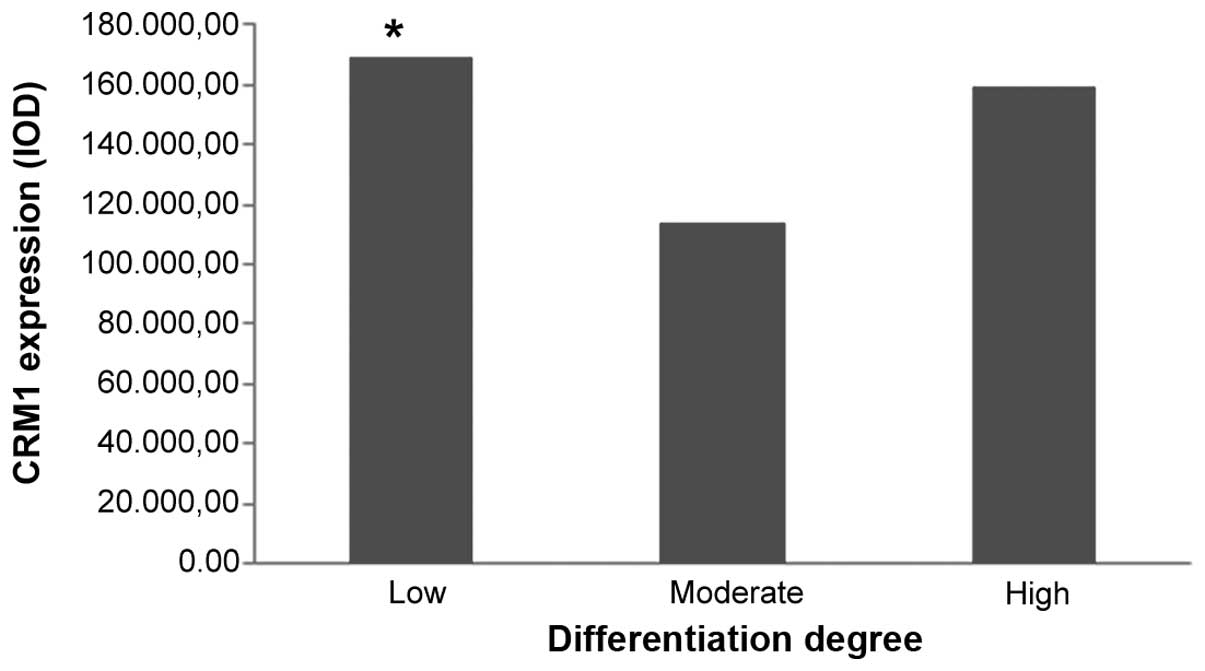
Analysis of CRM1 expression in tumor
tissues
As shown in Fig. 4,
positive CRM1 expression was found to be significantly correlated
with low differentiation (P=0.045). However, negative associations
were detected between positive CRM1 expression and other
parameters, including gender (P=0.566), age (P=0.570), cirrhosis
(P=0.457), HBsAg (P=0.412), HBeAg (P=0.322), AFP (P=0.308), CEA
(P=0.225), tumor diameter (P=0.428) and AJCC stage (all P>0.05;
Table III).
 | Table III.Association between CRM1 expression
of tumor tissues and clinical pathological features. |
Table III.
Association between CRM1 expression
of tumor tissues and clinical pathological features.
| Feature | No. | CRM1 expression
(IOD) in tumor tissues | T-value | P-value |
|---|
| Gender |
|
|
Male | 120 |
143,353.57±151,369.26 | 0.114 | 0.566 |
|
Female | 30 |
139,825.88±156,349.77 |
|
|
| Age, years |
|
|
≤55 | 77 |
129,275.10±150,039.60 | −1.102 | 0.570 |
|
>55 | 75 |
156,396.40±153,464.20 |
|
|
| Cirrhosis |
|
| + | 102 |
157,349.60±147,664.49 | 1.715 | 0.457 |
| − | 50 |
112,685.13±157,300.15 |
|
|
| HBsAg |
|
| + | 139 |
141,232.62±155,627.62 | −0.377 | 0.412 |
| − | 13 |
157,890.67±106,454.30 |
|
|
| HBeAg |
|
| + | 120 |
142,652.84±157,021.42 | 0.021 | 0.322 |
| − | 31 |
143,295.45±135,059.08 |
|
|
| AFP, µg/l |
|
|
≤400 | 120 |
135,903.39±147,779.25 | −0.996 | 0.308 |
|
>400 | 30 |
166,934.08±171,099.53 |
|
|
| CEA, µg/l |
|
| ≤5 | 116 |
135,737.99±150,342.95 | −1.244 | 0.225 |
|
>5 | 36 |
183,316.95±158,906.97 |
|
|
| Diameter, cm |
|
| ≤5 | 86 |
140,864.01±164,325.30 | −0.166 | 0.428 |
|
>5 | 66 |
144,994.05±135,065.09 |
|
|
|
Differentiation |
|
|
| 0.045a, 0.155b, 0.771c |
|
Low | 67 |
169,434.42±154,982.45 |
|
|
|
Moderate | 53 |
113,388.64±132,470.57 |
|
|
|
High | 32 |
159,589.03±136,877.62 |
|
|
| AJCC |
|
|
| 0.997d, 0.649e, 0.805f, |
| stage |
|
|
| 0.747g, 0.821h, 0.674i |
| I | 131 |
141,491.21±147,973.91 |
|
|
| II | 9 |
141,687.09±166,898.77 |
|
|
|
III | 10 |
164,408.01±208,434.76 |
|
|
| IV | 2 |
114,490.13±121,496.74 |
|
|
Analysis of CRM1 expression in
adjacent normal tissue
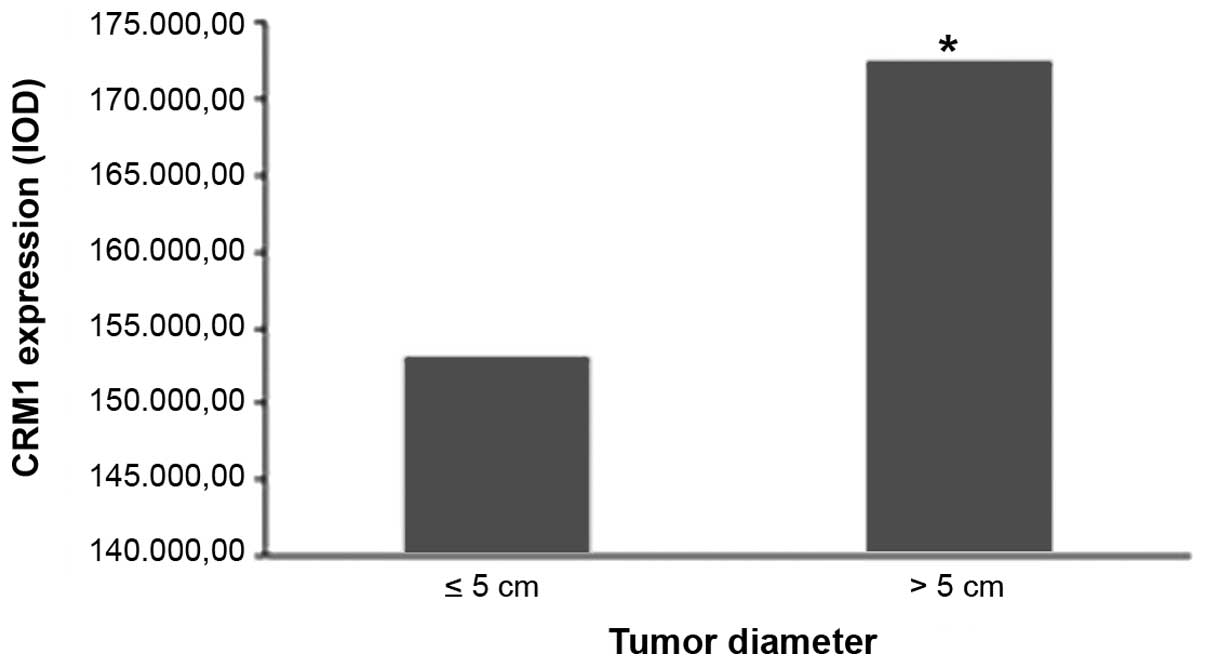
As shown in Fig. 5,
positive CRM1 expression was found to be significantly correlated
with the tumor diameter in adjacent normal tissues (P=0.004).
However, negative associations were detected between positive CRM1
expression and other parameters, including gender (P=0.383), age
(P=0.144), cirrhosis (P=0.394), HBsAg (P=0.257), HBeAg (P=0.125),
AFP (P=0.718), CEA (P=0.355), differentiation degree and AJCC stage
(all P>0.05; Table IV).
 | Table IV.Association between CRM1 expression
of adjacent normal tissues and clinical pathological features. |
Table IV.
Association between CRM1 expression
of adjacent normal tissues and clinical pathological features.
| Feature | Patients | CRM1 (IOD) | T-value | P-value |
|---|
| Gender |
|
|
Male | 83 |
172,651.86±137,014.50 | 1.073 | 0.383 |
|
Female | 18 |
133,104.73±162,584.74 |
|
|
| Age, years |
|
|
≤55 | 54 |
154,336.02±128,576.80 | −0.855 | 0.144 |
|
>55 | 47 |
178,549.88±156,105.03 |
|
|
| Cirrhosis |
|
| + | 65 |
169,193.31±140,717.66 | 0.340 | 0.394 |
| − | 36 |
159,122.89±145,626.71 |
|
|
| HBsAg |
|
| + | 94 |
166,820.07±144,761.11 | 0.314 | 0.257 |
| − | 7 |
149,271.89±100,829.23 |
|
|
| HBeAg |
|
| + | 19 |
217,854.03±174,268.98 | 1.802 | 0.125 |
| − | 82 |
153,497.11±131,531.72 |
|
|
| AFP, µg/l |
|
|
≤400 | 86 |
160,477.47±140,133.43 | −1.086 | 0.718 |
|
>400 | 14 |
204,903.30±153,643.40 |
|
|
| CEA, µg/l |
|
| ≤5 | 81 |
153,332.91±136,638.43 | −0.947 | 0.355 |
|
>5 | 20 |
192,052.42±145,804.10 |
|
|
| Diameter, cm |
|
| ≤5 | 65 |
153,098.93±115,767.30 | 0.658 | 0.004 |
|
>5 | 36 |
172,529.66±154,822.13 |
|
|
|
Differentiation |
|
|
| 0.653a, 0.790b, 0.529c |
|
Low | 44 |
168,498.02±137,486.15 |
|
|
|
Moderate | 35 |
153,890.49±139,782.16 |
|
|
|
High | 22 |
178,450.43±158,258.18 |
|
|
| AJCC stage |
|
|
| 0.850d, 0.690e, 0.665f |
| I | 91 |
167,146.51±144,340.17 |
|
|
| II | 5 |
179,673.43±153,475.79 |
|
|
|
III | 4 |
137,812.58±97,291.86 |
|
|
Discussion
Epidemiological investigations have shown that
primary liver cancer is connected with polygenic susceptibility
(37). The development of cancer is
a complicated process that includes cell proliferation,
inactivation of tumor-suppressor gene and activation of oncogene
mediated by numerous signaling cascades (9,37). CRM1,
a member of the karyopherin-β protein family, is a nuclear export
receptor that, upon binding with RanGTP, is able to recognize the
substrates rich in NES (21,38). The CRM1 signaling cascade is
important in mediating the expression of growth factors and
cytokines, serving a critical role in the malignant transformation
of cells (30–33,38–41). To
date, studies have demonstrated that CRM1 is strongly expressed in
various types of tumor cells, and it may promote malignant
transformation of mammal cells. In addition, its overexpression is
associated with cell carcinogenesis and poor prognosis of solid
tumor (42–45). In human glioma cells overexpressing
CRM1, tumor cell growth was promoted by the nuclear export of p27
protein (42). In lung cancer, CRM1
was found to increase tumor growth by changing the suppressor gene
p53 subcellular localization (43).
In the present study, due to limited availability of
normal human liver tissues, tumor tissues were collected and their
adjacent normal tissues were used as the control group. The current
study detected the CRM1 expression in tumor tissues and adjacent
normal tissues, and estimated the association of CRM1 expression
with various clinical and pathological features, including the
patient gender, age, cirrhosis statis, HBsAg, HBeAg, tumor
diameter, AFP, differentiation degree and AJCC stage. The results
aimed to provide further evidence on whether CRM1 serves an
important role in liver cancer invasion and metastasis, and whether
it is an important molecular mechanism underlying the progression
of liver cancer.
The results of the present study demonstrated that
although male patients >55 years of age had higher expression
levels of CRM1 in adjacent normal tissues compared with tumor
tissues, no significant difference was observed. The number of male
patients with liver cancer were higher than female patients, and
El-Serag et al (2) also
reported that the ratio of affected men to affected women was
between 2:1 and 4:1. Men are therefore more likely to develop liver
cancer than women, but whether liver cancer metastasis affects a
gender or age more than the other requires further study. The
results of the present study revealed that CRM1 was expressed
similarly in tumor and adjacent normal tissues. However, a higher
CRM1 expression in adjacent normal tissues compared with that in
tumor tissues was found to be significantly correlated with the
differentiation degree. In addition, in the tumor tissue group,
positive CRM1 expression was significantly correlated with the
differentiation degree, whereas in the adjacent normal tissue
group, positive CRM1 expression was observed to be significantly
correlated with the tumor diameter. A low degree of differentiation
and higher tumor diameter resulted in increased CRM1 expression in
adjacent normal tissues; thus, CRM1 may participate in tumor cell
metastasis and invasion to adjacent normal tissues. Numerous
studies have confirmed an increased expression of CRM1 in various
solid tumors (such as glioblastoma, pancreatic cancer, ovarian
cancer and cervical cancer) and hematologic malignancies (33,44).
Furthermore, previous studies revealed that the nuclear export of
221 types of NES-containing proteins was mediated by CRM1.
Moreover, numerous NES-containing proteins are critical signaling
molecules of tumor cell proliferation and malignant transformation
(21–52). It is also estimated that CRM1
inhibitors may effectively inhibit the proliferation of tumor cells
and accelerate apoptosis, while their efficacy may be enhanced when
combined with chemotherapy drugs (21).
In fact, HBV is a major cause of primary cancer of
liver (4). A study by Forgues et
al (52) has investigated the
interaction of the HBV X protein with the CRM1-dependent nuclear
export pathway and suggested that CRM1 may serve a role in HBV X
protein-mediated liver carcinogenesis. The oncogenic HBV X protein
contains a functional NES motif and mutations of this motif result
in nuclear redistribution of HBV X protein. In the present study,
the association between HBsAg/HBeAg and CRM1 expression in tumor
tissues and adjacent normal tissues was investigated. The results
showed that the CRM1 expression in adjacent normal tissues with
positive HBsAg was higher compared with that in the tumor tissue
group, but with no statistically significant difference observed.
However, CRM1 expression in adjacent normal tissues was
significantly higher compared with that in tumor tissues with a
negative HBeAg status. According to these findings, it is suggested
that liver cancer in patients with HBV infection, particularly
patients with negative HBeAg, is more likely to present invasion
and metastasis. E antigen is encoded by the pre-C gene, and a
longer duration of hepatitis B accompanied by a mutation to the
pre-C gene will result in the absence of E antigen in the serum.
However, the HBV DNA load will remain high, so HBeAg-negative
patients may experience more severe liver inflammation and are more
likely to develop liver cirrhosis and liver cancer as compared with
HBeAg-positive patients (53).
HBV-associated or hepatitis C virus-associated liver
fibrosis and cirrhosis are considered to be risk factors for the
development of primary liver cancer (54,55). The
majority of primary liver cancer cases (even up 80% of cases)
occurred in patients with HBV infection or cirrhosis in China
(56). Cirrhosis has a significantly
effect on the liver microenvironment, including increased
extracellular matrix proteins and cytokines, angiogenesis and
changes in the immune regulation mechanism. Changes in the liver
microenvironment also affect the environment of tumor cells. In the
present study, a negative association was detected between positive
CRM1 expression and cirrhosis in the tumor tissues and adjacent
normal tissues; however, CRM1 expression in patients with cirrhosis
was higher compared with that in non-cirrhotic patients in both
tissue groups. This increased trend may suggest that cirrhosis is a
dependent risk factor of invasion and metastasis of liver cancer.
The study by Pascale et al (57) revealed that the CRM1 expression in
liver fibrosis, cirrhosis and liver cancer increased by 2–2.9 times
when compared with the expression in normal livers of mice
(57). Therefore, overexpression of
CRM1 may be an important molecular mechanism of cirrhosis evolution
in liver cancer.
AFP and CEA are important serum markers of cancer
diagnosis, and are usually applied in combination (58). In the present study, patients with
high serum levels of AFP and CEA exhibited higher CRM1 expression
both in tumor tissues and adjacent normal tissues compared with
those with low levels of AFP and CEA. However, significant
differences were not observed. These results suggested that high
levels of AFP and CEA may be associated with the degree of
malignancy of the liver cancer and how prone it is to metastasis.
Ma et al (58) also reported
that AFP levels were predictive of the malignant features and
prognosis of liver cancer. However, the current study presented
certain limitations. Although the sample size of the study is not
particularly small, due to limited availability of fresh liver
samples, only immunohistochemical assay was used to assess the CRM1
expression. Studies with much larger sample sizes and novel
experimental methods are required to further investigate the
molecular mechanism of tumor invasion and metastasis.
In conclusion, CRM1 is suggested to serve an
important role in the invasion and metastasis of primary liver
cancer. CRM1 expression was found to be associated with the
differentiation degree and diameter of the tumor. A lower
differentiation degree and larger tumor diameter resulted in higher
CRM1 expression in adjacent normal tissues and increased the
possibility of invasion and metastasis. Furthermore, chronic
hepatitis B patients with negative HBeAg were at a high risk of
invasion and metastasis. Therefore, a high expression of CRM1 may
be an important molecular mechanism of cirrhosis evolution in liver
cancer.
Acknowledgements
The authors would like to thank their colleagues at
the Department of Pathology of The First Affiliated Hospital of
Dalian Medical University for the help and technical guidance
provided. This study was supported by The National Science
Foundation of China (grant no. 81273925).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
El-Serag HB: Hepatocellular carcinoma. N
Engl J Med. 365:1118–1127. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Di Bisceglie AM, Rustgi VK, Hoofnagle JH,
Dusheiko GM and Lotze MT: NIH conference. Hepatocellular carcinoma.
Ann Intern Med. 108:390–401. 1998. View Article : Google Scholar
|
|
5
|
But DY, Lai CL and Yuen MF: Natural
history of hepatitis-related hepatocellular carcinoma. World J
Gastroenterol. 14:1652–1656. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dubbelboer IR, Lilienberg E, Ahnfelt E,
Sjögren E, Axén N and Lennernäs H: Treatment of intermediate stage
hepatocellular carcinoma: A review of intrahepatic doxorubicin
drug-delivery systems. Ther Deliv. 5:447–466. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Villanueva A, Minguez B, Forner A, Reig M
and Liovet JM: Hepatocellular carcinoma: Novel molecular approaches
for diagnosis, prognosis, and therapy. Annu Rev Med. 61:317–328.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li WX, Chen LP, Sun MY, Li JT, Liu HZ and
Zhu W: 3′3-Diindolylmethane inhibits migration, invasion and
metastasis of hepatocellular carcinoma by suppressing FAK
signaling. Oncotarget. 6:23776–23792. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Villanueva A, Newell P, Chiang DY,
Friedman SL and Llovet JM: Genomics and signaling pathways in
hepatocellular carcinoma. Semin Liver Dis. 27:55–76. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kudo N, Khochbin S, Nishi K, Kitano K,
Yanagida M, Yoshida M and Horinouchi S: Molecular cloning and cell
cycle-dependent expression of mammalian CRM1, a protein involved in
nuclear export of proteins. J Biol Chem. 272:29742–29751. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nguyen KT, Holloway MP and Altura RA: The
XPO1 nuclear export protein in normal development and disease. Int
J Biochem Mol Biol. 3:137–151. 2012.PubMed/NCBI
|
|
12
|
Turner JG, Dawson J and Sullivan DM:
Nuclear export of proteins and drug resistance in cancer. Biochem
Pharmacol. 83:1021–1032. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Muqbil I, Bao B, Abou-Samra AB, Mohammad
RM and Azmi AS: Nuclear export mediated regulation of MicroRNAs:
Potential target for drug intervention. Curr Drug Targets.
14:1094–1100. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Siddiqui N and Borden KL: mRNA export and
cancer. Wiley Interdiscip Rev RNA. 3:13–25. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Santiago A, Li D, Zhao LY, Godsey A and
Liao D: p53 SUMOylation promotes its nuclear export by facilitating
its release from the nuclear export receptor CRM1. Mol Biol Cell.
24:2739–2752. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brodie KM and Henderson BR:
Characterization of BRCA1 protein targeting, dynamics, and function
at the centrosome: A role for the nuclear export signal, CRM1, and
Aurora A kinase. J Biol Chem. 287:7701–7716. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chan KS, Wong CH, Huang YF and Li HY:
Survivin withdrawal by nuclear export failure as a physiological
switch to commit cells to apoptosis. Cell Death Dis. 1:e572010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mariano AR, Colombo E, Luzi L, Martinelli
P, Volorio S, Bernard L, Meani N, Bergomas R, Alcalay M and Pelicci
PG: Cytoplasmic localization of NPM in myeloid leukemias is
dictated by gain-of-function mutations that create a functional
nuclear export signal. Oncogene. 25:4376–4380. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Henderson BR: Nuclear-cytoplasmic
shuttling of APC regulates beta-catenin subcellular localization
and turnover. Nat Cell Biol. 2:653–660. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ishizawa J, Kojima K, Hail N Jr, Tabe Y
and Andreeff M: Expression, function, and targeting of the nuclear
exporter chromosome region maintenance 1 (CRM1) protein. Pharmacol
Ther. 153:25–35. 2015.PubMed/NCBI
|
|
21
|
Xu D, Grishin NV and Chook YM: NESdb: A
database of NES-containing CRM1 cargoes. Mol Biol Cell.
23:3673–3676. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fabregat I: Dysregulation of apoptosis in
hepatocellular carcinoma cells. World J Gastroenterol. 15:513–520.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chiorazzi N: Cell proliferation and death:
Forgotten features of chronic lymphocytic leukemia B cells. Best
Pract Res Clin Haematol. 20:399–413. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fulda S and Debatin KM: Apoptosis pathways
in neuroblastoma therapy. Cancer Lett. 197:131–135. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Giovanni GL, Senapedis W, McCauley D,
Baloglu E, Shacham S and Festuccia C: Nucleo-cytoplasmic tansport
as a therapeutic target of cancer. J Hematol Oncol. 7:852014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hamamoto T, Uozumi T and Beppu T:
Leptomycins A and B, new antifungal antibiotics. III. Mode of
action of leptomycin B on Schizosaccharomyces pombe. J Antibiot
(Tokyo). 38:1573–1580. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Meissner T, Krause E and Vinkemeier U:
Ratjadone and leptomycin B block CRM1-dependent nuclear export by
identical mechanisms. FEBS Lett. 576:27–30. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mutka SC, Yang WQ, Dong SD, Ward SL, Craig
DA, Timmermans PB and Murli S: Identification of nuclear export
inhibitors with potent anticancer activity in vivo. Cancer
Res. 69:510–517. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Etchin J, Sanda T, Mansour MR, Kentsis A,
Montero J, Le BT, Christie AL, McCauley D, Rodig SJ, Kauffman M, et
al: KPT-330 inhibitor of CRM1 (XPO1)-mediated nuclear export has
selective anti-leukaemic activity in preclinical models of T-cell
acute lymphoblastic leukaemia and acute myeloid leukaemia. Br J
Haematol. 161:117–127. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gao W, Lu C, Chen L and Keohavong P:
Overexpression of CRM1: A characteristic feature in a transformed
phenotype of lung carcinogenesis and a molecular target for lung
cancer adjuvant therapy. J Thorac Oncol. 10:815–825. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang X, Cheng L, Yao L, Ren H, Zhang S,
Min X, Chen X, Zhang J and Li M: Involvement of chromosome region
maintenance 1 (CRM1) in the formation and progression of esophageal
squamous cell carcinoma. Med Oncol. 31:1552014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu X, Niu M, Xu X, Cai W, Zeng L, Zhou X,
Yu R and Xu K: CRM1 is a direct cellular target of the natural
anti-cancer agent plumbagin. J Pharmacol Sci. 124:486–493. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Walker CJ, Oaks JJ, Santhanam R, Neviani
P, Harb JG, Ferenchak G, Ellis JJ, Landesman Y, Eisfeld AK, Gabrail
NY, et al: Preclinical and clinical efficacy of XPO1/CRM1
inhibition by the karyopherin inhibitor KPT-330 in Ph+ leukemias.
Blood. 122:3034–3044. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Theise ND, Curado MP, Franceschi S, et al:
Hepatocellular carcinoma. Classification of tumours of the
digestive system (4th). Bosman FT, Carneiro F, Hruban RH and Theise
ND: IARC. (Lyon). 2010.205
|
|
35
|
Jain D: Tissue diagnosis of hepatocellular
carcinoma. J Clin Exp Hepatol. 4(Suppl 3): S67–S73. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fleming ID: AJCC/TNM cancer staging,
present and future. J Surg Oncol. 77:233–236. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Farazi PA and DePinho RA: Hepatocellular
carcinoma pathogenesis: From genes to environment. Nat Rev Cancer.
6:674–687. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Stade K, Ford CS, Guthrie C and Weis K:
Exportin 1 (Crm1p) is an essential nuclear export factor. Cell.
90:1041–1050. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ma HX, Shu QH, Pan JJ, Liu D, Xu GL, Li
JS, Ma JL, Jia WD, Yv JH and Ge YS: Expression of Kindlin-1 in
human hepatocellular carcinoma and its prognostic significance.
Tumor Biol. 36:4235–4241. 2015. View Article : Google Scholar
|
|
40
|
Yang X, Cheng L, Yao L, Ren H, Zhang S,
Min X, Chen X, Zhang J and Li M: Involvement of chromosome region
maintenance 1 (CRM1) in the formation and progression of esophageal
squamous cell carcinoma. Med Oncol. 31:1152014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
van der Watt PJ, Maske CP, Hendricks DT,
Parker ML, Denny L, Govender D, Birrer MJ and Leanner VD: The
Karyopherin proteins, Crm1 and Karyopherin beta1, are overexpressed
in cervical cancer and are critical for cancer cell survival and
proliferation. Int J Cancer. 124:1829–1840. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shen A, Wang Y, Zhao Y, Zou L, Sun L and
Cheng C: Expression of CRM1 in human gliomas and its significance
in p27 expression and clinical prognosis. Neurosurgery. 65:153–159;
discussion 159–160. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen L, Moore JE, Samathanam C, Shao C,
Cobos E, Miller Ms and Gao W: CRM1-dependent p53 nuclear
accumulation in lung lesions of a bitransgenic mouse lung tumor
model. Oncol Rep. 26:223–228. 2011.PubMed/NCBI
|
|
44
|
Noske A, Weichert W, Niesporek S, Röske A,
Buckendahl AC, Koch L, Sehouli J, Dietel M and Denkert C:
Expression of the nuclear export protein chromosomal region
maintenance/exportin 1/Xpo1 is a prognostic factor in human ovarian
cancer. Cancer. 112:1733–1743. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang WY, Yue L, Qiu WS, Wang LW, Zhou XH
and Sun YL: Prognostic value of CRM1 in pancreas cancer. Clin
Invest Med. 32:E3152009.PubMed/NCBI
|
|
46
|
Lapalombella R, Sun Q, Williams K,
Tangeman L, Jha S, Zhong Y, Goettl V, Mahoney E, Berglund C, Gupta
S, et al: Selective inhibitors of nuclear export showed that
CRM1/XPO1 is a target in chronic lymphocytic leukemia. Blood.
120:4621–4634. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tai YT, Landesman Y, Acharya C, Calle Y,
Zhong MY, Cea M, Tannenbaum D, Cagnetta A, Reagan M, Munshi AA, et
al: CRM1 inhibition induces tumor cell cytotoxicity and impairs
osteoclastogenesis in multiple myeloma: Molecular mechanisms and
therapeutic implications. Leukemia. 28:155–165. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kojima K, Kornblau SM, Ruvolo V, Dilip A,
Duvvuri S, Davis RE, Zhang M, Wang Z, Coombes KR, Zhang N, et al:
Prognostic impact and targeting of CRM1 in acute myeloid leukemia.
Blood. 121:4166–4174. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Schmidt J, Braggio E, Kortuem KM, Egan JB,
Zhu YX, Xin CS, Tiedemann RE, Palmer SE, Garbitt VM, McCauley D, et
al: genome-wide studies in multiple myeloma identify XPO1/CRM1 as a
critical target validated using the selective nuclear export
inhibitor KPT-276. Leukemia. 27:2357–2365. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Azmi AS, Al-Katib A, Aboukameel A,
McCauley D, Kauffman M, Shacham S and Aohammad RM: Selective
inhibitors of nuclear export for the treatment of non-Hodgkin's
lymphomas. Haematologica. 98:1098–1106. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Walker CJ, Oaks JJ, Santhanam R, Neviani
P, Harb JG, Ferenchak G, Ellis JJ, Landesman Y, Eisfeld AK, Gabrail
NY, et al: Preclinical and clinical efficacy of XPO1/CRM1
inhibition by the karyopherin inhibitor KPT-330 in Ph+ leukemias.
Blood. 122:3034–3044. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Forgues M, Marrogi AJ, Spillare EA, Wu CG,
Yang Q, Yoshida M and Wang XW: Interaction of the hepatitis B virus
X protein with the Crm1-dependent nuclear export pathway. J Biol
Chem. 276:22797–22803. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chuanxin Zou, Jiayan Nie, Shaojun Dai, et
al: Study of relationships between primary carcinoma of the liver
and chronic hepatitis B with positive/negative HBeAg. World Chinese
Journal of Digestology. 16:3696–3699. 2008.
|
|
54
|
Biselli M, Conti F, Gramenzi A, Frigerio
M, Cucchetti A, Fatti G, D'Angelo M, Dall'Agata M, Giannini EG, et
al: A new approach to the use of α-fetoprotein as surveillance test
for hepatocellular carcinoma in patients with cirrhosis. Br J
Cancer. 112:69–76. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ishizuka M, Kubota K, Kita J, Shimoda M,
Kato M, Mori S, Iso Y, Yamagishi H and Kojima M: Aspartate
aminotransferase-to-platelet ratio index is associated with liver
cirrhosis in patients undergoing surgery for hepatocellular
carcinoma. J Surg Res. 194:63–68. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang FS, Fan JG, Zhang Z, Gao B and Wang
HY: The global burden of liver disease: The major impact of China.
Hepatology. 60:2099–2108. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Pascale RM, Simile MM, Calvisi DF, Frau M,
Muroni MR, Seddaiu MA, Daino L, Muntoni MD, De Miglio MR,
Thorgeirsson SS and Feo F: Role of HSP90, CDC37, and CRM1 as
modulators of P16(INK4A) activity in rat liver carcinogenesis and
human liver cancer. Hepatology. 42:1310–1319. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ma WJ, Wang HY and Teng LS: Correlation
analysis of preoperative serum alpha-fetoprotein (AFP) level and
prognosis of hepatocellular carcinoma (HCC) after hepatectomy.
World J Surg Oncol. 11:2122013. View Article : Google Scholar : PubMed/NCBI
|