Introduction
Atrial fibrillation (AF) is the most common type of
sustained cardiac arrhythmia, affecting 1–2% of the general
population (1). The prevalence of AF
increases with age (2,3) and the condition is a known risk factor
for a number of cardiovascular diseases, including a five-fold risk
of stroke, three-fold incidence of congestive heart failure and an
increased rate of mortality (4).
Previous studies have found that the occurrence of
AF affects atrial activity, causing the atria to quiver
persistently or relapse into fibrillation soon after. Wijffels
et al (1) first described
this process as atrial electrical remodeling, observing that AF was
able to induce electrophysiological changes itself, which in turn
promoted further AF development and maintenance (‘AF begets AF’)
(5). The theory of atrial electrical
remodeling has become increasingly accepted and this
self-perpetuating process is considered to be an important
mechanism of AF maintenance.
Changes to a number of ion channel currents may
result in electrical remodeling, including the L-type calcium
current, the transient outward potassium current when downregulated
and the inward rectifier potassium current when upregulated
(6). Previous studies have found
that the hyperpolarization current (If) is associated
with atrial electrical remodeling in patients with AF (7–9). The
If current is a depolarizing sodium-potassium mixed
non-specific current with hyperpolarization-activated intracellular
cAMP regulatory characteristics, which is involved in the
depolarization of cardiac pacemaker cells (10). Increased If current
activity may lead to an increase in myocardial tissue
self-perpetuation, which commonly results in a tachyarrhythmia. The
molecular propagators of the If current are
hyperpolarization-activated cyclic nucleotide-gated (HCN) channels
(11). Therefore, the inhibition of
HCN channels has become a focus of study for the treatment of
arrhythmia (6). However, few studies
have investigated changes in the expression of HCN channel isoforms
in atrial cells following AF. The aim of the present study was to
investigate the molecular mechanisms of AF and atrial electrical
remodeling by comparing the mRNA and protein expression levels of
HCN channel isoforms (HCN1, 2 and 4) in left atrial tissues from a
canine model of AF.
Materials and methods
Animal model establishment
A total of 14 healthy beagle dogs (n=2 for the
preliminary experiment; n=12 for the remaining experiments)
weighing 10.1±2.9 kg and aged 1.5–2.5 years were divided into two
groups. All the dogs were implanted with pacemakers in a
subcutaneous neck pocket using a previously described procedure
(12,13). Following a 24-h recovery period,
seven dogs were subjected to continuous right atrial pacing at 400
bpm for 50 days (AF group). In the remaining seven dogs, the
pacemaker was not activated and a normal sinus rhythm heart beat
was maintained (sham control group). The pacemaker was programmed
to stimulate the atrium at 400 bpm, (cycle length, 150 msec) using
0.42 msec square wave pulses at twice-threshold current. The
surface electrocardiogram was verified after 24 h, and then weekly
to ensure continuous 1:1 atrial capture. The dogs were euthanized
with an overdose of α-chloralose (350 mg/kg). The heart was
removed, and the right atrial free wall and sinoatrial node were
isolated. The left atrial appendage tissues were separated (weight,
~200 mg), and the samples were rapidly frozen in liquid nitrogen
and stored at −80°C. All animal experiments were performed in
accordance with the protocols approved by the Animal Care Committee
of the Institute of Zoology at the Chinese Academy of Sciences
(Beijing, China).
RNA isolation
RNA was isolated from 0.1–1.0-g samples of left
atrial tissue using TRIzol reagent (Invitrogen Life Technologies,
Carlsbad, CA, USA), following which phenol-chloroform extraction
and isopropanol precipitation were conducted. Genomic DNA was
eliminated through incubation in DNase I (0.1 U/ml, 37°C) for 30
min, followed by acid-phenol-chloroform extraction. The RNA was
quantified by measuring the spectrophotometric absorbency at 260
nm, while the purity was confirmed by
A260/A280 ratio assessment. DNA integrity was
evaluated with ethidium bromide staining on a denaturing agarose
gel, and the RNA samples were stored at −80°C in RNAsecure
Resuspension Solution (Ambion Life Technologies, Carlsbad, CA,
USA).
Quantitative polymerase chain reaction
(qPCR)
A 1-µg sample of reference RNA was reverse
transcribed into cDNA using a reverse transcriptase kit (A3500;
Promega Corporation, Madison, WI, USA). The design and synthesis
details of the primers used are shown in Table I; the primers were synthesized by
Takara Bio, Inc. (Otsu, Japan). The cDNA sample was diluted five
times and 2 µl was added to the qPCR system (25 µl), which also
contained 12.5 µl SYBR Green qPCR Master Mix (Ruian BioTechnologies
Co., Ltd., Shanghai, China), 0.5 µl upstream primer (10 µM), 0.5 µl
downstream primer (10 µM) and 9.5 µl ddH2O. The reaction
was conducted as follows: Denaturation for 2 min at 95°C, followed
by 35 PCR cycles of denaturation for 10 sec at 95°C, annealing for
30 sec at 58, 56.5, 59 and 57°C for HCN1, HCN2, HCN4 and β-actin,
respectively, and a final extension for 40 sec at 72°C. The mRNA
expression levels were calculated for each HCN channel isoform
using the comparative Ct-method. The length of the target gene
products (HCN2, 143 bp; HCN4, 132 bp; β-actin, 152 bp) were
consistent with the theoretical values. PCR products were
visualized under UV light using ethidium bromide staining in 1.5%
agarose gel. Images were captured with an Olympus DP22 camera
(Olympus Corporation, Tokyo, Japan) and the band intensity was
determined using Quantity One software (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). A DNA MassMarker (100 ng; Takam Co., Japan) was
used to determine the size and quantity of the DNA bands and to
create standard curves in each experiment for absolute
quantification.
 | Table I.Primer sequences used for qPCR. |
Table I.
Primer sequences used for qPCR.
| Gene | Primer sequence | Product length
(bp) | Annealing temperature
(°C) |
|---|
| β-actin | F: 5
AAGGACCTGTATGCCAACACA 3 |
|
|
|
| R: 5
ATCCACACAGAATACTTGCGTT 3 | 152 | 57 |
| HCN1 | F: 5
GACGCTATGGGCTATGAGTTAC 3 |
|
|
|
| R: 5
AGTCCAGGTAGCCCTTTAGGT 3 | 199 | 58 |
| HCN2 | F: 5
ACTGCTGGGTCTCCATCAAC 3 |
|
|
|
| R: 5
CGTCAGCCAGATGTCCGT 3 | 143 | 56.5 |
| HCN4 | F: 5
CATCCAGTCCCTGGACTCGT 3 |
|
|
|
| R: 5
TGGTAGCGGTGCTCGTAGTAG 3 | 132 | 59 |
Western blot analysis
Total protein was extracted using a western blot kit
(bs-0542; Beijing Bioss Science and Technology Co., Ltd., Beijing,
China) according to the manufacturers instructions, and the protein
concentration was assessed using the bicinchoninic acid method.
SDS-PAGE was used to separate the total protein content of the
50-µg samples in each well. The separated protein samples were
transferred by electroporation onto a nitrocellulose membrane.
Following elution, 5% non-fat dry milk and Tris-buffered-saline
were used for blocking. The membranes were incubated overnight at
4°C with specific rabbit primary antibodies against HCN1 (cat. no.
ab19405), HCN2 (cat. no. ab136839), HCN4 (Abcam, Cambridge, UK;
cat. no. ab69054), SR-4 antibody (cat. no. sc-376158; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) and β-actin (Wuhan Boster
Biological Technology Ltd., Wuhan, China; cat. no. 10082003) at 1
ml/mg. Subsequently, the membranes were incubated for 1 h with a
horseradish peroxidase-conjugated secondary antibody (goat
anti-rabbit IgG; Wuhan Boster Biological Technology, Ltd.; cat. no.
BA1055). Finally, the membranes underwent visualization using a
3,3-diaminobenzidine colorimetric gel imaging system (Bio-Rad
Laboratories, Inc.) in order to quantify the hybridization
signal.
Statistical analysis
Measurement data are expressed as the mean ±
standard deviation. The t-test was used to compare between groups
or non-parametric test, the χ2 test was used in
categorical data. Associations between variables were detected
using linear correlation analysis. Statistical analysis was
performed with SPSS 16.0 statistical software (SPSS, Inc., Chicago,
IL, USA), where P<0.05 was considered to indicate a
statistically significant difference.
Results
mRNA expression
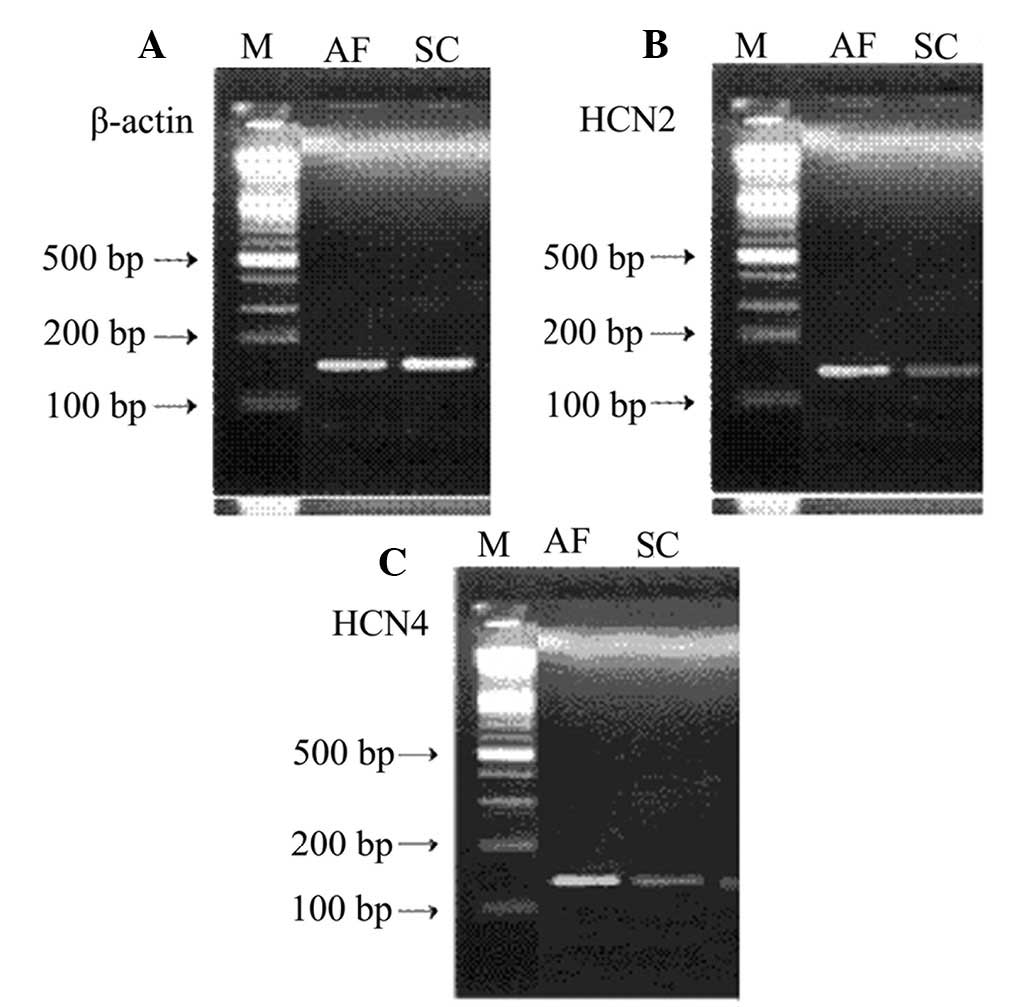
PCR products of the HCN channel isoforms are shown
in Table I. HCN1 mRNA expression was
not detected in the canine atrial muscle samples, which may be due
to very low levels of expression. Therefore, only the mRNA
expression levels of HCN2 and HCN4 were compared. The mRNA
expression levels of HCN2 and HCN4 showed a statistically
significant increase in the AF group when compared with the sham
control group (P<0.05; Table
II).
 | Table II.Comparison of mRNA expression levels
of HCN isoforms in the AF and SC groups. |
Table II.
Comparison of mRNA expression levels
of HCN isoforms in the AF and SC groups.
| Category | Dogs (n) | HCN2 | HCN4 |
|---|
| SC | 5 | 1.27±0.11 | 1.14±0.14 |
| AF | 7 |
2.23±0.22a |
2.97±0.21a |
| P-value | – | 0.03 | 0.01 |
Using a 100-bp DNA marker as a reference, the
lengths of the β-actin, HCN2 and HCN4 mRNA fragments were found to
be 152, 143 and 132 bp, respectively (Fig. 1).
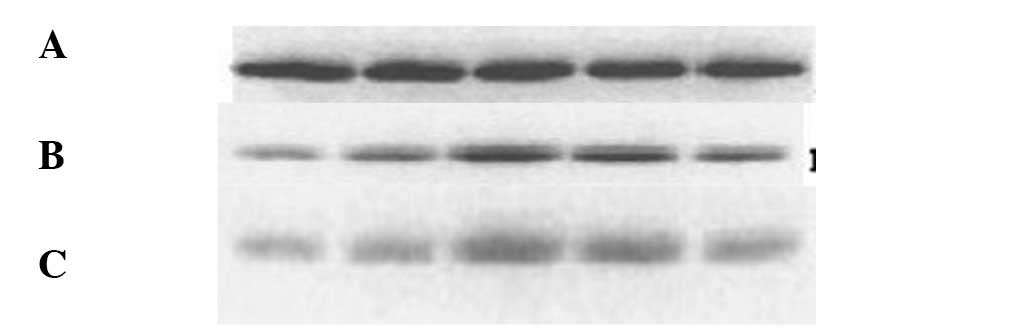
Protein expression
HCN1 protein expression was not detected in the
canine atrial muscle samples. Western blot analyses of the HCN2 and
HCN4 proteins are shown in Fig. 2,
where β-actin was used a reference to calculate the HCN2 and HCN4
protein expression levels. The protein expression levels of HCN2
and HCN4 showed a statistically significant increase in the AF
group when compared with dogs in the sham control group
(P<0.05).
Discussion
HCN channels are the molecular propagators of the
If current (6). There are
four HCN channel isoforms in mammals, HCN1-4. Configurations of HCN
channels in different species and tissues vary considerably. The
majority of HCN3 channels are located in central nervous system
tissues, while HCN1, HCN2 and HCN4 are expressed abundantly in the
heart tissue. The present study revealed that HCN1 expression was
not detectable in the atrial muscle of dogs in the AF or sham
control groups, which may have been due to very low expression
levels. The results were consistent with the findings of previous
studies, including the study by Stephen et al (14). It is widely accepted that HCN3 is
expressed primarily in the central nervous system and not in the
heart; however, the present study did not include the detection of
HCN3. Expression of HCN2 and HCN4 was observed in the atria of the
sham control and AF groups. Therefore, the results indicate that
the majority of HCN channels in canine atrial muscle are HCN4 and
HCN2 isoforms.
Cardiac sinus node pacemaker HCN channels are
proteins with important molecular functions (7). Under normal conditions, the expression
of HCN channels in pacemaker cells and the myocardium is relatively
low, and the If current is weak. A variety of
pathological factors may stimulate abnormal myocardial expression
of HCN channels or cause the development of structural
abnormalities. These changes may result in an increase or decrease
of the If current, which can in turn cause a variety of
arrhythmias. Previous studies have found that mutations in the HCN4
channels of pacemaker cells may cause the If current to
weaken and pacemaker cells to self-regulate their heart rate
(8–11). In addition, a previous study
demonstrated that expression of HCN4 and HCN2 channels in
ventricular myocytes increases the ventricular If
current density, which increases the risk of ventricular
arrhythmias (6). If
current and HCN channels have been intensively researched; however,
the association between HCN channels and atrial arrhythmias remains
unknown. Further studies into this association are necessary in
order to reveal the role played by HCN channels in cases of AF
electrical remodeling. Zorn-Pauly et al (12) found that increases in the
If current may cause atrial myocytes to function in a
similar manner to pacemaker cells, subsequently increasing the
local atrial automaticity, decreasing the effective atrial
refractory period and enhancing the risk of atrial arrhythmias. The
present study found that the mRNA and protein expression levels of
HCN2 and HCN4 in the AF group increased significantly when compared
with the sham control group. Increased expression of these HCN
channels in dogs with multiple organ failure and AF can increase
the If current, which may increase atrial automaticity,
decrease the effective atrial refractory period and enhance the
risk of atrial arrhythmias, with the possibility of causing atrial
electrical remodeling (13).
Robinson et al (7) found that increased atrial pacing may
lead to an ectopic pacemaker, which is able to progress into a
variety of heart conditions, including AF and potentially fatal
ventricular arrhythmias. Multiple organ failure can cause the
downregulation of a variety of potassium channels, which may be due
to the abnormal expression and function of gap junctions, which
produces spatial heterogeneity. It was hypothesized that multiple
organ failure in dogs with AF, and the subsequent upregulation of
HCN2 and HCN4 expression, may result in an ectopic pacemaker, which
may in turn progress into a clinically significant arrhythmia. This
abnormal pacing may cause cardiac dysfunction due to cellular
pathologies, including heart failure, renal failure and multiple
organ failure, and is affected by the expression of HCN channel
proteins.
Future studies should focus on multiple organ
failure in dogs and examine the range of pathological factors
affecting HCN channel expression.
References
|
1
|
Wijffels MC, Kirchhof CJ, Dorland R and
Allessie MA: Atrial fibrillation begets atrial fibrillation. A
study in awake chronically instrumented goats. Circulation.
92:1954–1968. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ma CS, Liu XP and Wang Y: Originating from
the pulmonary veins electrophysiological characteristics of focal
atrial fibrillation with radio frequency ablation. Zhonghua Xin Lu
Shi Chang Xue Za Zhi. 4:18–22. 2000.(In Chinese).
|
|
3
|
Gaborit N, Steenman M, Lamirault G, et al:
Human atrial ion channel and transporter subunit gene-expression
remodeling associated with valvular heart disease and atrial
fibrillation. Circulation. 112:471–481. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Baruscotti M, Bucchi A and Difrancesco D:
Physiology and pharmacology of the cardiac pacemaker (‘funny’)
current. Pharmacol Ther. 107:59–79. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Morillo CA, Klein GJ, Jones DL and
Guiraudon CM: Chronic rapid atrial pacing. Structural, functional,
and electrophysiological characteristics of a new model of
sustained atrial fibrillation. Circulation. 91:1588–1595. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Herrmann S, Stieber J and Ludwig A:
Pathophysiology of HCN channels. Pflugers Arch. 454:517–522. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Robinson RB and Siegelbaum SA:
Hyperpolarization-activated cation currents: from molecules to
physiological function. Annu Rev Physiol. 65:453–480. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schulze-Bahr E, Neu A, Friederich P, et
al: Pacemaker channel dysfunction in a patient with sinus node
disease. J Clin Invest. 111:1537–1545. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ueda K, Nakamura K, Hayashi T, et al:
Functional characterization of a trafficking-defective HCN4
mutation, D553N, associated with cardiac arrhythmia. J Biol Chem.
279:27194–27198. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Milanesi R, Baruscotti M, Gnecchi-Ruscone
T and DiFrancesco D: Familial sinus bradycardia associated with a
mutation in the cardiac pacemaker channel. N Engl J Med.
354:151–157. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nof E, Luria D, Brass D, et al: Point
mutation in the HCN4 cardiac ion channel pore affecting synthesis,
trafficking, and functional expression is associated with familial
asymptomatic sinus bradycardia. Circulation. 116:463–470. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zorn-Pauly K, Schaffer P, Pelzmann B, et
al: If in left human atrium: a potential contributor to atrial
ectopy. Cardiovasc Res. 64:250–259. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Stillitano F, Lonardo G, Zicha S, et al:
Molecular basis of funny current (If) in normal and failing human
heart. J Mol Cell Cardiol. 45:289–299. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Stephen J, Emerson B, Fox KA and
Dransfield I: The uncoupling of monocyte-platelet interactions from
the induction of proinflammatory signaling in monocytes. J Immunol.
191:5677–5683. 2013. View Article : Google Scholar : PubMed/NCBI
|