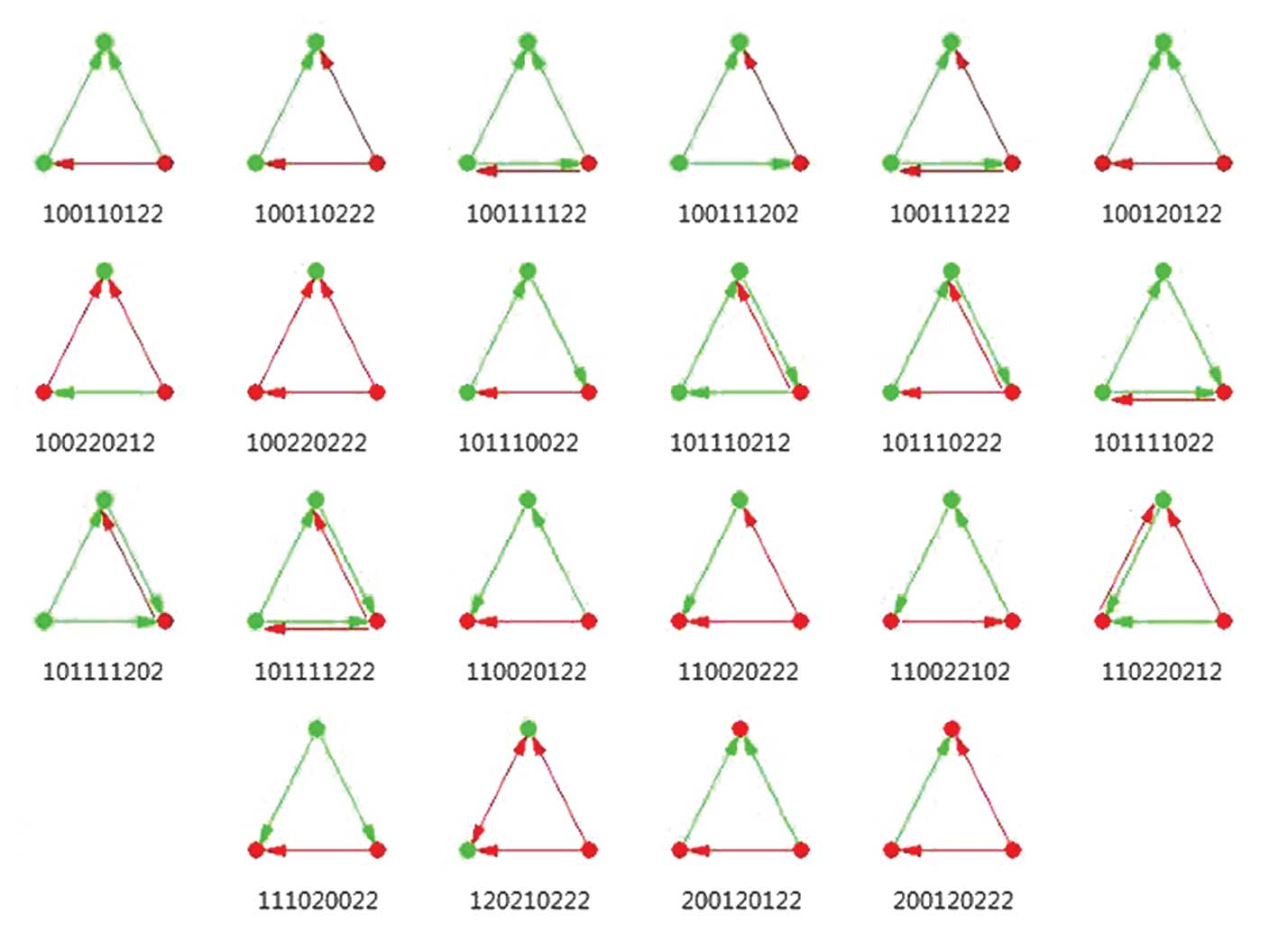
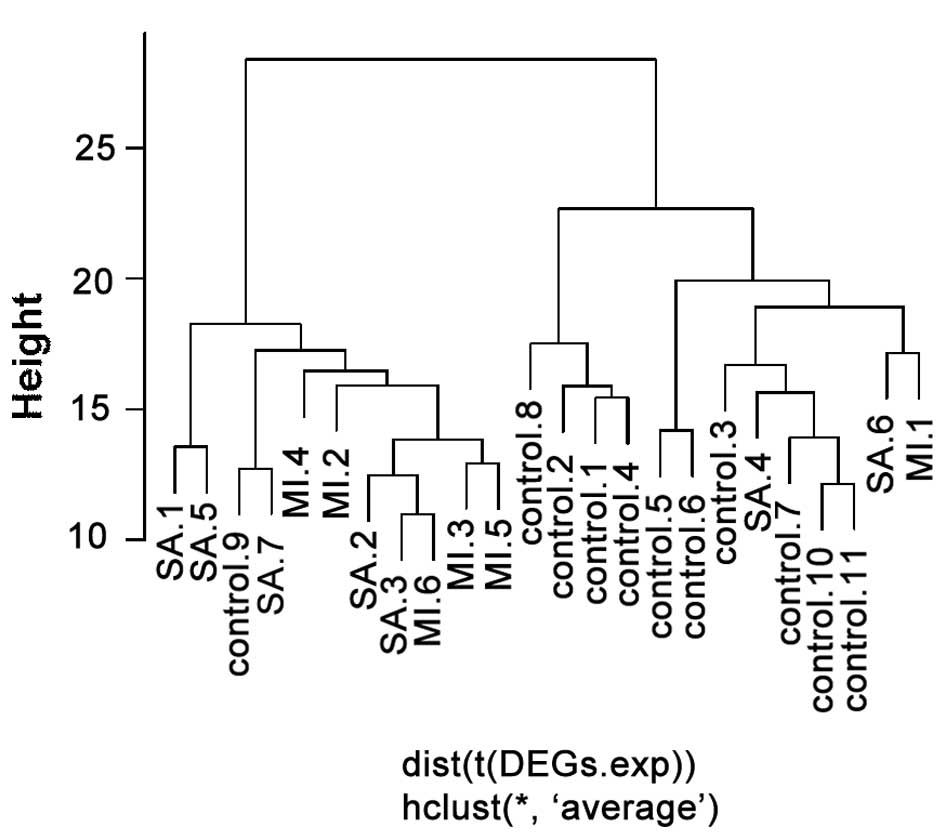
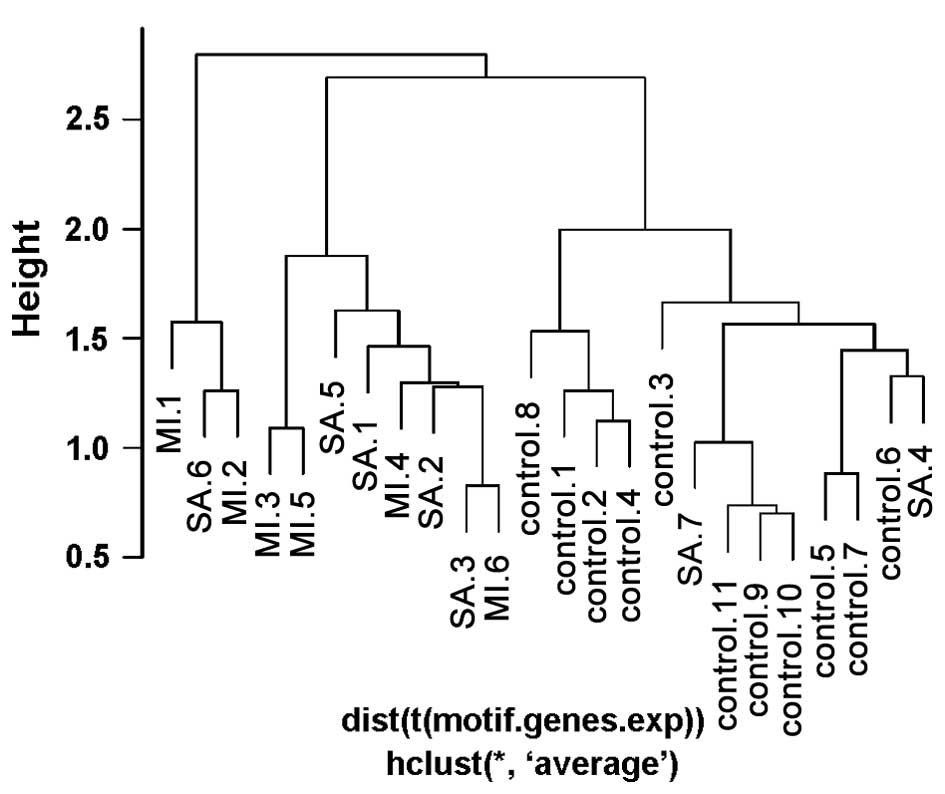
|
1.
|
Gaziano TA, Bitton A, Anand S,
Abrahams-Gessel S and Murphy A: Growing epidemic of coronary heart
disease in low- and middle-income countries. Curr Probl Cardiol.
35:72–115. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Jaarsma C, Leiner T, Bekkers SC, Crijns
HJ, Wildberger JE, Nagel E, Nelemans PJ and Schalla S: Diagnostic
performance of noninvasive myocardial perfusion imaging using
single-photon emission computed tomography, cardiac magnetic
resonance, and positron emission tomography imaging for the
detection of obstructive coronary artery disease: A meta-analysis.
J Am Coll Cardiol. 59:1719–1728. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Beltrame JF, Dreyer R and Tavella R:
Epidemiology of coronary artery disease. Coronary Artery Disease -
Current Concepts in Epidemiology, Pathophysiology, Diagnostics and
Treatment. Gaze D: (Rijeka). InTech. 1–30. 2012.
|
|
4.
|
Semmlow J and Rahalkar K: Acoustic
detection of coronary artery disease. Annu Rev Biomed Eng.
9:449–469. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Chen L, Qu X, Cao M, Zhou Y, Li W, Liang
B, Li W, He W, Feng C, Jia X and He Y: Identification of breast
cancer patients based on human signaling network motifs. Sci Rep.
3:33682013.PubMed/NCBI
|
|
6.
|
Doncic A and Skotheim JM: Feedforward
regulation ensures stability and rapid reversibility of a cellular
state. Mol Cell. 50:856–868. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Albert I and Albert R: Conserved network
motifs allow protein-protein interaction prediction.
Bioinformatics. 20:3346–3352. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Itzkovitz S, Levitt R, Kashtan N, Milo R,
Itzkovitz M and Alon U: Coarse-graining and self-dissimilarity of
complex networks. Phys Rev E Stat Nonlin Soft Matter Phys.
71:0161272005. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Kalir S, McClure J, Pabbaraju K, Southward
C, Ronen M, Leibler S, Surette MG and Alon U: Ordering genes in a
flagella pathway by analysis of expression kinetics from living
bacteria. Science. 292:2080–2083. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Wingrove JA, Daniels SE, Sehnert AJ,
Tingley W, Elashoff MR, Rosenberg S, Buellesfeld L, Grube E, Newby
LK, Ginsburg GS and Kraus WE: Correlation of peripheral-blood gene
expression with the extent of coronary artery stenosis. Circ
Cardiovasc Gene. 1:31–38. 2008. View Article : Google Scholar
|
|
11.
|
Sinnaeve PR, Donahue MP, Grass P, Seo D,
Vonderscher J, Chibout SD, Kraus WE, Sketch M Jr, Nelson C and
Ginsburg GS: Gene expression patterns in peripheral blood correlate
with the extent of coronary artery disease. PLoS One. 4:e70372009.
View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. P Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar
|
|
13.
|
Ferrari F, Bortoluzzi S, Coppe A, Sirota
A, Safran M, Shmoish M, Ferrari S, Lancet D, Danieli GA and
Bicciato S: Novel definition files for human GeneChips based on
GeneAnnot. BMC Bioinformatics. 8:4462007. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Gerstein MB, Kundaje A, Hariharan M, Landt
SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et
al: Architecture of the human regulatory network derived from
ENCODE data. Nature. 489:91–100. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Peri S, Navarro JD, Kristiansen TZ,
Amanchy R, Surendranath V, Muthusamy B, Gandhi TK, Chandrika KN,
Deshpande N, Suresh S, et al: Human protein reference database as a
discovery resource for proteomics. Nucleic Acids Res. 32:D497–D501.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Wernicke S and Rasche F: FANMOD: A tool
for fast network motif detection. Bioinformatics. 22:1152–1153.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Hummel M, Meister R and Mansmann U:
GlobalANCOVA: Exploration and assessment of gene group effects.
Bioinformatics. 24:78–85. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Aoki KF and Kanehisa M: Using the KEGG
database resource. Curr Protoc Bioinformatics Unit.
1:1.12.1–1.12.43. 2005.
|
|
19.
|
Dimmer EC, Huntley RP, Alam-Faruque Y,
Sawford T, O'Donovan C, Martin MJ, Bely B, Browne P, Mun Chan W and
Eberhardt R: The UniProt-GO annotation database in 2011. Nucleic
Acids Res. 40:D565–D570. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:32003.
View Article : Google Scholar
|
|
21.
|
Larsson O, Wahlestedt C and Timmons JA:
Considerations when using the significance analysis of microarrays
(SAM) algorithm. BMC Bioinformatics. 6:1292005. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Bennett MR: Apoptosis of vascular smooth
muscle cells in vascular remodelling and atherosclerotic plaque
rupture. Cardiovasc Res. 41:361–368. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Kockx MM: Apoptosis in the atherosclerotic
plaque quantitative and qualitative aspects. Arterioscler Thromb
Vasc Biol. 18:1519–1522. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Bennett MR, Evan GI and Schwartz SM:
Apoptosis of human vascular smooth muscle cells derived from normal
vessels and coronary atherosclerotic plaques. J Clin Invest.
95:22661995. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Chinnaiyan AM, Tepper CG, Seldin MF,
O'Rourke K, Kischkel FC, Hellbardt S, Krammer PH, Peter ME and
Dixit VM: FADD/MORT1 is a common mediator of CD95 (Fas/APO-1) and
tumor necrosis factor receptor-induced apoptosis. J Biol Chem.
271:4961–4965. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Chinnaiyan AM, O'Rourke K, Tewari M and
Dixit VM: FADD, a novel death domain-containing protein, interacts
with the death domain of Fas and initiates apoptosis. Cell.
81:505–512. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Schneider P, Thome M, Burns K, Bodmer J-L,
Hofmann K, Kataoka T, Holler N and Tschopp J: TRAIL receptors 1
(DR4) and 2 (DR5) signal FADD-dependent apoptosis and activate
NF-kappaB. Immunity. 7:831–836. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Imanishi T, McBride J, Ho Q, O'Brien KD,
Schwartz SM and Han DK: Expression of cellular FLICE-inhibitory
protein in human coronary arteries and in a rat vascular injury
model. Am J Pathol. 156:125–137. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Minami R, Shimada M, Yokosawa H and
Kawahara H: Scythe regulates apoptosis through modulating
ubiquitin-mediated proteolysis of the Xenopus elongation factor
XEF1AO. Biochem J. 405:495–501. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Witztum JL and Steinberg D: Role of
oxidized low density lipoprotein in atherogenesis. J Clin Invest.
88:1785–1792. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
de Nigris F, Youssef T, Ciafré S, Franconi
F, Anania V, Condorelli G, Palinski W and Napoli C: Evidence for
oxidative activation of c-Myc-dependent nuclear signaling in human
coronary smooth muscle cells and in early lesions of Watanabe
heritable hyperlipidemic rabbits: Protective effects of vitamin E.
Circulation. 102:2111–2117. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Goff DC, Bertoni AG, Kramer H, Bonds D,
Blumenthal RS, Tsai MY and Psaty BM: Dyslipidemia prevalence,
treatment, and control in the Multi-Ethnic Study of Atherosclerosis
(MESA): Gender, ethnicity, and coronary artery calcium.
Circulation. 113:647–656. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Sellak H, Choi C, Browner N and Lincoln
TM: Upstream stimulatory factors (USF-1/USF-2) regulate human
cGMP-dependent protein kinase I gene expression in vascular smooth
muscle cells. J Biol Chem. 280:18425–18433. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Naukkarinen J, Gentile M, Soro-Paavonen A,
Saarela J, Koistinen HA, Pajukanta P, Taskinen M-R and Peltonen L:
USF1 and dyslipidemias: Converging evidence for a functional
intronic variant. Hum Mol Genet. 14:2595–2605. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Laurila P-P, Naukkarinen J, Kristiansson
K, Ripatti S, Kauttu T, Silander K, Salomaa V, Perola M, Karhunen
PJ, Barter PJ, et al: Genetic association and interaction analysis
of USF1 and APOA5 on lipid levels and atherosclerosis. Arterioscler
Thromb Vasc Biol. 30:346–352. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Rosenquist TH, Bennett GD, Brauer PR,
Stewart ML, Chaudoin TR and Finnell RH: Microarray analysis of
homocysteine-responsive genes in cardiac neural crest cells in
vitro. Dev Dyn. 236:1044–1054. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
McCully KS: The biomedical significance of
homocysteine. J Sci Explor. 15:5–20. 2001.
|