Introduction
The Candida species causes nosocomial bloodstream
infections under certain circumstances (1). Candida albicans is a type of
Candida species that may act as an opportunistic pathogen in
immunocompromised or immunosuppressed patients (2). The incidence of candidemia is 1.1–24
cases/100,000 individuals, and the associated mortality is >30%,
even when patients are treated with antifungal agents (1,3,4). Therefore, elucidating the molecular
mechanism underlying C. albicans infection and developing
novel clinical approaches is urgently required.
In recent years, the mechanism underlying C.
albicans infection has been clarified. Sun et al
(5) demonstrated that Ssa1
plays a key role in the ability of C. albicans to damage
host cells via binding to host endothelial cell cadherins and
inducing host cell endocytosis in the models of oropharyngeal
candidiasis. Furthermore, a previous study demonstrated that
endothelial cells respond to infection with C. albicans by
synthesizing interleukin (IL)-8 in vitro (6). Müller et al (7) suggested that activation of the p38
mitogen-activated protein kinase (MAPK) cascade is important for
Candida-induced expression of chemokine (C-X-C Motif) ligand
8/IL-8 in endothelial cells. Several studies have demonstrated that
the pathogenicity of C. albicans is induced by hyphae and
epithelial cell damage (8,9). Notably, Moyes et al (10) demonstrated that the MAPK/MKP1/c-Fos
signaling pathway is important for the formation of C.
albicans hyphae in oral epithelial cells. However, the
molecular mechanism underlying the host immune response and
pathogen recognition is complex, and therefore our understanding of
C. albicans infection is not fully complete.
Gene expression microarray analysis is used to
observe changes in gene expression levels in various types of
disease (11,12). Müller et al (7) provided the microarray data of GSE7355
(accession no.), and analyzed the differentially-expressed genes
(DEGs) of human umbilical vein endothelial cells (HUVECs) following
exposure to C. albicans. In addition, they investigated the
nuclear factor (NF)-κB and p38 MAPK signaling pathways in C.
albicans infection. However, the interaction between DEGs was
not analyzed, and a protein-protein interaction (PPI) network was
not constructed.
To fully understand the HUVEC response to C.
albicans, in the present study the microarray profile of HUVECs
infected with C. albicans were analyzed and compared to a
control. The DEGs between the two groups were screened, and a gene
ontology (GO) function package was used to perform GO and pathway
enrichment analysis of the DEGs. The extraction of the correlations
among the DEGs were then carried out using the Kyoto Encyclopedia
of Genes and Genomes (KEGG). Finally, a PPI network was
constructed.
Materials and methods
Analysis of microarray data
The gene expression data GSE7355 (7) was downloaded from the National Center
of Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) using the GPL96
platform of Affymetrix Human Genome U133A Arrays. A total of 8
samples were used in the present study, including 4 samples from
untreated HUVEC monolayers (GSM177134, GSM177140, GSM17177141 and
GSE177142) that served as the control group, and 4 samples from
HUVECs exposed to C. albicans (GSM177136, GSM177137,
GSM17177138 and GSE177139) that served as the experimental group.
Raw data were downloaded for further analysis.
Data preprocessing and identification
of DEGs
The Affy package (http://www.bioconductor.org/packages/release/bioc/html/affy.html)
(13) of Bioconductor (http://bioconductor.org/) was used to calculate the
gene expression levels. Subsequently, a robust multiarray average
algorithm (13) was used to perform
the quartile data normalization. A t-test was carried out using the
Limma package (http://www.bioconductor.org/packages/release/bioc/html/limma.html)
(14) and applied to screen for DEGs
between the two groups. P<0.05 and |log FC|>0.58 were
selected as the criterion for DEGs.
GO and pathway enrichment
analysis
Frequently, GO is used to conduct the functional
enrichment analysis for large-scale genes (15). To identify the functions of the DEGs
between the control and experimental samples, GO enrichment
analysis was performed. In addition, KEGG pathway enrichment
analysis was carried out for the DEGs, and bioinformatics databases
containing all types of biochemistry signaling pathways were
assessed (16). The GOFunction
package (http://www.bioconductor.org/packages/release/bioc/html/GOFunction.html)
of Bioconductor was used to perform the GO and pathway enrichment
analysis. A P<0.05 and gene counts ≥2 were considered as the
cut-off value. Furthermore, the correlation among DEGs was
extracted according to the interactions of the genes in the
KEGG.
Construction of a protein-protein
interaction (PPI) network
The Search Tool for the Retrieval of Interacting
Genes (STRING) database (http://string-db.org/) (17) is an online database that provides
information on the interaction between proteins. In the present
study, the STRING database was used to screen functional
interactions between DEGs. A combined score >4 were regarded as
the threshold. According to the criterion, Cytoscape (http://cytoscapeweb.cytoscape.org/) (18) was then used to display the PPI
network.
Results
Identification of DEGs
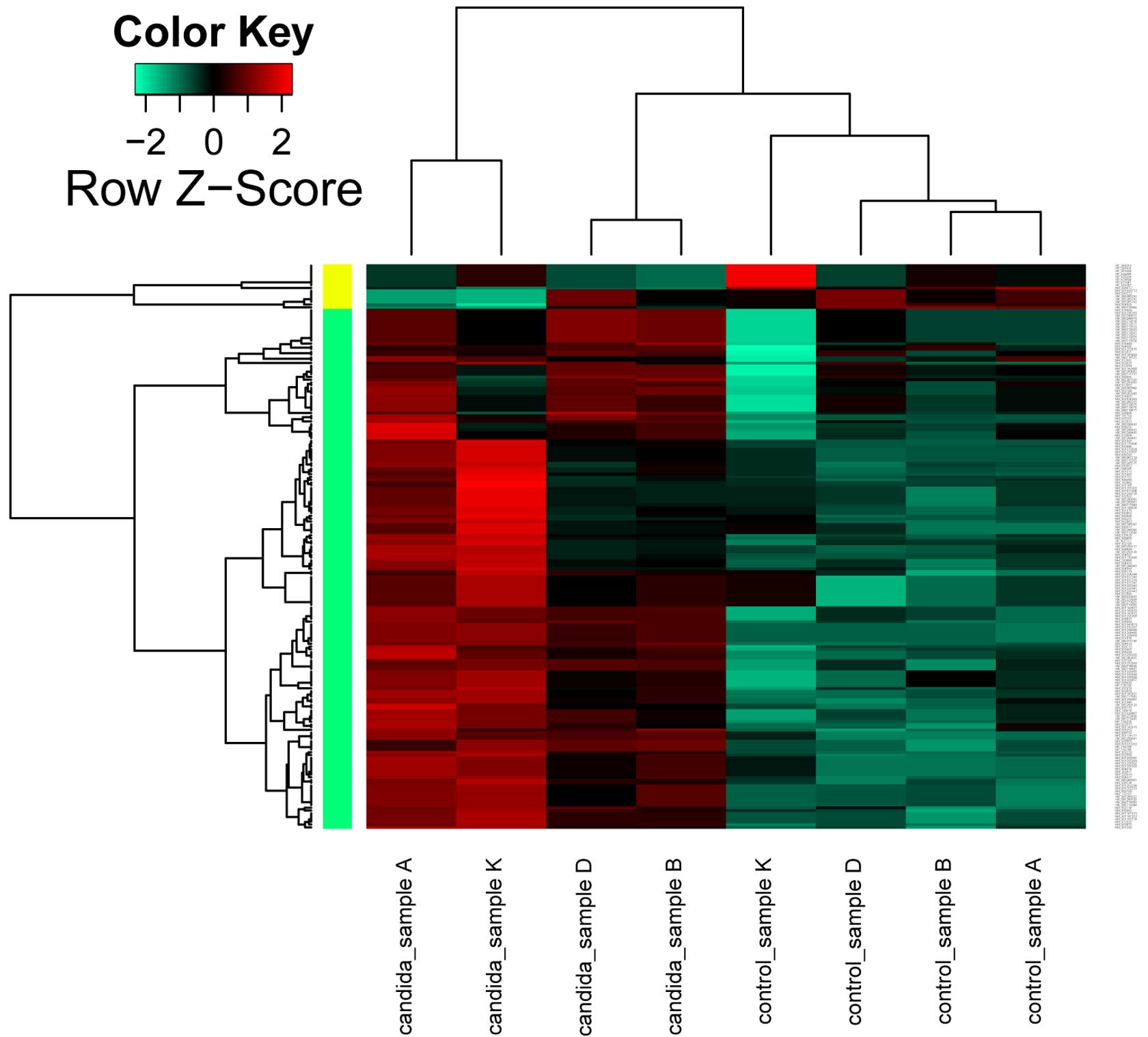
Compared with the untreated HUVEC samples, a total
of 77 DEGs were identified, including 69 upregulated DEGs
corresponding to 187 transcripts, and 8 downregulated DEGs
corresponding to 16 transcripts in the candida-infected HUVEC
samples. The cluster heat map of 77 DEGs is presented in Fig. 1.
GO and pathway enrichment
analysis
The GOfunction package was used to identify GO
functions and signaling pathways for the significant DEGs. Notably,
DEGs from the Candida-infected HUVEC samples were
significantly enriched in response to external biological process
stimuli (P<1.00E-15), chemokine activity (P=5.58E-08) and
cytokine activity of molecular function (P=4.17E-07; Table I). In addition, DEGs were
significantly enriched in the nodulation-like receptor signaling
pathway (P=1.82E-08), toll-like receptor signaling pathway
(P=1.00E-02) and MAPK signaling pathway (P=2.30E-02). The results
of the pathway enrichment analysis are displayed in Table II.
 | Table I.Gene ontology functional enrichment
analysis of DEGs. |
Table I.
Gene ontology functional enrichment
analysis of DEGs.
| Gene ontology | Function | Total genes
(n) | Enriched DEGs
(n) | P-value |
|---|
| BP | Response to
external stimulus | 1,941 | 41 | 1.00E-15 |
|
| Response to
stress | 3,341 | 45 | 7.33E-15 |
|
| Cell migration | 1,015 | 26 | 5.02E-14 |
|
| Response to
stimulus | 7,662 | 63 | 2.77E-13 |
|
| Cell motility | 1,095 | 26 | 2.94E-13 |
| CC | Extracellular
space | 1,212 | 19 | 2.67E-07 |
|
| I-κB/NF-κB
complex | 5 | 3 | 6.61E-07 |
|
| Bcl-3/NF-κB2
complex | 2 | 2 | 1.66E-05 |
|
| Side of
membrane | 300 | 8 | 3.10E-05 |
|
| Cell surface | 645 | 10 | 2.90E-04 |
| MF | Protein
binding | 8,384 | 61 | 5.89E-09 |
|
| Chemokine
activity | 46 | 6 | 5.58E-08 |
|
| Chemokine receptor
binding | 56 | 6 | 1.87E-07 |
|
| Cytokine
activity | 210 | 9 | 4.17E-07 |
|
| Binding | 12,580 | 70 | 2.53E-06 |
 | Table II.KEGG signaling pathway analysis of
DEGs. |
Table II.
KEGG signaling pathway analysis of
DEGs.
| Name | Total genes
(n) | Enriched DEGs
(n) | P-value |
|---|
| NOD-like receptor
signaling pathway | 58 | 8 | 1.82E-08 |
| Rheumatoid
arthritis | 91 | 8 | 6.56E-07 |
| Malaria | 51 | 6 | 3.35E-06 |
| Cytokine-cytokine
receptor interaction | 265 | 11 | 8.15E-06 |
| Osteoclast
differentiation | 128 | 7 | 7.98E-05 |
| African
trypanosomiasis | 35 | 4 | 1.86E-04 |
| Pathways in
cancer | 326 | 10 | 2.87E-04 |
| Chemokine signaling
pathway | 189 | 7 | 0.000877 |
| Chagas disease
(American trypanosomiasis) | 104 | 5 | 0.001629 |
| Epithelial cell
signaling in Helicobacter pylori infection | 68 | 4 | 0.00236 |
| Toll-like receptor
signaling pathway | 102 | 4 | 0.010011 |
| Amoebiasis | 106 | 4 | 0.011421 |
| RIG-I-like receptor
signaling pathway | 71 | 3 | 0.021015 |
| Leishmaniasis | 72 | 3 | 0.021802 |
| MAPK signaling
pathway | 268 | 6 | 0.023041 |
| B cell receptor
signaling pathway | 75 | 3 | 0.02426 |
| Small cell lung
cancer | 85 | 3 | 0.033497 |
| Bladder cancer | 42 | 2 | 0.047575 |
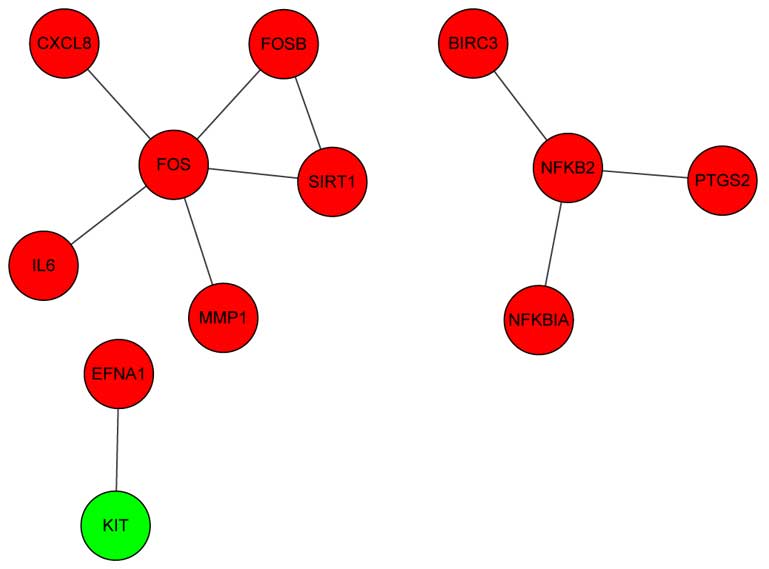
The interaction network between the DEGs extracted
from the KEGG, including 12 nodes and 10 edges, is shown in
Fig. 2. In this network, FBJ murine
osteosarcoma viral oncogene homolog (FOS) and nuclear factor κ
light polypeptide gene enhancer in B cells 2 (NFKB2; p49/p100) had
the highest connectivity degrees.
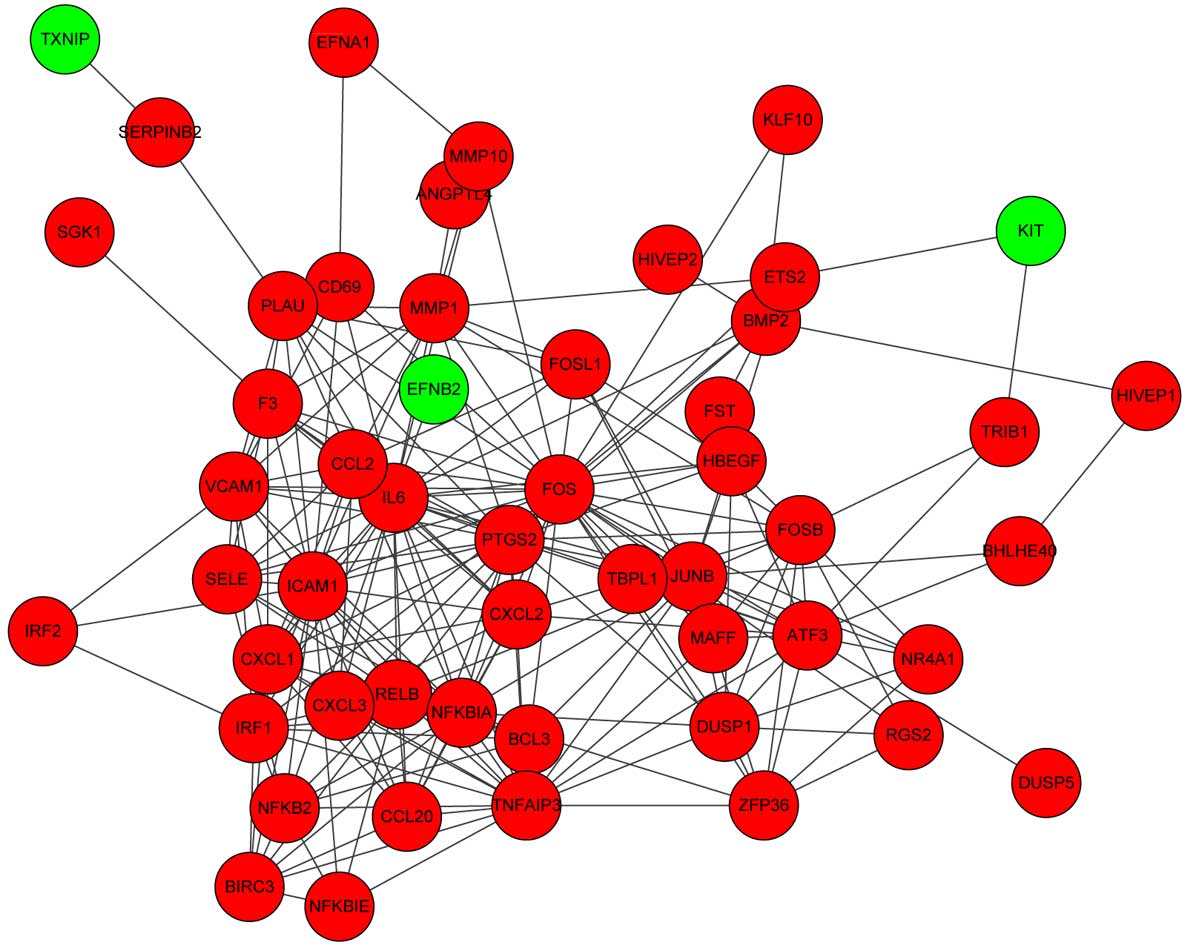
Construction of the PPI network
In the present study, 52 nodes and 226 edges were
used to construct the PPI network (Fig.
3). Notably, several nodes exhibited higher connectivity
degrees: FOS (degree, 30), IL-6 (degree, 26), intercellular
adhesion molecule 1 (degree, 22) and prostaglandin-endoperoxide
synthase 2 (prostaglandin G/H synthase and cyclooxygenase; degree,
21).
Discussion
The present study examined the gene expression data
GSE7355 and investigated the HUVEC reaction patterns to the fungal
pathogen C. albicans. In total, 77 DEGs were identified,
including 69 upregulated DEGs corresponding to 187 transcripts and
8 downregulated DEGs corresponding to 16 transcripts in C.
albicans-infected HUVEC samples. Notably, DEGs such as FOS and
IL-6 were significantly enriched in the toll-like receptor
signaling pathway, whereas NFKB2 was significantly enriched in the
MAPK signaling pathway. In addition, FOS and IL-6 exhibited high
connectivity degrees in the PPI network.
Toll-like receptors are the primary innate
recognition system for microbial invaders in vertebrates, and are
responsible for the immune response to microbial pathogens
(19). Accumulating evidence
suggests that the activation of toll-like receptors is important
for Candida infection (20,21).
Notably, Zakikhany et al (22) suggested that hypha formation was
crucial for the pathogenicity of C. albicans as well as the
proinflammatory responses of mucosal tissues, which protect against
subsequent fungal infection mediated by toll-like receptor 4
signaling (23). In the current
study, FOS and IL-6 were significantly enriched in the toll-like
receptor signaling pathway. FOS activation is mediated by
hypha-associated surface moieties including MKP1 (24). Previously, a study demonstrated that
hypha formation is dependent on the MAPK response, constituted by
the activation of FOS and MKP1 (8).
Moyes et al (10) reported
that the MAPK/MKP1/FOS signaling pathway is important for the
formation of C. albicans hyphae in oral epithelial cells.
Furthermore, the results of the present study demonstrated that FOS
was upregulated, results which were concordant with those of Moyes
et al (25) who demonstrated
that FOS expression levels gradually increased with the progression
of C. albicans infection in vaginal epithelial cells. In
addition, hypha formation dependent on FOS activation and cell
damage can induce the production of cytokines (10). IL-6 has been reported to protect
against Candida infection (26). The present study demonstrated that
IL-6 expression levels were upregulated, results which were
concordant with those of Mostefaoui et al (27). Mostefaoui et al (27) demonstrated that IL-6 mRNA expression
levels were significantly upregulated in human oral mucosa tissue
following infection with C. albicans. These findings
demonstrate that FOS and IL-6 may have important roles in C.
albicans infection via the toll-like receptor signaling
pathway.
Infection of epithelial cells with C.
albicans causes the activation of NF-κB, as well as a MAPK
signaling response, which further induces a pro-inflammatory
response (28). Furthermore, a
previous study suggested that MAPK signaling pathways may served as
targets for antifungal therapy (29). Notably, another investigation
demonstrated that the MAPK signaling pathway enables human
epithelial tissues to regulate innate immune responses against the
hyphae of C. albicans (25).
In the present study, NFKB2 expression levels were demonstrated to
be upregulated and significantly enriched in the MAPK signaling
pathway. NFKB2 is a member of the NF-κB signal transduction pathway
which has important roles in inflammatory and immune responses
(30). Furthermore, fungal infection
may be responsible for the release of chemotactic molecules in
innate immune effector cells (31).
Therefore, these data suggested that NFKB2 may have important roles
in C. albicans infection via the MAPK signaling pathway.
In conclusion, the results of the present study
further elucidated the mechanism underlying the effects of C.
albicans infection in HUVECs. The screened DEGs, including FOS,
IL-6 and NFKB2 may be important genes for the pathogenesis of C.
albicans infection in HUVECs, and these genes may serve as
therapeutic targets to treat patients infected with C.
albicans. However, this study presented some limitations. The
most important limitation was that the study was conducted using
bioinformatics methods, but the results have not been further
demonstrated through experiments. Therefore, further investigation
using animal experiments should be considered.
Acknowledgements
The present study was supported by the Youth Science
and Technology Research Foundation of Shanxi (grant no.
2012021035-3) and the Natural Science Foundation of Shanxi (grant
no. 2012011045-3).
References
|
1
|
Wisplinghoff H, Bischoff T, Tallent SM,
Seifert H, Wenzel RP and Edmond MB: Nosocomial bloodstream
infections in US hospitals: Analysis of 24, 179 cases from a
prospective nationwide surveillance study. Clin Infect Dis.
39:309–317. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Alby K, Schaefer D and Bennett RJ:
Homothallic and heterothallic mating in the opportunistic pathogen
Candida albicans. Nature. 460:890–893. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gudlaugsson O, Gillespie S, Lee K, Vande
Berg J, Hu J, Messer S, Herwaldt L, Pfaller M and Diekema D:
Attributable mortality of nosocomial candidemia, revisited. Clin
Infect Dis. 37:1172–1177. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stevens DA: Combination immunotherapy and
antifungal chemotherapy. Clin Infect Dis. 26:1266–1269. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sun JN, Solis NV, Phan QT, Bajwa JS,
Kashleva H, Thompson A, Liu Y, Dongari-Bagtzoglou A, Edgerton M and
Filler SG: Host cell invasion and virulence mediated by Candida
albicans Ssa1. PLoS Pathog. 6:e10011812010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Orozco AS, Zhou X and Filler SG:
Mechanisms of the proinflammatory response of endothelial cells to
Candida albicans infection. Infect Immun. 68:1134–1141.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Müller V, Viemann D, Schmidt M, Endres N,
Ludwig S, Leverkus M, Roth J and Goebeler M: Candida
albicans triggers activation of distinct signaling pathways to
establish a proinflammatory gene expression program in primary
human endothelial cells. J Immunol. 179:8435–8445. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Moyes DL, Runglall M, Murciano C, Shen C,
Nayar D, Thavaraj S, Kohli A, Islam A, Mora-Montes H, Challacombe
SJ and Naglik JR: A biphasic innate immune MAPK response
discriminates between the yeast and hyphal forms of Candida
albicans in epithelial cells. Cell Host Microbe. 8:225–235.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jayatilake J, Samaranayake L, Lu Q and Jin
L: IL-1alpha, IL-1ra and IL-8 are differentially induced by
Candida in experimental oral candidiasis. Oral Dis.
13:426–433. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Moyes DL, Murciano C, Runglall M, Kohli A,
Islam A and Naglik JR: Activation of MAPK/c-Fos induced responses
in oral epithelial cells is specific to Candida albicans and
Candida dubliniensis hyphae. Med Microbiol Immunol.
201:93–101. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sørlie T, Perou CM, Tibshirani R, Aas T,
Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen
T, et al: Gene expression patterns of breast carcinomas distinguish
tumor subclasses with clinical implications. Proc Natl Acad Sci
USA. 98:10869–10874. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Beer DG, Kardia SL, Huang CC, Giordano TJ,
Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al:
Gene-expression profiles predict survival of patients with lung
adenocarcinoma. Nat Med. 8:816–824. 2002.PubMed/NCBI
|
|
13
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. Nat
Genet. 25:25–29. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41D808–D815. (Database Issue)2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Takeda K, Kaisho T and Akira S: Toll-like
receptors. Ann Rev Immunol. 21:335–376. 2003. View Article : Google Scholar
|
|
20
|
Gantner BN, Simmons RM, Canavera SJ, Akira
S and Underhill DM: Collaborative induction of inflammatory
responses by dectin-1 and Toll-like receptor 2. J Exp Med.
197:1107–1117. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Netea MG, Van der Graaf CA, Vonk AG,
Verschueren I, Van der Meer JW and Kullberg BJ: The role of
toll-like receptor (TLR) 2 and TLR4 in the host defense against
disseminated candidiasis. J Infect Dis. 185:1483–1489. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zakikhany K, Naglik JR, Schmidt-Westhausen
A, Holland G, Schaller M and Hube B: In vivo transcript profiling
of Candida albicans identifies a gene essential for
interepithelial dissemination. Cell Microbiol. 9:2938–2954. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Weindl G, Naglik JR, Kaesler S, Biedermann
T, Hube B, Korting HC and Schaller M: Human epithelial cells
establish direct antifungal defense through TLR4-mediated
signaling. J Clin Invest. 117:3664–3672. 2007.PubMed/NCBI
|
|
24
|
Gow NA and Hube B: Importance of the
Candida albicans cell wall during commensalism and
infection. Curr Opin Microbiol. 15:406–412. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Moyes DL, Murciano C, Runglall M, Islam A,
Thavaraj S and Naglik JR: Candida albicans yeast and hyphae
are discriminated by MAPK signaling in vaginal epithelial cells.
PLoS One. 6:e265802011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dongari-Bagtzoglou A and Fidel P Jr: The
host cytokine responses and protective immunity in oropharyngeal
candidiasis. J Dent Res. 84:966–977. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mostefaoui Y, Bart C, Frenette M and
Rouabhia M: Candida albicans and Streptococcus
salivarius modulate IL-6, IL-8 and TNF-alpha expression and
secretion by engineered human oral mucosa cells. Cell Microbiol.
6:1085–1096. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kempe S, Kestler H, Lasar A and Wirth T:
NF-κB controls the global pro-inflammatory response in endothelial
cells: evidence for the regulation of a pro-atherogenic program.
Nucleic Acids Res. 33:5308–5319. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
de Herrero Dios C, Román E, Monge RA and
Pla J: The role of MAPK signal transduction pathways in the
response to oxidative stress in the fungal pathogen Candida
albicans: implications in virulence. Curr Protein Pept Sci.
11:693–703. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vallabhapurapu S and Karin M: Regulation
and function of NF-κB transcription factors in the immune system.
Annu Rev Immunol. 27:693–733. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yumoto H, Nakae H, Fujinaka K, Ebisu S and
Matsuo T: Interleukin-6 (IL-6) and IL-8 are induced in human oral
epithelial cells in response to exposure to periodontopathic
Eikenella corrodens. Infect Immun. 67:384–394.
1999.PubMed/NCBI
|