Introduction
Pseudomonas aeruginosa is not only a common
conditional pathogen but also one of major pathogenic bacteria that
may cause outbreak of nosocomial infections. However, increasing
imipenem-resistant Pseudomonas aeruginosa (IRPA) results in
difficulties (1) in clinical
treatment. Previous studies showed that metal MBL β-lactamase (MBL)
could split the β-lactam antibiotic β-lactamase ring to make
antibiotics ineffective on bacteria (2). It is one of the important drug
resistance mechanisms for the bacteria. MBL-mediated IRPA was
identified and confirmed and includes seven types - IMP, VIM, GIM,
SPM, SIM, AIM and NDM-1, of which IMP and VIM have the closest
clinical relationships (3).
The micropore proteins OprC, OprD and OprE are major
parts of the outer membrane protein (OMP) of Pseudomonas
aeruginosa. They are also specialized pathways for small
molecule hydrophilic substances to enter into the bacteria. OprD2
protein is a specific pathway for imipenem entering into the
Pseudomonas aeruginosa. It has ligand specificity with
loci-specific binding of the imipenem. It has no cross resistance
(4) with other β-lactam antibiotics.
Previous findings have shown a decrease in OMP OprD2 expression and
deficiency is a main cause for Pseudomonas aeruginosa having
resistance to carbapenems, leading to a decrease in OMP
permeability, resulting in Pseudomonas aeruginosa having
resistance to carbapenems in the end (5).
The present study aimed to investigate the mechanism
of IRPA from the angle of MBL generation and OMP OprD2 deficiency.
It has great relevance to clinically reasonable and effective use
of antibiotics.
Materials and methods
Bacterial isolation and
identification
A total of 200 specimens derived from each clinical
department of The First Affiliated Hospital of Guangxi Medical
University (Guangxi, China) from January, 2015 to January, 2016
were selected and the Pseudomonas aeruginosa standard strain
ATCC27853 was taken as a quality control strain while the IRPA
acted as the control. They were validated by the VITEK-2 full-auto
bacteria identification/drug susceptibility analysis system
(Biomerieux, Marcy l'Étoile, France), as well as the drug
sensitivity test. Based on 2012 standard constituted by American
Clinical and Laboratory Standard Association as references, the
minimum inhibitory concentration (MIC) values of IRPA were
collected after being tested by agar plate double dilution
method.
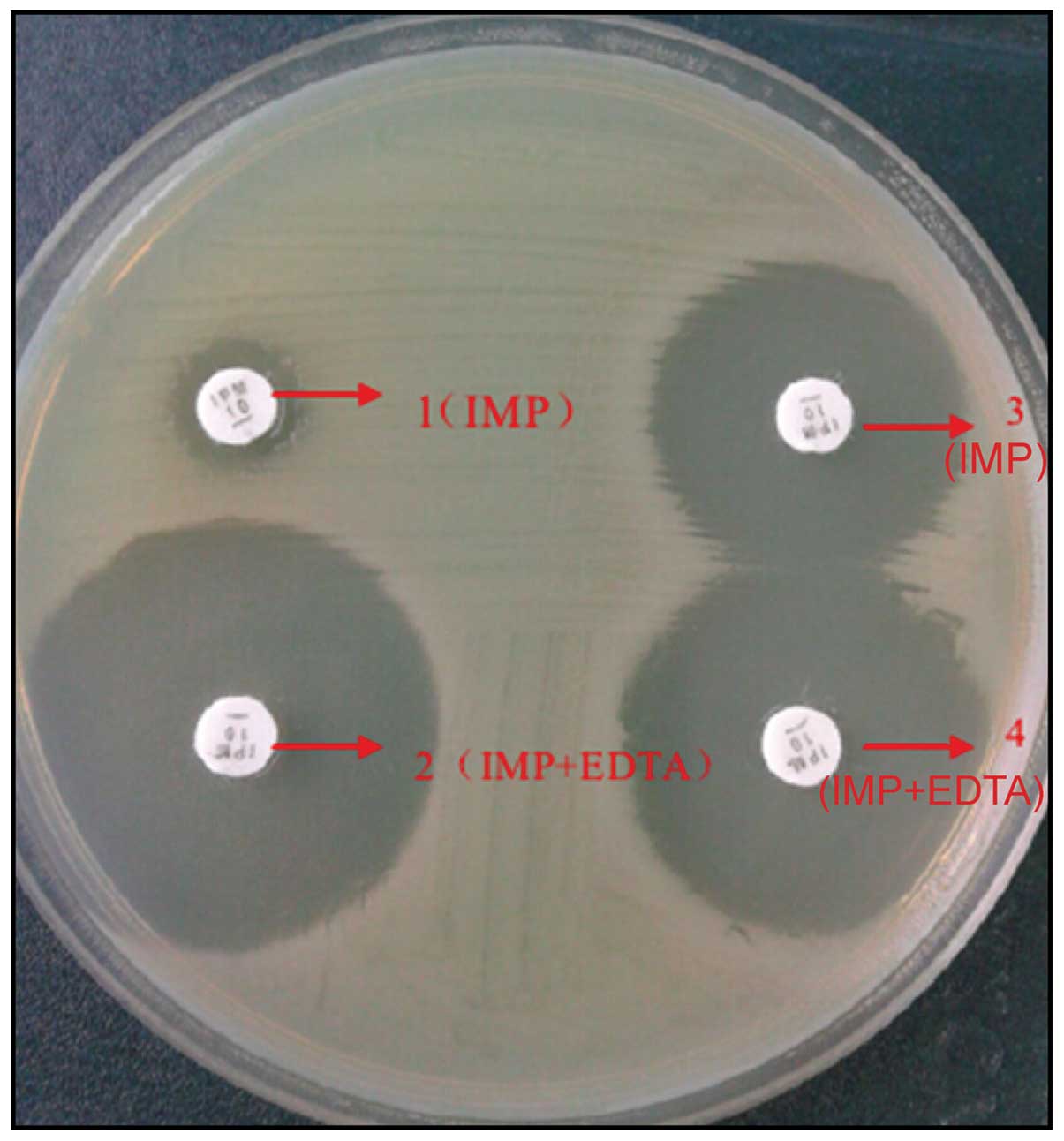
MBL phenotype screened by Kirby-Bauer
diffusion method
Experimental strains were produced to 0.5 M
bacterial suspension by using sterilized saline water and were
evenly coated on a Müller-Hinton (M-H) plate in length of 9 cm. Two
10 µg IMP drug sensitivity test papers (Merck Millipore, Billerica,
MA, USA) were pasted of which one with 10 µl EDTA in concentration
of 500 mmol/l. The M-H plate with pasted drug sensitivity test
papers was placed into a constant temperature incubation under 35°C
for 18–24 h. Subsequently, the diameter of inhibition zone of IMP
drug sensitivity test papers with EDTA and only with IMP was
tested. The diameter of inhibition zone of IMP papers with EDTA was
≥5 mm than only with IMP paper, which was the MBL phenotype
-positive strain.
Polymerase chain reaction (PCR)
testing metalloenzyme genes IMP-1, VIM-1, SPM and OprD2
PCR Premix Taq regent and primer synthesis (Takara
Bio, Dalian, China), electrophoresis and DNA marker (Beijing
TransGen Biotech Co., Ltd., Beijing, China), and PCR (Bio-Rad,
Berkeley, CA, USA) were used in this study. To extract total DNA, a
single colony was taken from blood agar and placed in 500 µl of
tri-distilled water at 100°C for 10 min, in a refrigerated air
dryer under 4°C at 8,000 × g for 10 min. The bacterial DNA in
supernatant (100 µl) was drawn and removed into another sterile
centrifuge tube and stored at −20°C for standby.
The reaction mixture consists of 2X Taq MasterMix
(25 µl), upstream and downstream primers (10 µM) for each 2 µl,
template DNA (4 µl), ddH2O (17 µl) to make up final
volume of 50 µl. The PCR conditions for IMP-1, VIM-1 and SPMs were
pre-denaturation at 94°C for 4 min, denaturation at 94°C for 30
sec, annealing at 56°C for 30 sec and extension at 72°C for 1 min
for 35 cycles and final extension at 72°C for 10 min. The PCR
conditions for OprD2 PC were pre-denaturation at 94°C for 5 min,
denaturation at 94°C for 30 sec, annealing at 55°C for 30 sec and
extension at 72°C for 1 min for total of 35 cycles and re-extension
at 72°C for 10 min. PCR products were separated by gel
electrophoresis (AGE), 5 µl of PCR products was electrophoresed
with a voltage of 110 V for 40 min. The gel image was captured with
UV light, and the sequences were logged in GenBank for comparison
and analysis. The primers used are listed in Table I.
 | Table I.Primer sequences used in this
study. |
Table I.
Primer sequences used in this
study.
| Gene | Primer sequence | Band length, bp |
|---|
| IMP-1 | F:
5′-CTACCGCAGCAGAGTCTTTG-3′ | 587 |
|
| R:
5′-AACCAGTTTTGCCTTACCAT-3′ |
|
| VIM-1 | F:
5′-AGTGGTGAGTATCCGACAG-3′ | 260 |
|
| R:
5′-ATGAAAGTGCGTGGAGAC-3′ |
|
| SPM | F:
5′-GCGTTTTGTTTGTTGCTC-3′ | 786 |
|
| R: 5′-
TTGGGGATGTGAGACTAC-3′ |
|
| OprD2 | F:
5′-GCGCATCTCCAAGACCATG-3′ | 193 |
|
| R:
5′-GCCACGCGATTTGACGGAG-3′ |
|
Western blotting for OprD2 protein
expression
Total protein was extracted based on the
specifications of bacterial membrane protein extraction kits and
the protein concentration was tested using the BCA protein titer
kits. It was prepared to 5.0 mg/ml and stored at −80°C. OMP was
separated by SDS-PAGE electrophoresis method (Beijing Liuyi
Instrument Factory, Beijing, China) for staining and decolorizing
using Coomassie Brilliant Blue fast staining liquid after
electrophoresis. The results were analyzed by gel-imaging system
(Syngene, Frederick, MD, USA).
OprD2 position confirmed by sodium
salicylate inhibition test
The imipenem-sensitive Pseudomonas aeruginosa
was respectively inoculated in 2 M-H plates, 1 with 32 mmol/l of
sodium salicylate and another without sodium salicylate. The
imipenem drug susceptibility discs were, respectively, pasted in
two M-H plates, to incubate at 37°C overnight to observe the size
of IMP inhibition zone. The OMP was extracted from IMP-sensitive
strains in the MH plate with or without sodium salicylate for
SDS-PAGE for electrophoresis. The OMP D2 location was determined
based on features of OMP D2 being suppressed by salicylic acid,
namely the OMP D2 grown in the M-H plate with sodium salicylate was
suppressed by salicylic acid and not expressed. During
electrophoresis, the protein bands were missing around 45 kDa. The
OMP D2 grown in the M-H plate without sodium salicylate was not
suppressed and the protein bands turned up around 45 kDa during
electrophoresis.
Statistical analysis
Data analysis was carried out using the SPSS 19.0
software (SPSS, Inc., Chicago, IL, USA). Quantitative data were
expressed as mean ± standard deviation. The comparison among groups
was carried out using ANOVA. The quantitative data were expressed
by case number or percentage (%). Comparison among groups was
performed by χ2 test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Comparisons of MIC value
A total of 80 portions (40%) of MB-positive strains
were screened from 200 specimens. MIC value of IRPA was
significantly higher than quality control bacteria and control
bacteria. The difference was of statistical significance
[(265.4±43.2) vs. (0.9±0.2), (1.2±0.3) µg/ml, F=152.342,
P<0.001], as shown in Fig. 1.
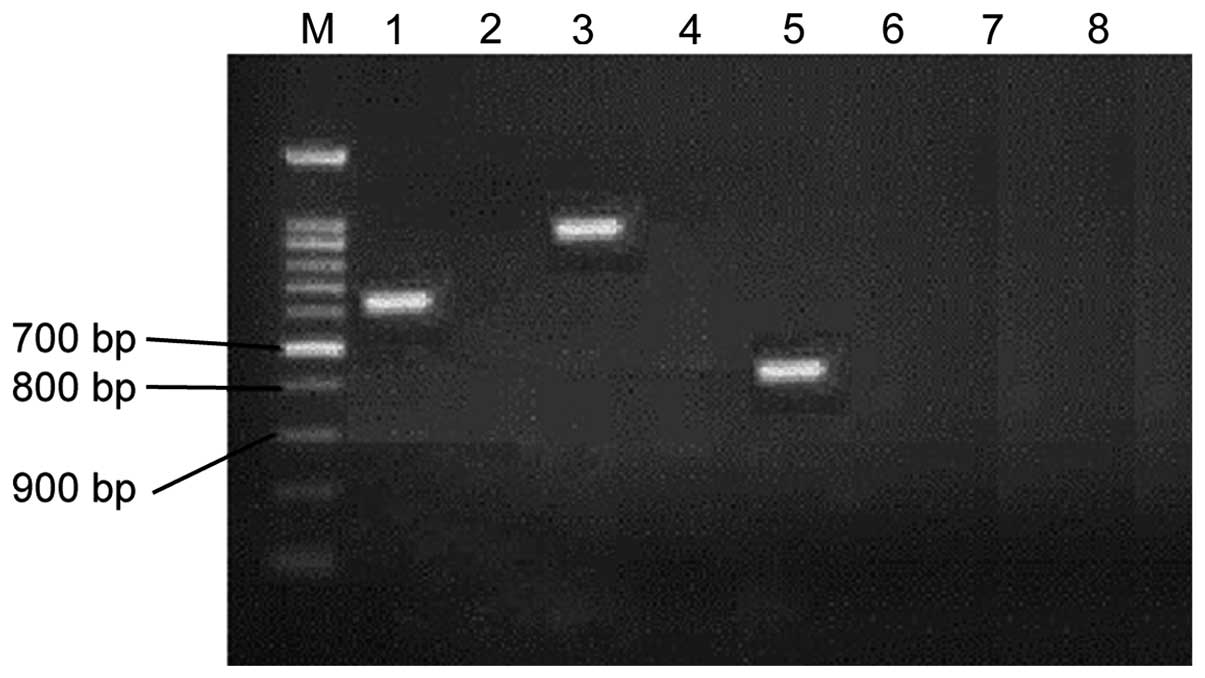
MBL gene test
A total of 35 cases with IMP-1 positivity were
screened out from 80 portions of MBL positive strains, 20 with
VIM-1 positive, 16 with SPM positive, 5 with 2 positive genes and 4
with 3 positive genes, as shown in Fig.
2.
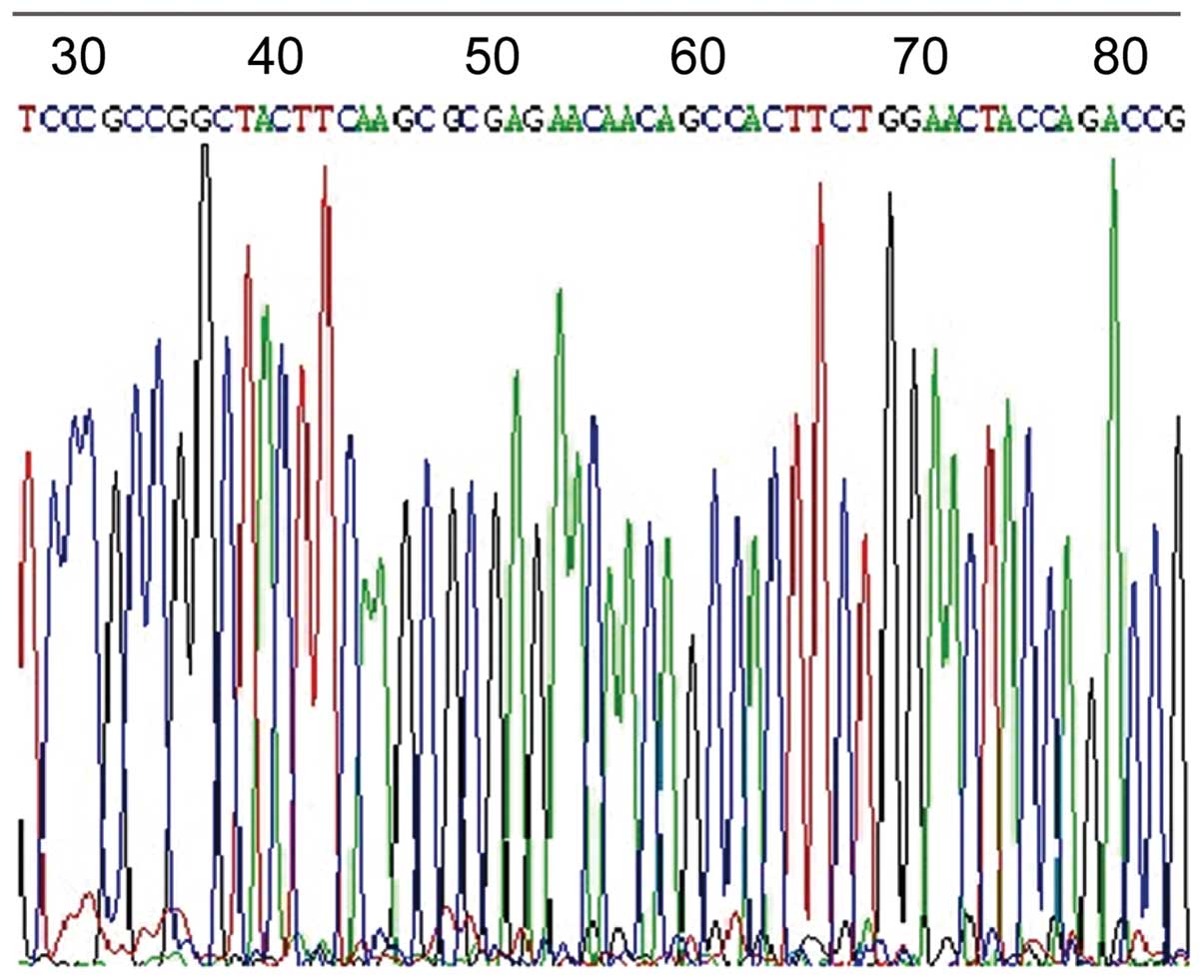
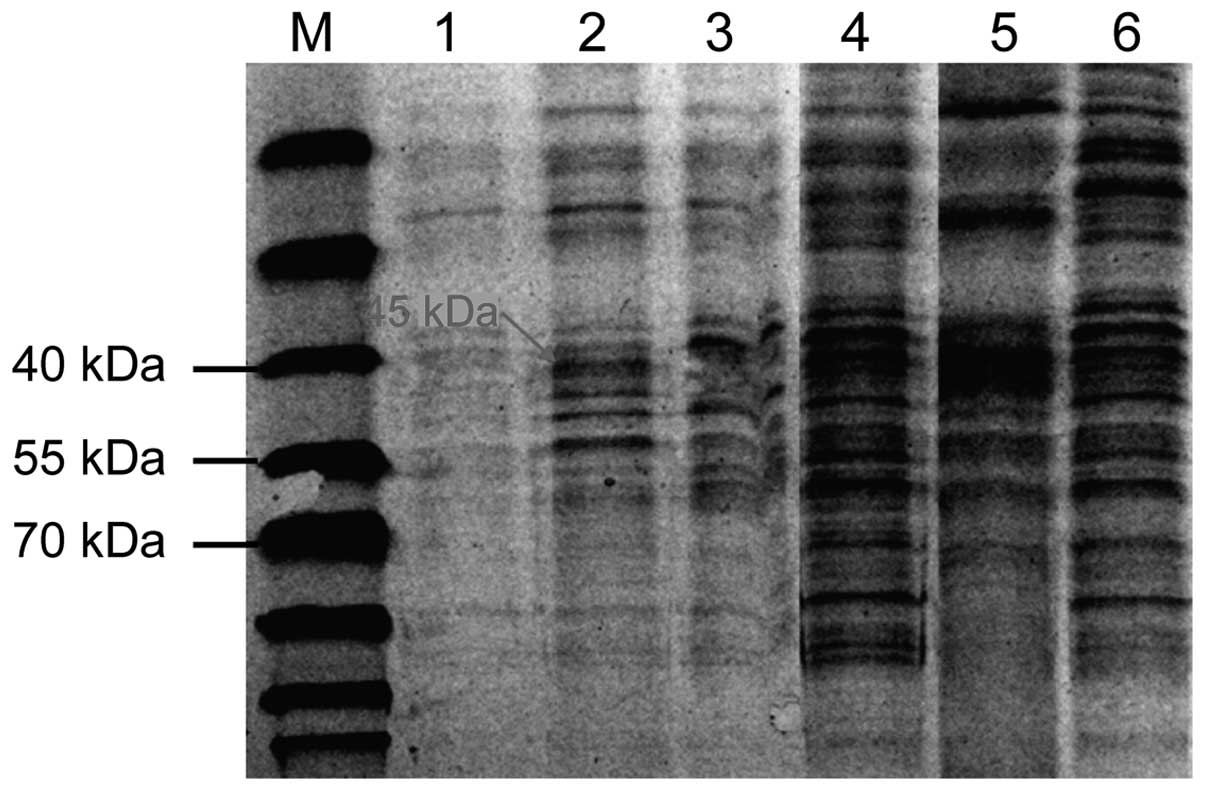
OprD2 gene and protein expression
A total of 150 portions (75%) of OprD2 deficiencies
were screened out of 200 specimens. Gene sequencing is shown in
Fig. 3. The OprD2 protein bands
appeared at 45 kDa in the standard strains and sensitive strains,
and no OprD2 protein bands appeared in OprD2 deficiency strains.
The results were in accordance with gene detection, as shown in
Fig. 4.
Discussion
Imipenem is stable to a lot of β-lactamase or super
spectrum β-lactamase and it has strong affinity in penicillin
binding proteins. Additionally, it has no cross resistance with the
third generation of cephalosporin, with powerful bacterial
activities. Therefore, it has become a common antibiotic in
treating gram-negative bacilli, especially Pseudomonas
aeruginosa infection (6). It has
been widely used clinically, but in the presence of other
antibiotics the detection rate of drug resistance also gradually
increased (7). This study has
concluded that MBL positive rate was 40%. The MIC value of IRPA was
significantly higher than quality control bacteria and control
bacteria. In total, 35 cases were screened out with IMP-1 positive,
20 with VIM-1 positive, 16 with SPM positive, 5 with 2 positive
genes and 4 with 3 positive genes.
The structural gene OprD of coding OprD2 protein was
located in the chromosome of bacteria. The changes of encoding gene
sequences may cause OprD2 protein expression reduction or even
missing and result in the decrease of OprE protein expression
volume. When OprD2 was missing or its expression was decreased, it
lead to permeability changes of bacterial outer membrane and then
blocking the carbapenems entering into the bacteria, showing
carbapenems resistance clinically (8). Additionally, OprD2 protein is able to
form a specific locus by binding bacteria and carbapenems. It is
one and only porin that allows antibiotics go through smoothly in
current studies, especially for Pseudomonas aeruginosa
(9). Therefore, loss or reduction in
OMP OprD2 expression is an important mechanism leading to
Pseudomonas aeruginosa resistance to carbapenem antibiotics.
The present study suggests that if the rate of missing OprD2
proteins is 75%, the rate of missing MBL positive strains could
reach up to 90%. The results of protein electrophoresis and gene
sequencing were the same. Due to different regions and antibiotic
habits, the detection rate of missing OprD2 protein can vary
(10). However, there was still
OprD2 protein expression positivity reported in MBL-positive
strains, which indicated that missing OMP OprD2 was a major
mechanism that causes IRPA, but it was not the single one. Some
studies reported that pure OprD2 gene missing only has low level of
IMP resistance while OprD2 missing can produce significant
resistance to carbapenems antibiotics only if it acts together with
other factors, such as producing chromosome-mediated OprD2 enzyme
(11).
Some studies have suggested that the active efflux
pump may play an important role in resistant Pseudomonas
aeruginosa (12). It can
positively expel antibiotics out of the bacteria. If the expression
quantity of efflux pump increases, the antibiotic output also goes
up resulting in bacteria resistant to this antibiotic. The
MexAB-OprM efflux system is the most common and most important
active efflux pump in Pseudomonas aeruginosa. Its regulatory
gene MexAB-OprM operon may be affected by upstream repressor gene
MexR coded MexR protein. Its substrate is relatively extensive
mainly including β-lactamase inhibitor, β-lactam, tetracyclines,
chloramphenicol, quinolones, novobiocin, macrolides, sulfonamides
and carbapenems. It usually causes multidrug resistance (13).
In conclusion, missing OMP OprD2 and MBL phenotype
positivity may be the important mechanisms for IRPA.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (no. 81260663) and the Health
Department Fund Project of Guangxi Province (no. z2013045).
References
|
1
|
Strateva T and Yordanov D: Pseudomonas
aeruginosa - a phenomenon of bacterial resistance. J Med Microbiol.
58:1133–1148. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Aktaş Z, Satana D, Kayacan C, Can B,
Gönüllü N and Küçükbasmacı O: Antibiotic susceptibility rates and
beta-lactam resistance mechanisms of Pseudomonas aeruginosa
strains. Mikrobiyol Bul. 46:386–397. 2012.(In Turkish). PubMed/NCBI
|
|
3
|
Bahar MA, Jamali S and Samadikuchaksaraei
A: Imipenem-resistant Pseudomonas aeruginosa strains carry
metallo-beta-lactamase gene bla(VIM) in a level I Iranian burn
hospital. Burns. 36:826–830. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Riera E, Cabot G, Mulet X, García-Castillo
M, del Campo R, Juan C, Cantón R and Oliver A: Pseudomonas
aeruginosa carbapenem resistance mechanisms in Spain: Impact on the
activity of imipenem, meropenem and doripenem. J Antimicrob
Chemother. 66:2022–2027. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Epp SF, Köhler T, Plésiat P,
Michéa-Hamzehpour M, Frey J and Pechère JC: C-terminal region of
Pseudomonas aeruginosa outer membrane porin OprD modulates
susceptibility to meropenem. Antimicrob Agents Chemother.
45:1780–1787. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Clissold SP, Todd PA and Campoli-Richards
DM: Imipenem/cilastatin. A review of its antibacterial activity,
pharmacokinetic properties and therapeutic efficacy. Drugs.
33:183–241. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Davodian E, Sadeghifard N, Ghasemian A and
Noorbakhsh S: Presence of blaPER-1 and blaVEB-1 beta-lactamase
genes among isolates of Pseudomonas aeruginosa from South West of
Iran. J Epidemiol Glob Health. Mar 1–2016.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li H, Luo YF, Williams BJ, Blackwell TS
and Xie CM: Structure and function of OprD protein in Pseudomonas
aeruginosa: From antibiotic resistance to novel therapies. Int J
Med Microbiol. 302:63–68. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shen J, Pan Y and Fang Y: Role of the
outer membrane protein OprD2 in carbapenem-resistance mechanisms of
Pseudomonas aeruginosa. PLoS One. 10:e01399952015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shen JL and Fang YP: Detection of
drug-resistance mechanism of Pseudomonas aeruginosa developing from
a sensitive strain to a persister during carbapenem treatment.
Genet Mol Res. 14:6723–6732. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
El-Mahdy TS: Expression of ampC, oprD, and
mexA, outer membrane protein analysis and carbapenemases in
multidrug resistant clinical isolates of Pseudomonas aeruginosa
from Egypt. J Chemother. 26:379–381. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chuanchuen R, Wannaprasat W and Schweizer
HP: Functional characterization of MexXY and OpmG in aminoglycoside
efflux in Pseudomonas aeruginosa. Southeast Asian J Trop Med Public
Health. 39:115–122. 2008.PubMed/NCBI
|
|
13
|
Choudhury D, Ghosh A, Chanda D Dhar, Das
Talukdar A, Choudhury M Dutta, Paul D, Maurya AP, Chakravorty A and
Bhattacharjee A: Premature termination of MexR leads to
overexpression of MexAB-OprM efflux pump in Pseudomonas aeruginosa
in a tertiary referral hospital in India. PLoS One.
11:e01491562016. View Article : Google Scholar : PubMed/NCBI
|