Introduction
Phosphatases of the regenerating liver [PRLs; also
known as the protein tyrosine phosphatase type IVA (PTP4A) family]
were originally identified as immediate-early genes in the
regenerating liver (1). The PRL
family is a group of protein tyrosine phosphatases (PTPs) and plays
a role in the development and metastasis of various cancers,
including colorectal, prostate, breast, gastric and liver cancers,
and particularly in metastatic cancers (2,3). The PRL
family comprises three genes: PRL-1, PRL-2 and PRL-3. The
overexpression of the PRL family has been frequently reported in
various cancers, especially in metastatic cancers (4–8).
Overexpression of PRLs in normal cells has been found to promote
proliferation, migration, and invasion (4,8,9) whereas the reduction of PRLs in cancer
cells using small interfering RNA (siRNA) has been shown to inhibit
cell motility and metastatic characteristics in a mouse model
(10).
PRLs affect a number of signaling pathways
associated with cell growth and cancer development. During
tumorigenesis, PRLs have been found to modulate integrin
β1-extracellular signal-regulated kinase 1/2, phosphoinositide
3-kinase/AKT, keratin 8, C-terminal Src kinase, Rho GTPase,
cyclin-dependent kinase 2, p53 and FK506-binding protein 8 (FKBP8)
signaling pathways (9,11–19).
Although it is important to elucidate the role of
PRLs in cancer progression and the signaling pathways they affect,
a major challenge to the analysis of the detailed signaling
mechanism of PRLs is the lack of a physiologically relevant
substrate and knowledge of its regulation by physical interaction.
Several PRL-interacting proteins such as activating transcription
factor-7, β-subunit of geranylgeranyl transferase-II, cadherin 22,
ezrin, elongation factor 2, keratin 8, integrin-α1, PRL-1 (trimer),
PRL-3 (oligomer) and FKBP8 have been reported (1,11,16,20–27).
PRL family members have been identified to be useful
biomarkers and therapeutic targets in cancer as well as in
metastatic cancer due to the aforementioned properties (1,3,27). However, little is known about the
proteins that bind to PRL and regulate PRL function or are
regulated by PRL. Therefore, in the present study, to screen for
novel PRL-interacting proteins, yeast two-hybrid methodology was
applied using PRL-1 and PRL-3 as bait. The identification of
PRL-binding proteins may be useful in providing a novel insight
into the mechanisms of tumorigenesis and other diseases, and might
eventually lead to the development of more effective therapies.
Materials and methods
Cell culture, plasmid and
reagents
HEK293T, HeLa and U2OS cell lines (American Type
Culture Collection, Manassas, VA, USA) were cultured under an
atmosphere of 5% CO2 at 37°C in Dulbecco's modified
Eagle's medium (Hyclone; GE Healthcare Life Sciences, Logan, UT,
USA) supplemented with 10% fetal bovine serum (Hyclone; GE
Healthcare Life Sciences), 100 IU/ml penicillin, and 100 µg/ml
streptomycin (Gibco-BRL; Thermo Fisher Scientific, Inc., Waltham,
MA, USA). cOmplete™ Mini Protease Inhibitor Cocktail tablets and
Phosphatase Inhibitor Cocktail tablets were obtained from Roche
Applied Science (Penzberg, Germany). Antibody against high
availability (HA) probe was purchased from Santa Cruz
Biotechnology, Inc. (Dallas, TX, USA; cat. no. SC-805) and
antibodies against Flag® M2 (cat. no. F3165) and β-actin
(cat. no. A5441) were purchased from Sigma-Aldrich (Merck
Millipore, Darmstadt, Germany).
Flag-PRL-1 and Flag-PRL-3 (12,19,28) were
digested with restriction enzymes (EcoRI/XhoI) and
cloned into the yeast expression vector pLexA (Clontech
Laboratories, Inc., Mountainview, CA, USA) to form pLexA-PRL-1 and
pLexA-PRL-3, respectively. The authenticity and correct orientation
of the cloned sequence were then confirmed by restriction digestion
and polymerase chain reaction (PCR).
Two cDNA clones encoding FKBP8 and SELPLG from
pJG4-5 (Clontech Laboratories, Inc.) were inserted into a pcHA
vector (Addgene vector database) to express their proteins in
mammalian cells. Prey genes were digested with restriction enzymes
(EcoRI/XhoI) and cloned into the mammalian expression vector pcHA.
Insertion of the prey genes were confirmed by restriction enzyme
digestion and nucleotide sequencing.
PCR
The DNA used for the PCR was obtained from bacterial
plasmid DNA (Bioneer Corporation, Daejeon, Korea). PCR was
performed with the following primer pairs: PRL-1 forward,
5′-TACACACAATCCAACCAATG-3′, and reverse,
5′-AATTAATGCTAGGGCAACAA-3′, and PRL-3 forward,
5′-TCATTGAGGACCTGAAGAAG-3′, and reverse,
5′-CTCAGCCAGTCTTCCACTAC-3′. PCR pre-mix was used for the reaction
(Bioneer Corporation). In each reaction, 20 µl final reaction
mixture contained 10 µl Premix Taq, 0.8 ml PCR forward
primer (10 mm), 0.8 ml reverse primer (10 mm), 2 µl DNA (100 ng/µl)
and dH2O. Subsequently, the reaction mixture was
incubated at 95°C for 5 min, followed by 40 cycles of 95°C for 15
sec and 60°C for 45 sec with 20 cycles. 1.5% agarose gel was used
for electrophoresis of the PCR product.
Screening of a HeLa library and
selection of proteins interacting with PRL-1 and PRL-3
The cDNA from a HeLa library (Clontech Laboratories,
Inc.) was sub-cloned into pJG4-5 vectors (Clontech Laboratories,
Inc.) for yeast two-hybrid screening. The EGY48 yeast strain
(Clontech Laboratories, Inc.) was transformed with pLexA-PRL-1 or
pLexA-PRL-3 by a small-scale yeast transformation protocol
(28) and plated onto synthetic
defined (SD)/-Trp1 (without yeast gene Trp1) medium (Sigma-Aldrich)
and grown at 30°C for 2–4 days. Selected clones were grown in 2 ml
yeast extract peptone dextrose medium containing ampicillin at 30°C
overnight with shaking. The yeast strain expressing LexA-PRL-1 or
PRL-3 bait protein was transformed with the HeLa cDNA library fused
to the GAL-4 activation domain by the lithium acetate method
(large-scale yeast transformation protocol) (28). The cDNA library was screened using a
yeast two-hybrid system (Matchmaker LexA two-hybrid system;
Clontech Laboratories, Inc.) to detect interacting proteins,
according to the manufacturer's protocol. Positive clones were
selected and assayed for lacZ reporter activity using a filter
β-galactosidase assay with X-Gal. Plasmids from positive yeast
clones were isolated and transformed into competent cells. Plasmids
isolated from competent cells were transformed into XL1-blue
competent cells (Agilent Technologies, Inc.- Santa Clara, CA, USA)
for analysis of the insert size and for sequencing. The interaction
between LexA-PRl-1 or PRL-3 and positive clones was confirmed by
small-scale yeast transformation.
DNA sequences were determined (Bioneer Corporation)
and nucleotide sequence databases were searched for homologous
sequences by Basic Local Alignment Search Tool (BLAST) analysis
(https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Transfection, immunoprecipitation and
immunoblot analysis
PRL-1 or PRL-3 expression vectors were transfected
into each cell line (HEK293T, HeLa and U2OS) using Lipofectamine
Plus (Gibco-BRL; Thermo Fisher Scientific, Inc.), using the
manufacturer's protocol. After 48 h, the cells were washed and
lysed with lysis buffer containing 150 mM NaCl, 0.1% Nonidet P-40
and 50 mM Tris-Cl (pH 7.4). Detergent-insoluble materials were
removed via centrifugation (1,000 × g), and the clear
lysates were incubated with anti-Flag® M2 antibody
(1:500) and Protein G Plus Agarose beads for 4 h (Santa Cruz
Biotechnology, Inc.). The beads were washed three times with lysis
buffer (29). For immunoblotting,
coprecipitates or whole cell extracts were resolved via 10% sodium
dodecyl sulfate-polyacrylamide gel electrophoresis, and
subsequently transferred to nitrocellulose membranes. The membranes
were immunoblotted with anti-HA (1:10,000) and
anti-Flag® M2 (1:2,000) antibodies and then developed
with an enhanced chemiluminescence detection system (Thermo Fisher
Scientific, Inc.).
Immunofluorescence analysis
U2OS cells (50,000) were plated on coverslips
pretreated with 0.1% gelatin in 12-well dishes, then transfected
with indicated expression vectors (HA-SELPLG, HA-FKBP8 and/or
Flag-PRL-1) and incubated for 2 days. The transfected cells were
washed with phosphate-buffered saline (PBS), fixed for 20 min in 4%
(w/v) paraformaldehyde, permeabilized for 10 min at room
temperature with PBS containing 0.3% (v/v) Triton X-100, and
further incubated for 10 min in 1% bovine serum albumin
(Sigma-Aldrich). Samples were subsequently incubated for 1 h with
primary antibodies anti-HA (1:10,000) and anti-Flag® M2
(1:2,000), washed three times with PBS, and then incubated with
Alexa Fluor 488-conjugated goat antibody against mouse IgG and
Alexa Fluor 594-conjugated goat antibody against rabbit IgG
(Molecular Probes; Thermo Fisher Scientific, Inc.). The coverslips
were mounted on glass slides in Vectashield medium (Vector
Laboratories, Inc., Burlingame, CA, USA). Images were acquired
using a Leica 6000 microscope (Leica Microsystems, Inc., Buffalo
Grove, IL, USA). For DAPI staining, 1 ml DAPI (3 µM) in staining
buffer (100 mM Tris, pH 7.4, 150 mM NaCl, 1 mM CaCl2,
0.5 mM MgCl2, 0.1% Nonidet P-40) was added to each cell
sample and incubated for 15 min at room temperature.
Dual-luciferase assay
HeLa cells were transfected with pRGC-luc (28), along with each expression vector
(HA-SELPLG, HA-FKBP8, Flag-PRL-1 and/or Flag-PRL-3) as indicated
using Lipofectamine Plus. The cells were lysed, and the luciferase
activity was evaluated using a dual luciferase assay kit (Promega
Corporation, Madison, WI, USA). The data were normalized to the
expression levels of a cotransfected Renilla luciferase
activity reporter control.
Functional classification, pathway analysis and
protein interaction network. The 12 identified proteins were sorted
by pathway and the Gene Ontology (GO) categories using the DAVID
database. SELPLG was selected in the Biocarta pathway. For the
network of the PRL-1, PRL-3 and prey proteins, the cellular protein
interaction network was constructed based on the screened proteins
in this study and in the STRING database.
Results
Screening of interacting proteins with
PRL-1 or 3 using a yeast two-hybrid system
The PRL family plays a significant role in the
development and cancer metastasis, and shares a high degree of
sequence similarity. Notably, PRL-3 has >75% amino-acid sequence
similarity to PRL-1, with a conserved function (1,27,30).
To screen novel PRL-interacting proteins, human
PRL-1 and PRL-3 were used as bait in a yeast two-hybrid system.
Flag-PRL-1 and Flag-PRL-3 were digested with restriction enzymes
(EcoRI/XhoI) and the inserts were cloned into the
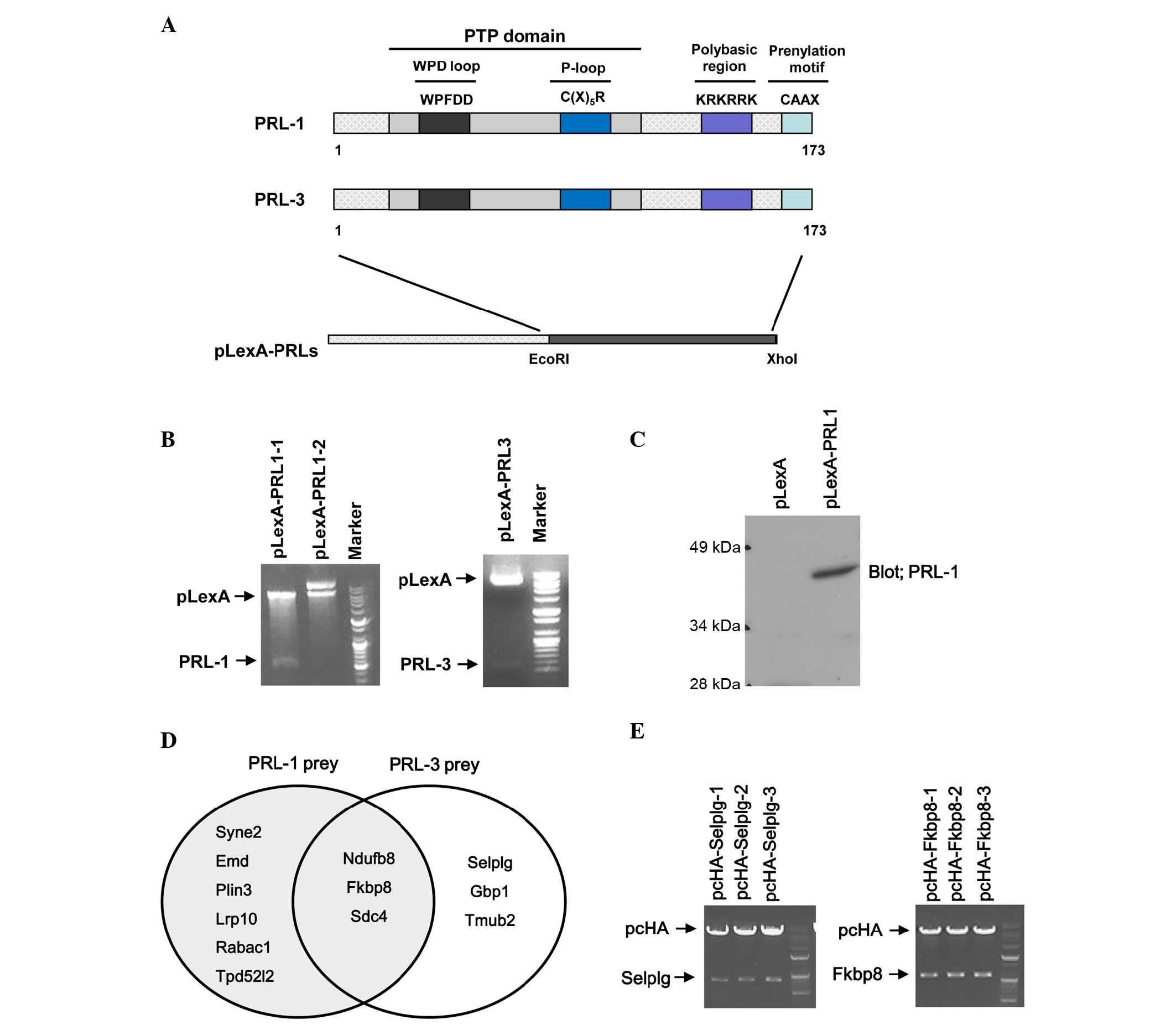
yeast expression vector pLexA (Fig.
1A). To confirm the cloning, PCR products of full length PRL-1
and PRL-3 from pLexA-PRL-1 and pLexA-PRL-3 were identified by
nucleotide electrophoresis (data not shown). In addition, the
inserts of PRL-1 and PRL-3 from pLexA-PRL-1 and pLexA-PRL-3 were
investigated by nucleotide electrophoresis following digestion with
same restriction enzymes (Fig. 1B).
Also, the sequence and the orientation of the inserts were
confirmed by sequencing analysis. Finally, the expression of the
PRL-1 bait in yeast EGY48 was confirmed by western blotting
(Fig. 1C).
A HeLa cDNA library was transformed in yeast EGY48
strains transformed with pLexA-PRL-1 or pLexA-PRL-3 bait vector
expressing PRL-1 or PRL-3 and cultured at 30°C for 2–4 days until
colonies appeared. Finally 38 blue colonies were observed on
SD/-His/-Leu/-Trp/X-Gal plates, the colonies were inoculated in
SD/-Leu/-Trp liquid medium and the plasmids were extracted.
Purified plasmids were retransformed in yeast EGY48 strains
containing pLexA-PRL-1 or PRL-3 bait vector and blue colonies were
observed again on SD/-His/-Leu/-Trp/X-Gal plates (data not shown).
Plasmids isolated from yeast were transformed into XL1-blue
competent cells for further analysis of the insert size and for
sequencing. Inserted fragments of library plasmids were mostly
between 500 and 2,000 bp in size. Identity of the prey was
determined by performing BLAST search analysis. The results of the
BLAST search against the human gene database indicated that 12
genes interact with PRL-1 or PRL-3: Synaptic nuclear envelope
protein 2 (SYNE2), emerin (EMD), mannose 6-phosphate
receptor-binding protein 1 (perilipin 3; PLIN3), low-density
lipoprotein receptor-related protein 10 (LRP10), Rab acceptor 1
(RABAC1), tumor protein D52-like 2 (TPD52L2), selectin P ligand
(SELPLG), guanylate binding protein 1 (GBP1), transmembrane and
ubiquitin-like domain-containing 2 (TMUB2), NADH:ubiquinone
oxidoreductase subunit B8 (NDUFB8), syndecan 4 (SDC4) and FKBP8
(Table I) were identified. Among
them, 9 prey proteins were isolated from screening using PRL-1 bait
and 6 prey proteins were obtained from screening using PRL-3 bait.
There were 3 prey proteins, namely NDUFB8, FKBP8 and SDC4, that
were identified from both PRL-1 and PRL-3 baits (Fig. 1D).
 | Table I.List of the identified preys from
screening. |
Table I.
List of the identified preys from
screening.
| Prey no. | Bait | Symbol | Full name | No. of clones |
|---|
| 1 | PRL-1 | SYNE2 | Synaptic nuclear
envelope protein 2 | 4 |
| 2 | PRL-1 | EMD | Emerin | 2 |
| 3 | PRL-1 | PLIN3 | Mannose 6-phosphate
receptor-binding protein 1 | 4 |
| 4 | PRL-1 | LRP10 | Low-density
lipoprotein receptor-related protein 10 | 2 |
| 5 | PRL-1 | RABAC1 | Rab acceptor 1 | 2 |
| 6 | PRL-1 | TPD52L2 | Tumor protein
D52-like 2 | 3 |
| 7 | PRL-3 | SELPLG | Selectin P
ligand | 4 |
| 8 | PRL-3 | GBP1 | Guanylate binding
protein 1 | 2 |
| 9 | PRL-3 | TMUB2 | Transmembrane and
ubiquitin-like domain-containing 2 | 2 |
| 10 | PRL-1, PRL-3 | NDUFB8 | NADH:ubiquinone
oxidoreductase subunit B8 | 4 |
| 11 | PRL-1, PRL-3 | FKBP8 | FK506-binding
protein 8 | 6 |
| 12 | PRL-1, PRL-3 | SDC4 | Syndecan 4 | 3 |
In vivo binding and
colocalization
From among the 12 candidate genes interacting with
PRL-1 or PRL-3, two cDNA clones encoding for FKBP8 and SELPLG were
inserted into pcHA vector to express their proteins in mammalian
cells. Prey genes were digested with restriction enzymes
(EcoRI/XhoI) and cloned into the mammalian expression
vector pcHA. Insertion of the prey genes was confirmed by
restriction enzyme digestion and nucleotide sequencing (Fig. 1E).
To confirm their binding in a yeast-independent
interaction assay, coimmunoprecipitation experiments were
performed. HEK293T cells were co-transfected with Flag-PRL-1 and
HA-FKBP8 or HA-SELPLG constructs, and cell extracts were then
subjected to immunoprecipitation with anti-Flag antibody, followed
by immunoblotting analysis with an anti-HA antibody. HA-tagged
FKBP8 and SELPLG were detected in anti-Flag-PRL-1
immunoprecipitates (Fig. 2A).
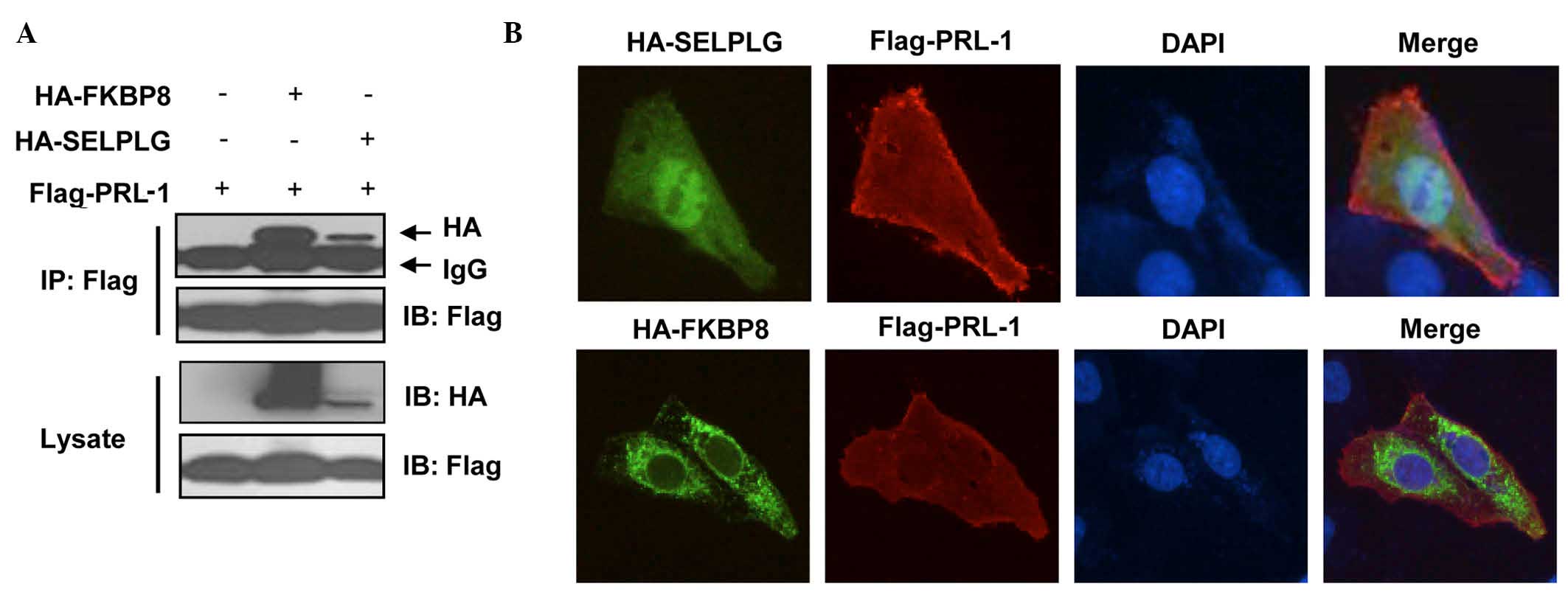 | Figure 2.In vivo binding and
colocalization. (A) FKBP8 and SELPLG interact with PRL-1.
Flag-PRL-1 and/or HA-FKBP8 or HA-SELPLG were transfected into
HEK293T cells. The cells were treated with MG132 for 4 h prior to
harvesting, and 48 h later, the cells were prepared for co-IP and
western blot analysis. (B) Colocalization of FKBP8 or SELPLG with
PRL-1. Flag-PRL-1 and HA-FKBP8 or HA-SELPLG were transfected in
U2OS cells. Then, 48 h later, the cells were prepared for
immunofluorescence analysis. Images were acquired using a Leica
6000 microscope (magnification, ×200). FKBP8, FK506-binding protein
8; SELPLG, selectin P ligand; PRL-1, phosphatase of regenerating
liver 1; HA, high availability; DAPI,
4′,6-diamidino-2-phenylindole; IP, immunoprecipitation; IB,
immunoblotting. |
The localization of bait proteins and prey proteins
was then examined. U2OS cells were transfected with Flag-PRL-1, and
HA-FKBP8 or HA-SELPLG. Localization of FLAG tagged-PRL-1 was
visualized with anti-FLAG primary antibody and Fluor 488-conjugated
goat antibody against mouse IgG and localization of HA-tagged preys
was visualized with anti-HA antibody and Alexa Fluor 594-conjugated
goat antibody against rabbit IgG.
In cells, PRLs are typically associated with the
plasma membrane and early endosome (1,27,30). An
important mechanism responsible for this localization is
prenylation, a post-translational lipid modification that commonly
targets proteins to membranes (3,27,30).
Fig. 2B and Table II show that PRL-1 localization is
observed in the endosome, early endosome, endoplasmic reticulum,
spindle, cytoskeleton, plasma membrane, microtubule cytoskeleton
and intracellular non-membrane-bounded organelle. SELPLG is visible
in the membrane fraction, insoluble fraction, plasma membrane, and
is integral to the plasma membrane while FKBP38 is observed in the
mitochondrial envelope, endoplasmic reticulum membrane, plasma
membrane, endomembrane system and nuclear envelope-endoplasmic
reticulum network (Fig. 2B and
Table II). The expression of SELPLG
and FKBP38 appears to be partially colocalized with PRL-1. In the
presence of preys, changes in the localization of PRL-1 were not
observed, suggesting that the expression of these preys does not
affect the prenylation and localization of PRL-1.
 | Table II.Analysis of the cellular components
associated with the identified proteins, based on the cellular
components gene ontology categories of DAVID. |
Table II.
Analysis of the cellular components
associated with the identified proteins, based on the cellular
components gene ontology categories of DAVID.
| Gene | Cellular
components |
|---|
| FKBP8 | Mitochondrial
envelope, endoplasmic reticulum membrane, plasma membrane, nuclear
envelope-endoplasmic reticulum network |
| NDUFB8 | Mitochondrion,
mitochondrial envelope, endoplasmic reticulum, integral to
membrane, NADH dehydrogenase complex |
| RABAC1 | Golgi apparatus,
plasma membrane, synaptic vesicle, integral to membrane, cell
junction, membrane-bounded vesicle, synapse |
| EMD | Nuclear envelope,
endoplasmic reticulum, spindle, cytoskeleton, endomembrane system,
microtubule cytoskeleton, nuclear membrane |
| GBP1 | Plasma membrane,
internal side of plasma membrane, plasma membrane part |
| LRP10 | Coated pit,
endomembrane system, integral to membrane, intrinsic to
membrane |
| PLIN3 | Endosome, Golgi
apparatus, lipid particle, plasma membrane, internal side of plasma
membrane, monolayer-surrounded lipid storage body |
| SELPLG | Cell fraction,
membrane fraction, insoluble fraction, plasma membrane, intrinsic
to plasma membrane |
| SYNE2 | Nuclear envelope,
cytoskeleton, plasma membrane, endomembrane system, integral to
membrane, nuclear membrane |
| SDC4 | Golgi apparatus,
plasma membrane, adherens junction, focal adhesion, cell surface,
cell-substrate junction, membrane raft, anchoring junction |
| TMUB2 | Integral to
membrane, intrinsic to membrane |
| TPD52L2 | Perinuclear region
of cytoplasm |
| PRL-1 | Endosome,
endoplasmic reticulum, spindle, cytoskeleton, plasma membrane,
microtubule cytoskeleton |
| PRL-3 | Endosome, early
endosome, plasma membrane |
SELPLG and FKBP8 inhibit the functions
of PRL-1 and PRL-3
Having verified the binding of FKBP8 and SELPLG with
PRL-3 protein, the next important question is whether FKBP8 and
SELPLG affect the functions of PRL-1 and PRL-3 in cells. The roles
of PRL-1 and PRL-3 are associated with the downregulation of p21
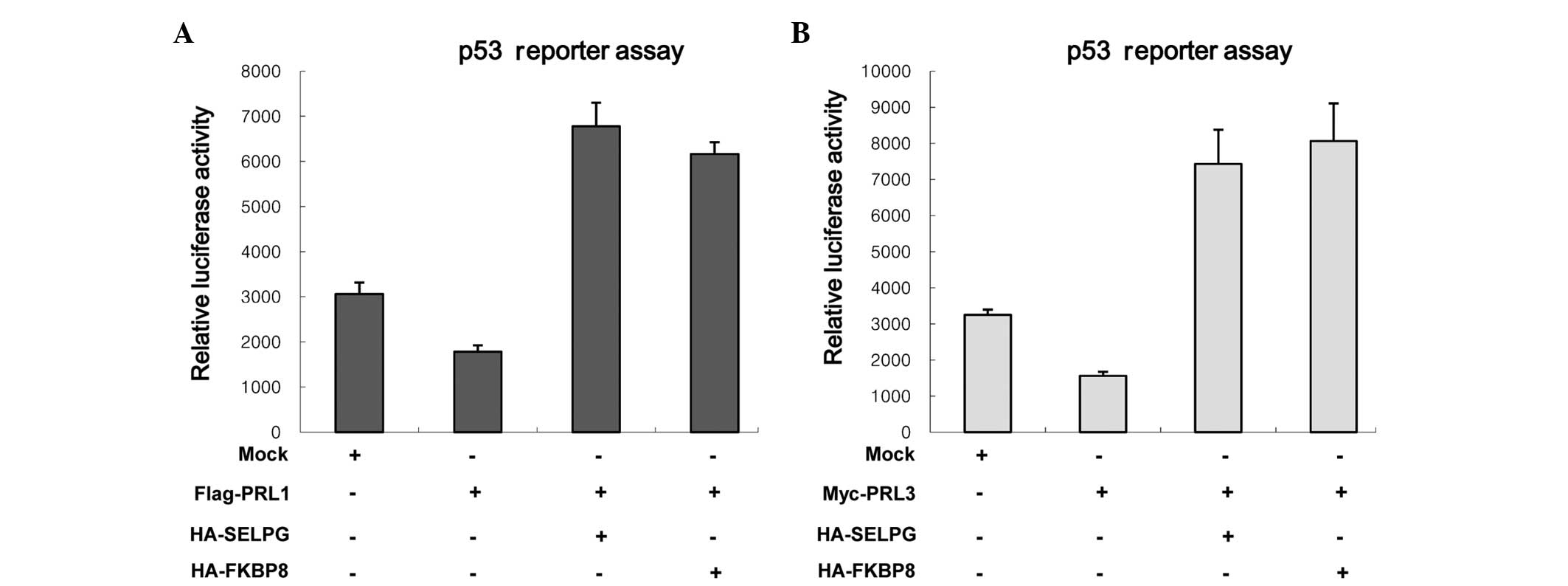
transcription as well as the activity of p53 (28). Therefore, the effects of two prey
proteins on the downregulation of p53 reporter activities mediated
by PRL-1 and PRL-3 were investigated. HeLa cells were transfected
with each prey protein and/or Flag-PRL-1 (or Myc-PRL-3) and
p53-luciferase reporter (pRGC-luc) (Fig.
3). When p53-luc was transfected with PRL-1 or PRL-3,
inhibition of luciferase activity was observed (Fig. 3A), as shown previously (28). However, SELPLG and FKBP8 markedly
attenuated the PLR-1-mediated p53-luc inhibition (Fig. 3A). Also, similar results were
observed when SELPLG and FKBP8 were introduced with PRL-3 (Fig. 3B). These findings reveal that SELPLG
and FKBP8 inhibit the ability of PRL-1 and PRL-3 to reduce p53
reporter activity and imply that SELPLG and FKBP8 inhibit the
cellular functions of PRL-1 and PRL-3.
Functional classification, pathway
analysis and protein interaction network
The identified proteins were sorted according to
pathways and GO categories using the DAVID bioinformatics resource.
Pathways for SELPLG were identified using the BioCarta pathway
database (data not shown). Pathways for NDUFB8, EMD, SELPLG and
SDC4 were identified using KEGG pathway analysis and contained
oxidative phosphorylation, Alzheimer's disease, Parkinson's
disease, Huntington's disease, hypertrophic cardiomyopathy,
arrhythmogenic right ventricular cardiomyopathy, dilated
cardiomyopathy, cell adhesion molecules, adhesion and diapedesis of
granulocytes, cells and molecules involved in local acute
inflammatory response, and extracellular matrix (ECM)-receptor
interaction (Table III). Among the
12 proteins, there were 9 proteins involved in diverse biological
processes including vesicle transport, protein folding, cell
proliferation, apoptosis, immune response, cell fate specification
and metabolic process (Table IV).
Cellular component data showed that the localizations of the 12
proteins mostly or partly matched with those of PRL-1 or PRL-3
(Table II).
 | Table III.Signal pathway analysis of the
identified proteins, based on the pathway categories of DAVID. |
Table III.
Signal pathway analysis of the
identified proteins, based on the pathway categories of DAVID.
| Gene | Signaling
pathway |
|---|
| NDUFB8 | Oxidative
phosphorylation, Alzheimer's disease, Parkinson's disease,
Huntington's disease |
| EMD | Hypertrophic
cardiomyopathy, arrhythmogenic right ventricular cardiomyopathy,
dilated cardiomyopathy |
| SELPLG | Cell adhesion
molecules, adhesion and diapedesis of granulocytes, cells and
molecules involved in local acute inflammatory response |
| SDC4 | ECM-receptor
interaction, cell adhesion molecules |
 | Table IV.Biological process analysis of the
identified proteins, based on the biological process gene ontology
categories of DAVID. |
Table IV.
Biological process analysis of the
identified proteins, based on the biological process gene ontology
categories of DAVID.
| Gene | Biological
process |
|---|
| FKBP8 | Cell fate
specification, regionalization, protein folding, apoptosis,
smoothened signaling pathway, pattern specification process,
dorsal/ventral pattern formation, neural tube patterning and
development, regulation of BMP signaling pathway, chordate
embryonic development |
| NDUFB8 | Oxidative
phosphorylation, mitochondrial electron transport, NADH to
ubiquinone, phosphorus metabolic process, energy derivation by
oxidation of organic compounds, phosphorylation, cellular
respiration, oxidation reduction |
| EMD | Muscle system
process, muscle contraction, nucleus organization, nuclear envelope
organization, muscle organ development, endomembrane organization,
membrane organization, nuclear envelope reassembly |
| GBP1 | Immune
response |
| LRP10 | Lipid transport,
endocytosis, membrane invagination, lipid localization, membrane
organization, vesicle-mediated transport |
| PLIN3 | Vesicle-mediated
transport |
| SELPLG | Cell motion,
leukocyte adhesion, cell-cell adhesion, cell migration, biological
adhesion, cellular |
|
| extravasation, cell
motility, leukocyte migration, leukocyte tethering or rolling,
localization of cell |
| SDC4 | Regulation of
muscle contraction, regulation of phosphate metabolic process,
regulation of phosphorylation, positive regulation of catalytic
activity, regulation of kinase activity, regulation of system
process, regulation of molecular function, regulation of
transferase activity |
| TPD52L2 | Regulation of cell
proliferation |
| PRL-1 | Protein amino acid
dephosphorylation, phosphate metabolic process, cell cycle,
regulation of cell migration, regulation of locomotion, regulation
of cell motion |
| PRL-3 | Protein amino acid
dephosphorylation, phosphorus metabolic process, phosphate
metabolic process |
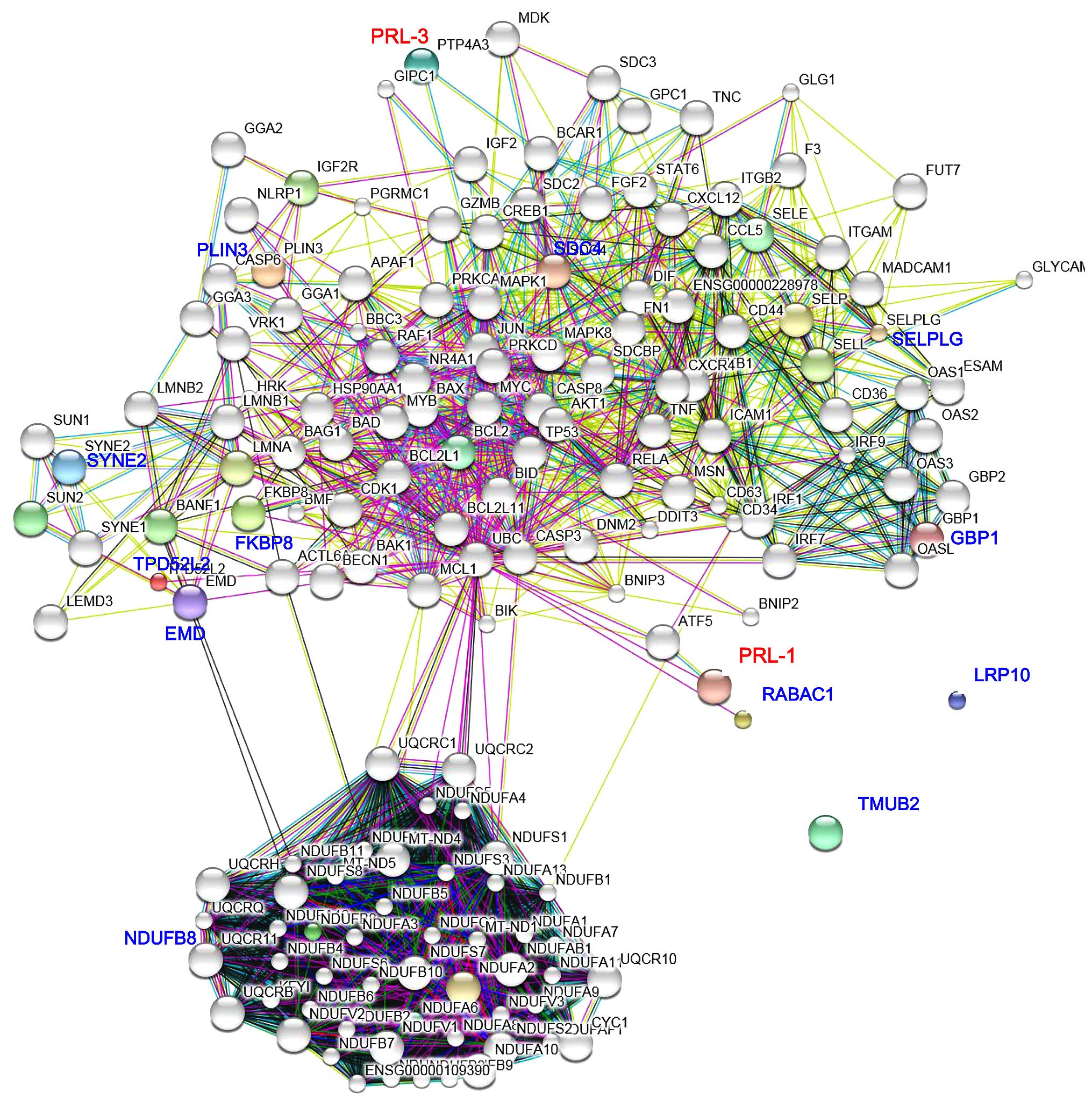
A PRL-1 and PRL-3-prey proteins interaction network
was constructed using the STRING database (Fig. 4). SDC4, PLIN3, SYNE2, TPD52L2, EMD
and FKBP8 were indicated to by the most closely-related and
specific node proteins associated with PRL-3, whereas SELPLG, GBP1,
RABAC1 and NDUFB8 were the most remarkable node proteins associated
with PRL-1. TMUB2 and LRP10 did not show any indirect interactions
with PRL-1 or PRL-3 (Fig. 4). These
notable node proteins appear to be particularly important in the
regulation and organization of PRL-1 and PRL-3 in the prey proteins
interaction network.
Discussion
The PRL family comprises a group of PTPs that play
an important role in the development and metastasis of various
types of cancer (12). The family
members, which include PRL-1, PRL-2 and PRL-3, share a high degree
of sequence similarity and show similar functional characteristics.
It has been reported that several signaling pathways involved in
cell growth and cancer development are affected (regulated by) PRLs
(3,4). However, the mechanisms by which PRLs
regulate signaling or interact with direct binding partners to
mediate their effects remains to be clearly elucidated.
In the present study, 12 proteins interacting with
PRL-1 or PRL-3 were identified using a yeast two-hybrid system. GO
biological process data indicated that these proteins are mostly
associated with nuclear envelope organization, endomembrane
organization and nucleus organization (Table IV). Cellular components data suggest
that they are located at membrane parts, integral to membrane,
intrinsic to membrane, envelope, nuclear membrane, contractile
fiber part, myofibril, organelle membrane and nuclear envelope
(Table II). Molecular functions of
6 genes were classified as protein binding (data not shown). They
were also found to be involved in various signaling pathways such
as oxidative phosphorylation, Alzheimer's disease, hypertrophic
cardiomyopathy, ECM-receptor interaction and cell adhesion
molecules in KEGG pathways (Table
III).
FKBP8 is a member of the FKBP family of proteins,
and is widely expressed in cancer cell lines (31,32). In
cancer, FKBP8 has potential antitumor effects via the regulation of
anti-invasive syndecan 1, proinvasive matrix metalloproteinase 9
(33,34), mechanistic target of rapamycin,
Rheb-GTP (35) and PRL-3 (28). Results of our previous study showed
that FKBP8 binds to PRL-3, and suppresses PRL-3-mediated p53
activity and cell proliferation (28). The present study also provided
evidence that FKBP8 binds to PRL-1, and suppresses the function of
PRL-1, in addition to that of PRL-3.
SELPLG is a glycoprotein that acts as a
counter-receptor for the cell adhesion molecules P-, E- and
L-selectin expressed on myeloid cells and T lymphocytes (36). In leukocyte trafficking during
inflammation, SELPLG tethers leukocytes to activating platelets or
selectin-expressing endothelia. SELPLG requires post-translational
modification by tyrosine sulfation and addition of the
sialyl-Lewis-x tetrasaccharide for its high-affinity binding
activity. Aberrant expression of and polymorphisms in the SELPLG
gene are associated with defects in the innate and adaptive immune
response.
In the present study, 12 potential PRL-1/3 binding
proteins were identified, including 11 novel binding partners and a
known binding partner, FKBP8. SELPLG and FKBP8 proteins were shown
to directly bind to PRL-1 and inhibit the downregulation of p53
reporter activities mediated by PRL-3 and PRL-1. These results
demonstrate that SELPLG and FKBP8 may be regulators of the
oncogenic proteins PRL-1 and PRL-3 and can have a marked impact on
cell proliferation.
It is possible that the 12 PRL-binding proteins
positively or negatively regulate PRL function (FKBP8 and SELPLG)
or may be regulated by PRLs. In regard to this hypothesis, further
studies are underway to reveal those mechanisms.
In conclusion, multiple PRLs binding proteins were
screened using a yeast two-hybrid system. The identified proteins
are associated with diseases including Alzheimer's disease,
Parkinson's disease, Huntington's disease, hypertrophic
cardiomyopathy, arrhythmogenic right ventricular cardiomyopathy and
dilated cardiomyopathy, suggesting that the PRL family may be
involved in diverse diseases as well as cancer. Furthermore, these
findings may provide valuable information for better understanding
the interactions between the PRL family and target proteins, and
revealing new biological functions of PRLs.
Acknowledgements
This study was supported by the National Research
Foundation of Korea (NRF) grant funded by the Korean Government
(2013-R1A1A1007596).
References
|
1
|
Al-Aidaroos AQ and Zeng Q: PRL-3
phosphatase and cancer metastasis. J Cell Biochem. 111:1087–1098.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Achiwa H and Lazo JS: PRL-1 tyrosine
phosphatase regulates c-Src levels, adherence, and invasion in
human lung cancer cells. Cancer Res. 67:643–650. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stephens BJ, Han H, Gokhale V and Von Hoff
DD: PRL phosphatases as potential molecular targets in cancer. Mol
Cancer Ther. 4:1653–1661. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saha S, Bardelli A, Buckhaults P,
Velculescu VE, Rago C, St Croix B, Romans KE, Choti MA, Lengauer C,
Kinzler KW and Vogelstein B: A phosphatase associated with
metastasis of colorectal cancer. Science. 294:1343–1346. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang Q, Holmes DI, Powell SM, Lu QL and
Waxman J: Analysis of stromal-epithelial interactions in prostate
cancer identifies PTPCAAX2 as a potential oncogene. Cancer Lett.
175:63–69. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Parker BS, Argani P, Cook BP, Liangfeng H,
Chartrand SD, Zhang M, Saha S, Bardelli A, Jiang Y, St Martin TB,
et al: Alterations in vascular gene expression in invasive breast
carcinoma. Cancer Res. 64:7857–7866. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Miskad UA, Semba S, Kato H and Yokozaki H:
Expression of PRL-3 phosphatase in human gastric carcinomas: Close
correlation with invasion and metastasis. Pathobiology. 71:176–184.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu X, Zeng H, Zhang X, Zhao Y, Sha H, Ge
X, Zhang M, Gao X and Xu Q: Phosphatase of regenerating liver-3
promotes motility and metastasis of mouse melanoma cells. Am J
Pathol. 164:2039–2054. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang H, Quah SY, Dong JM, Manser E, Tang
JP and Zeng Q: PRL-3 down-regulates PTEN expression and signals
through PI3K to promote epithelial-mesenchymal transition. Cancer
Res. 67:2922–2926. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kato H, Semba S, Miskad UA, Seo Y, Kasuga
M and Yokozaki H: High expression of PRL-3 promotes cancer cell
motility and liver metastasis in human colorectal cancer: A
predictive molecular marker of metachronous liver and lung
metastases. Clin Cancer Res. 10:7318–7328. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mizuuchi E, Semba S, Kodama Y and Yokozaki
H: Down-modulation of keratin 8 phosphorylation levels by PRL-3
contributes to colorectal carcinoma progression. Int J Cancer.
124:1802–1810. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Min SH, Kim DM, Heo YS, Kim YI, Kim HM,
Kim J, Han YM, Kim IC and Yoo OJ: New p53 target, phosphatase of
regenerating liver 1 (PRL-1) downregulates p53. Oncogene.
28:545–554. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fiordalisi JJ, Keller PJ and Cox AD: PRL
tyrosine phosphatases regulate rho family GTPases to promote
invasion and motility. Cancer Res. 66:3153–3161. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Werner SR, Lee PA, DeCamp MW, Crowell DN,
Randall SK and Crowell PL: Enhanced cell cycle progression and down
regulation of p21 (Cip1/Waf1) by PRL tyrosine phosphatases. Cancer
Lett. 202:201–211. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liang F, Liang J, Wang WQ, Sun JP, Udho E
and Zhang ZY: PRL3 promotes cell invasion and proliferation by
down-regulation of Csk leading to Src activation. J Biol Chem.
282:5413–5419. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peng L, Jin G, Wang L, Guo J, Meng L and
Shou C: Identification of integrin alpha1 as an interacting protein
of protein tyrosine phosphatase PRL-3. Biochem Biophys Res Commun.
342:179–183. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hinds PW: Too much of a good thing: The
Prl-3 in p53's oyster. Mol Cell. 30:260–261. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Basak S, Jacobs SB, Krieg AJ, Pathak N,
Zeng Q, Kaldis P, Giaccia AJ and Attardi LD: The
metastasis-associated gene Prl-3 is a p53 target involved in
cell-cycle regulation. Mol Cell. 30:303–314. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Min SH, Kim DM, Heo YS, Kim HM, Kim IC and
Yoo OJ: Downregulation of p53 by phosphatase of regenerating liver
3 is mediated by MDM2 and PIRH2. Life Sci. 86:66–72. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Forte E, Orsatti L, Talamo F, Barbato G,
De Francesco R and Tomei L: Ezrin is a specific and direct target
of protein tyrosine phosphatase PRL-3. Biochim Biophys Acta.
1783:334–344. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jeong DG, Kim SJ, Kim JH, Son JH, Park MR,
Lim SM, Yoon TS and Ryu SE: Trimeric structure of PRL-1 phosphatase
reveals an active enzyme conformation and regulation mechanisms. J
Mol Biol. 345:401–413. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li M, Brooks CL, Wu-Baer F, Chen D, Baer R
and Gu W: Mono-versus polyubiquitination: Differential control of
p53 fate by Mdm2. Science. 302:1972–1975. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pascaru M, Tanase C, Vacaru AM, Boeti P,
Neagu E, Popescu I and Szedlacsek SE: Analysis of molecular
determinants of PRL-3. J Cell Mol Med. 13:3141–3150. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Peters CS, Liang X, Li S, Kannan S, Peng
Y, Taub R and Diamond RH: ATF-7, a novel bZIP protein, interacts
with the PRL-1 protein-tyrosine phosphatase. J Biol Chem.
276:13718–13726. 2001.PubMed/NCBI
|
|
25
|
Si X, Zeng Q, Ng CH, Hong W and Pallen CJ:
Interaction of farnesylated PRL-2, a protein-tyrosine phosphatase,
with the beta-subunit of geranylgeranyltransferase II. J Biol Chem.
276:32875–32882. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sun JP, Wang WQ, Yang H, Liu S, Liang F,
Fedorov AA, Almo SC and Zhang ZY: Structure and biochemical
properties of PRL-1, a phosphatase implicated in cell growth,
differentiation, and tumor invasion. Biochemistry. 44:12009–12021.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bessette DC, Qiu D and Pallen CJ: PRL
PTPs: Mediators and markers of cancer progression. Cancer
Metastasis Rev. 27:231–252. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Choi MS, Min SH, Jung H, Lee JD, Lee TH,
Lee HK and Yoo OJ: The essential role of FKBP38 in regulating
phosphatase of regenerating liver 3 (PRL-3) protein stability.
Biochem Biophys Res Commun. 406:305–309. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Min SH, Lau AW, Lee TH, Inuzuka H, Wei S,
Huang P, Shaik S, Lee DY, Finn G, Balastik M, et al: Negative
regulation of the stability and tumor suppressor function of Fbw7
by the Pin1 prolyl isomerase. Mol Cell. 46:771–783. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Peng L, Xing X, Li W, Qu L, Meng L, Lian
S, Jiang B, Wu J and Shou C: PRL-3 promotes the motility, invasion,
and metastasis of LoVo colon cancer cells through PRL-3-integrin
beta1-ERK1/2 and-MMP2 signaling. Mol Cancer. 8:1102009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kang CB, Feng L, Chia J and Yoon HS:
Molecular characterization of FK-506 binding protein 38 and its
potential regulatory role on the anti-apoptotic protein Bcl-2.
Biochem Biophys Res Commun. 337:30–38. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bulgakov OV, Eggenschwiler JT, Hong DH,
Anderson KV and Li T: FKBP8 is a negative regulator of mouse sonic
hedgehog signaling in neural tissues. Development. 131:2149–2159.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fong S, Mounkes L, Liu Y, Maibaum M,
Alonzo E, Desprez PY, Thor AD, Kashani-Sabet M and Debs RJ:
Functional identification of distinct sets of antitumor activities
mediated by the FKBP gene family. Proc Natl Acad Sci USA.
100:14253–14258. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rosner M, Hofer K, Kubista M and
Hengstschläger M: Cell size regulation by the human TSC tumor
suppressor proteins depends on PI3K and FKBP38. Oncogene.
22:4786–4798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bai X, Ma D, Liu A, Shen X, Wang QJ, Liu Y
and Jiang Y: Rheb activates mTOR by antagonizing its endogenous
inhibitor, FKBP38. Science. 318:977–980. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Luan SL, Boulanger E, Ye H, Chanudet E,
Johnson N, Hamoudi RA, Bacon CM, Liu H, Huang Y, Said J, et al:
Primary effusion lymphoma: Genomic profiling revealed amplification
of SELPLG and CORO1C encoding for proteins important for cell
migration. J Pathol. 222:166–179. 2010. View Article : Google Scholar : PubMed/NCBI
|