Introduction
Human fibroblast growth factor 21 (hFGF-21), which
is a member of the FGF family, is known to be a regulator of the
metabolic system and can increase glucose uptake and GLUT1 mRNA
expression (1,2). For example, there is a weak
cause-effect relationship between hFGF-21 and ketogenesis in humans
(3,4). In addition, previous studies reported
that hFGF-21 regulates glucose and lipid metabolism via its
interactions with the peroxisome proliferator-activated receptor
alpha and gamma pathways (5–7). However, the role of hFGF-21 in the
metabolic system remains unclear. In the present study, an
anti-hFGF-21 monoclonal antibody (mAb) was prepared and
characterized, in order to provide a novel method for researching
the molecular mechanisms by which hFGF-21 acts in the metabolic
system.
Previous studies identified that elevated serum
levels of hFGF-21 were associated with numerous diseases, including
diabetes (8–10). In addition, a previous study revealed
an association between hFGF-21 serum levels and long term diabetic
complications, such as nephropathy and carotid atheromatosis
(11). Furthermore, increased serum
levels of hFGF-21 have been found to be associated with obesity
(12). Additionally, elevated serum
hFGF-21 levels were discovered in patients with ischemic heart
disease (1). Conversely, improvement
in the symptoms of patients with diseasesassociated with elevated
hFGF-21 serum levels is typically accompanied with reduced serum
levels of hFGF-21, for example, weight loss was accompanied by a
decrease in hFGF-21 levels in humans with obesity-related metabolic
complications (13). and hFGF-21
levels decreased in diabetic patients following therapy with
insulin or oral agents (14). In the
current study, anti-hFGF-21 mAb (clone 2D8) was produced, purified
and characterized in vitro. Then, new lipoprotein A (NlpA)
-based bacterial display library technology was used for epitope
screening. In addition, the in vitro bioactivity of the mAb
was determined using a glucose uptake assay and by measuring
glucose transporter 1 (GLUT1) mRNA expression. There is a lack of
relevant previous studies on the anti-hFGF-21 mAb and its
bioactivity. The present study identified that the mAb prepared
could specifically detect serum levels of hFGF-21 and thus has
potential as a prognostic factor to indicate the development of
hFGF-21-related diseases. In addition, it could be used for future
research into hFGF-21, which may identify therapeutic targets for
the treatment of hFGF-21-associated diseases.
Materials and methods
Ethics statement
All experiments in the present study were approved
by the Northeast Agricultural University Provincial Experimental
Animal Management Committee (Harbin, China) and were performed in
accordance with the guidelines of this committee.
Chemicals and reagents
Freund's adjuvant (complete), incomplete Freund's
adjuvant, bovine serum albumin (BSA) and
3,3′,5,5′-tetramethylbenzidine (TMB) were purchased from
Sigma-Aldrich (Merck Millipore, Darmstadt, Germany). Horseradish
peroxidase (HRP)-conjugated goat anti-mouse secondary antibody
(A21010) was purchased from Abbikine, Inc. (Redlands, CA, USA). SAB
Clonotyping System-HRP (5300–05) was purchased from SouthernBiotech
(Birmingham, AL, USA). Fluorescein isothiocyanate (FITC) Antibody
Labeling Kit (53027) was purchased from Thermo Fisher Scientific,
Inc., (Waltham, MA, USA). Glucose Assay Kit (0105102), which
utilzizes the GOD-PAP method was purchased from Sichuan Maccura
Biotechnology Co., Ltd (Chengdu, China). Other reagent grade
chemicals were purchased from Sigma-Aldrich (Merck Millipore). DNA
maker2000, λEcoT14 DNA Marker, and prestained
protein MW Marker were purchased from Fermentas (Thermo Fisher
Scientific, Inc.). All polymerase chain reaction (PCR) primers
(Tables I and II) were synthesized by Invitrogen (Thermo
Fisher Scientific, Inc.).
 | Table I.Primer sequences for polymerase chain
reaction amplification of different hFGF-21 segments. |
Table I.
Primer sequences for polymerase chain
reaction amplification of different hFGF-21 segments.
| Segment | Primer sequence
(5′-3′) |
|---|
| PU | F:
GGCCCAGCCGGCCCACCCCATCCCTGACTCC |
|
| R:
GGCCCCCGAGGCCTTACTCAGGGTCAAAGTGGAGCGAT |
| PD | F:
GGCCCAGCCGGCCGCCTGCAGCTTCCGGGAG |
|
| R:
GGCCCCCGAGGCCTCAGGAAGCGTAGCTGGGGC |
| PD1 | F:
GGCCCAGCCGGCCGCCTGCAGCTTCCGGGAG |
|
| R:
GGCCCCCGAGGCCTTAGTTCCCTGGCAGGTGCAGC |
| PD2 | F:
GGCCCAGCCGGCCAAGTCCCCACACCGGG |
|
| R:
GGCCCCCGAGGCCTTATCCGGGTGGCTCCGGGAG |
| PD3 | F:
GGCCCAGCCGGCCATCCTGGCCCCCCAGCC |
|
| R:
GGCCCCCGAGGCCTCAGGAAGCGTAGCTGGGGC |
| PD4 | F:
GGCCCAGCCGGCCTACCAGTCCGAAGCCCACG |
|
| R:
GGCCCCCGAGGCCTTAGAAGCGAGCTGGTCCTC |
| PD5 | F:
GGCCCAGCCGGCCCTGCCACTACCAGGCCTGC |
|
| R:
GGCCCCCGAGGCCTTACAGAGGGTCCGAGGAGC |
 | Table II.Primer sequences for the quantitative
polymerase chain reaction. |
Table II.
Primer sequences for the quantitative
polymerase chain reaction.
| Gene name | Primer sequences
(5′-3′) |
|---|
| β-actin | F:
GAGACCTTCAACACCCC |
|
| R:
GTGGTGGTGAAGCTGTAGCC |
| GLUT1 | F:
CCATCCACCACACTCACCAC |
|
| R:
GCCCAGGATCAGCATCTCAA |
Cell culture
Sp2/0 lymphocytes, 3T3-L1 adipocytes and DH5α
Escherichia coli were lab stocks. DH5α (MLCC3002) was
purchased from Miaolingbio Bioscience & Technology Co., Ltd.,
(Wuhan, China; −80°C). Sp2/0 (CC-Y2093), which were cultured in
RPMI-1640 supplemented with 10% fetal bovine serum (FBS) at 37°C in
an atmosphere containing 5% CO2, and 3T3-L1 adipocytes
(CC-Y2002), which were cultured in Dulbecco's modified Eagle medium
(DMEM) supplemented with 10% FBS at 37°C, 5% CO2, were
both purchased from Enzyme Research Biotechnology Co., Ltd.,
(Beijing, China). RPMI-1640 (CM0302) was purchased from You Kang
Biotechnology Co., Ltd., (Beijing, China). DMEM (PM150310) was
purchased from Procell (Wuhan, China).
hFGF-21 expression and
purification
Whole hFGF-21 protein was expressed and purified
during previous studies conducted in our laboratory (2).
Experimental animals
Six female and six male BALB/c mice (age, 6–8 weeks
old; weight, 11–13 g) were purchased from Harbin Veterinary
Research Institute (Harbin, China), and housed in starter batteries
with access to water and commercial feed.
Anti-hFGF-21 mAb (clone 2D8)
production
BALB/c mice (female, n=3) were immunized with 100 µg
hFGF-21 (as 400 ml of 1:1 hFGF-21: Freund's adjuvant), after
2-weeks of feeding, followed by second immunization with 100 µg
hFGF-21 (400 ml of 1:1 hFGF-21: incomplete Freund's adjuvant),
third immunization was the same as the second immunization and was
performed 2-weeks later. Prior to hybridoma production, the mice
received a booster immunization of 100 µg hFGF-21 in
phosphate-buffered saline (PBS; pH 7.5), and separated eyeball
blood samples as positive serum. BALB/c mice (female, n=3) under
the same rearing conditions were used to obtain negative serum by
separating eyeball blood sampling. The establishment methods of
hybridoma were performed according to previously described methods
(15).
Indirect ELISA was performed to screen for
individual clones secreting hFGF-21 mAb. Prior to cell plating, the
96-well plates were coated with 20 µg/ml hFGF-21 (100 µl) and
incubated at 4°C overnight. Then, plates were washed three times
with washing buffer (0.05% Tween-20 in PBS), blocked with 5%
skimmed milk in PBS for 2 h at 37°C, followed by washing (as
previously described). Hybridomas cultures (100 µl) as primary
antibodies (positive serum as positive control, negative serum as
negative control) were added to the wells and incubated for 1 h at
37°C. Following washing (as described above), plates were incubated
with HRP-conjugated goat anti-mouse secondary antibody (1:15,000;
100 µl) for 1 h at 37°C, followed by washing (as previously
described). The chromogenic reagent TMB (100 µl) was added to the
wells and following 20 min the reaction was stopped with 2 mol/l
H2SO4 (50 µl), and the plates
spectroscopically analyzed at 450 nm using a microplate reader
(Bio-Rad, Hercules, CA, USA). hFGF-21-positive clones were then
subcloned and rescreened (as previously described).
Purification, classification and
SDS-PAGE analysis of anti-hFGF-21 mAb
For large-scale production of the hFGF-21 mAb, a
single highly positive subclone hybridoma cells (2D8) were injected
into the peritoneal cavity of BALB/c mice (male, n=3) and allowed
to proliferate prior to collection of fluid from ascites, the Sp2/0
cells were injected into the peritoneal cavity of BALB/c mice
(male, n=3) and allowed to proliferate prior to collection of fluid
from ascites as negative control. The fluid was centrifuged (12,000
× g, 10 min; 4°C) and the supernatant purified using a
Protein A-Sepharose (GalaxyBio, Beijing, China). The concentration
of purified mAb was measured by a spectrophotometer, and it was
used as the source of the 2D8 hFGF-21 mAb used in the present
study. The mAb was further analyzed using SDS-PAGE (resolving gel,
15%; stacking gel, 5%; 1 h at 200 V; 10 µl sample; 10 ng). The
class and subclass of the mAb was determined using the mouse mAb
isotyping kit.
Titer measurements
With the exception of to the purified mAbs (gradient
dilution from 250×20-250×213; 100 µl)
replaced with hybridomas cultures (100 µl) as primary antibodies,
(purified Sp2/0 ascites replaced hybridomas cultures as negative
control), the other operating methods were the same as indirect
ELISA, as previously described. Each sample was tested in
triplicate and the results presented as the mean ± standard
deviation. The maximum dilution ratio of the positive well value
and negative control greater than or equal to 2.1 is the titer of
the mAb.
Western blot analysis
The hFGF-21 protein and negative control (BSA) were
used analyzed by western blot. Once the quality of hFGF-21 and BSA
were consistent, samples (10 ng) were loaded onto a 12% gel,
subjected to SDS-PAGE and blotted on to a polyvinylidene difluoride
membrane. The membrane was blocked with 5% skimmed milk in
tris-buffered saline with Tween-20 (TBST; 10 mmol/l pH 7.5
Tris-HCl; 150 mmol/l NaCl) at 4°C overnight. Following washing
three times for 15 min in TBST, the membrane was incubated with the
mAb (1:500 dilution) for 1 h at room temperature. Following washing
(as described above), the membrane was incubated with
HRP-conjugated goat anti-mouse secondary antibody (1:7,500
dilution) at room temperature for 1 h. The membrane was then washed
again (as described above) and blots were visualized by Super
Signal West Femto Maximum Sensitivity Substrate (34095; Thermo
Fisher Scientific, Inc.) and SmartChem™ II image analysis system
(SG2011060; Sagecreation, China).
Analysis of cross-reactivity
Indirect ELISA was performed to determine the
cross-reactivity between hFGF-21 and mouse FGF-21 (mFGF-21)
produced in our laboratory (16). In
the ELISA, 20 µg/ml of hFGF-21, mFGF-21 or BSA (negative control)
were used as the coating antigen and 2.5 µg/ml of the mAb (100 µl)
was used as the primary antibody. All other steps of the ELISA were
as previously described. All assays were performed in triplicate
and the results presented as the mean ± standard deviation.
APEx bacterial display analysis to
identify the anti-hFGF-21 mAb epitope
Anchored periplasmic expression (APEx) bacterial
display vectors containing the new lipoprotein A (NlpA) sequence
(CDQSSS) plus 6 amino acids, obtained via previous studies
published by our laboratory (17–19).
hFGF-21 sequence was obtained from our previous study (2).
For the first round of screening, the hFGF-21
polypeptide sequence was divided into 2 segments that included
polymerase chain reaction upper sequence (PU: 1–91 amino acids
upstream of hFGF-21) and polymerase chain reaction downstream
sequence (PD: 92–181 amino acids downstream of hFGF-21), and then
cloned (primers described in Table
I) into the APEx. The cloning vector was constructed from the
secretory expression vector pET27b(+) as previously described
(20). The vectors formed (APEx-PU
and APEx-PD) were analyzed using the restriction enzyme Sfi
I, and 1% gel analysis was performed to determine the size of PU
and PD.
Vectors were transformed into DH5α E. coli,
which were cultured in LB media (Amp 100 µg/ml; 37°C for 4 h). When
the optical density (OD) reading of the culture at 600 nm
(OD600) reached between 0.3 and 0.4, 0.25 mmol/l of
isopropyl-β-D-thiogalactoside (IPTG) was added. The E. coli
were grown at 37°C for 4 h to induce the expression of PU or PD.
Then, bacterial cells (1 ml) were collected, centriguged (12,000 ×
g; 5 min; 4°C), washed twice with PBS and resuspended in 350
µl of ice-cold suspension solution (0.75 mol/l sucrose, 0.1 mol/l
Tris-HCl pH 8.0) supplemented with 10 mg/ml lysozyme (35 µl;
Shanghai Yi Sheng Biotechnology Co., Ltd., Shanghai, China). Cells
were treated with 1 mmol ice-cold EDTA (700 µl) and 0.5 mol
MgCl2 (50 µl), and centrifuged again as previously
described. Following this, cell pellets were centrifuged as
previously described, resuspended in 90 µl PBS, and incubated with
2 mg/ml FITC-labeled anti-hFGF-21 mAb (4 µl) and 10 µl 1% BSA at
4°C for 1 h. Then, the cells were washed three times with 0.5%
PBS-Tween and resuspended in 500 µl of PBS for
fluorescence-activated cell sorting (FACS) analysis at 488 nm. The
negative control was treated the same, except that it was not
incubated with the FITC-labeled mAb.
For the second round of screening, the PD was
divided into 5 overlapping segments, which included PD1
(amino acids 92–121 of hFGF-21), PD2 (amino acids
122–151 of hFGF-21), PD3 (amino acids 152–181 of
hFGF-21), PD4 (amino acids 107–136 of hFGF-21) and
PD5 (amino acids 137–166 of hFGF-21). The 5 segments
were cloned and cloned into the APEx bacterial display vector to
produce APEx-PD1, APEx-PD2,
APEx-PD3, APEx-PD4 and APEx-PD5.
The steps described in the first screening were then performed.
3T3-L1 stimulation with hFGF-21 and
anti-hFGF-21 mAb
3T3-L1 adipocytes (1×106 per ml) were
seeded into 96-well plates and incubated at 37°C in a 5%
CO2, and 95% humidified atmosphere overnight. Then, the
cells were starved for 12 h in serum-free DMEM, followed by
stimulation with hFGF-21 (1,000 nmol/l) and anti-hFGF-21 mAb (0.0,
0.075, 0.15, 0.3, 0.6 or 1.2 µg/ml) for 24 h. Controls were not
stimulated.
Glucose uptake assay
Glucose uptake of 3T3-L1 adipocytes was measured
following stimulation using the glucose uptake assay kit, according
to the manufacturer's instructions. Absorbance was recorded at 490
nm and the glucose consumption rate was calculated. Each sample was
tested in triplicate and the results presented as the mean ±
standard deviation.
Measuring GLUT1 mRNA expression
RNA isolation and reverse transcription (RT)
Following stimulation of 3T3-L1 adipocytes (as
described above), total RNA was isolated using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions. Total RNA was subsequently converted
to cDNA using 10 µl oligo-dT primers (500 µg/ml) and 10 µl dNTP (10
mmol/l each) in 100 µl ddH2O at 65°C for 5 min, followed
by cycling at 42°C for 50 min with 40 µl 5X first-strand buffer, 20
µl 0.1 MDTT, 10 µl RNase inhibitor (40 U/µl) and 10 µl M-MLV
reverse transcriptase (200 U/µl). The reaction was terminated by
heating at 70°C for 15 min.
Quantitative PCR (qPCR)
qPCR was performed using the Thermal Cycler Dice
Real Time PCR System (Takara Biotechnology Co., Ltd., Dalian,
China), according to the manufacturer's instructions. The PCR
reaction (20 µl final volume) contained 10 µl SYBR Premix Ex Taq
(2X), 2 µl cDNA (as described above), 0.4 µl of each primer
(Table II, 10 µmol/l), 0.4 µl ROX
Reference Dye II (50X) 25 µmol and 6.8 µl of double distilled
H2O. PCR was then performed as follows: 95°C for 10 sec;
40 cycles of 95°C for 5 sec; and 60°C for 34 sec. Each sample was
tested in triplicate. The mean cycle quantification (Cq) was
calculated for GLUT1 and β-actin. The quantity of GLUT1 mRNA copies
was normalized to β-actin [ΔCq = Cq (GLUT1) - Cq (β-actin)]. The
ΔΔCq value was calculated as: ΔΔCq = ΔCq (stimulated sample) - ΔCq
(non-stimulated sample). The relative quantity of GLUT1 mRNA copies
in the stimulated sample compared with the non-stimulated sample
was calculated using the 2−ΔΔCq method (21). The results were presented as the mean
± standard deviation.
Statistical analysis
Data was analyzed using a two-tailed Student's
t-test. All analyses were performed using SPSS software (version
13.0; SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
Preparation and characterization of
the anti-hFGF-21 mAb (clone 2D8)
Secretory hybridoma cells were cloned, followed by
recloning three times to increase monoclonality of the produced
immunoglobulins (Igs). One stable hybridoma, designated 2D8 was
obtained. The concentration of the mAbs produced following
purification was 2.3 mg/ml. As expected, the heavy chain of the
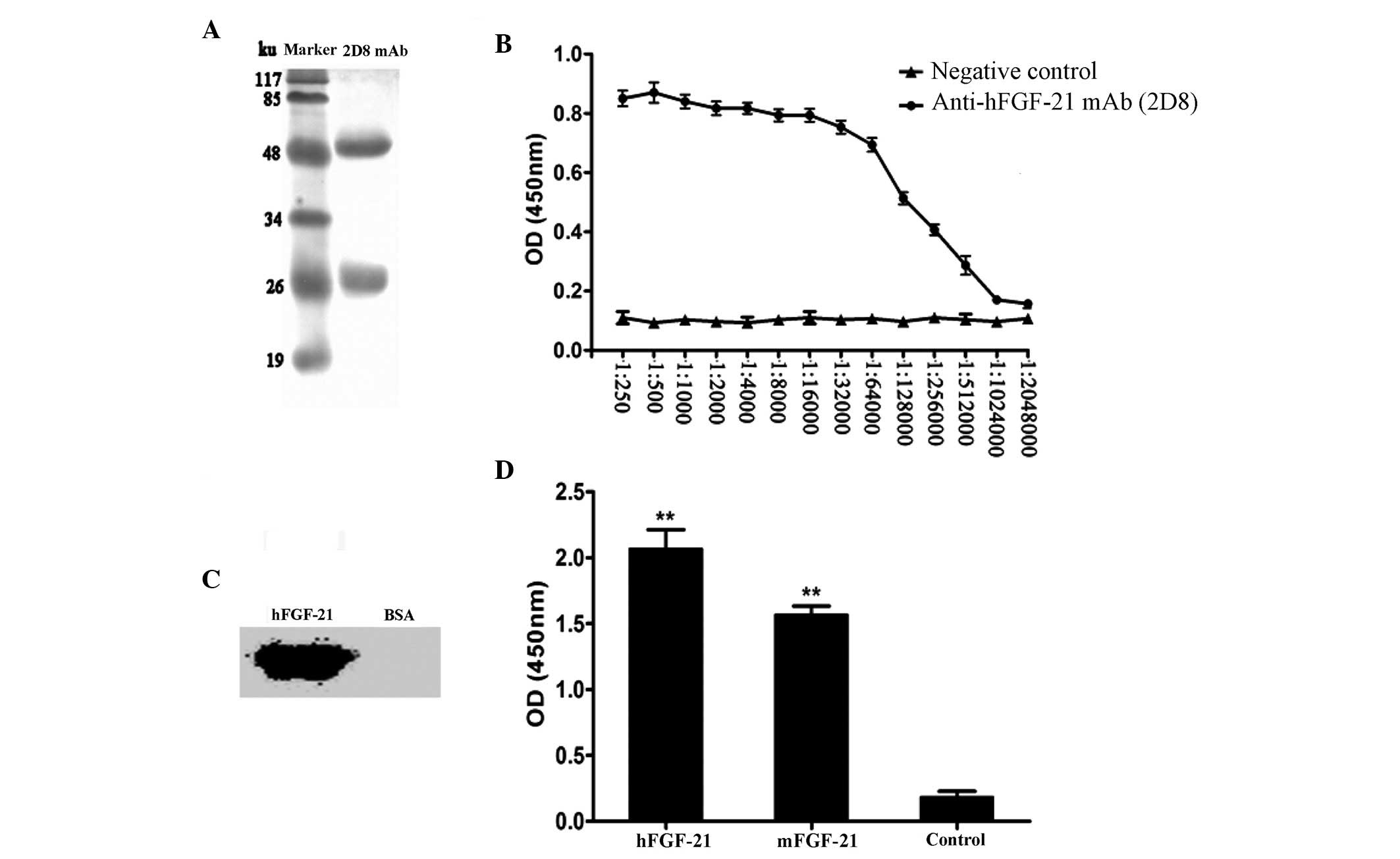
mAbs was 50 ku, and light chain was 25 ku (Fig. 1A). The isotype of the mAb was
determined to be IgG2b. The titer of the mAb was
1:1.024×106 (Fig. 1B).
Western blot analysis revealed that hFGF-21 could be detected by
the mAb, whereas the negative control (BSA) had no reaction with
the mAb, indicating good specificity (Fig. 1C). Cross-reactivity analysis
identified that compared with the negative control (BSA), the mAb
reacted significantly with mFGF-21 and hFGF-21 (P<0.01; Fig. 1D), which have ~80% amino acid
sequence homology (16). The
reaction with hFGF-21 reaction was stronger compared with the
mFGF-21 reaction (Fig. 1D), which is
consistent with previous published data (16).
Identification of the anti-hFGF-21 mAb
(clone 2D8) epitope
Clones of different hFGF-21 segments showed that PU
and PD were both 273 base pairs (bp) in length (Fig. 2A), and that the 5 different segments
of PD were all 90 bp (Fig. 2B). The
results of restriction enzyme analysis identified that each segment
was successfully cloned into the APEx bacterial display vector
(Fig. 2C and D). The first round of
FACS (Fig. 3A-C) showed that,
compared with the negative control (Fig.
3A), the mAb could not bind to the PU segment (Fig. 3B), whereas it could strongly bind to
the PD segment (Fig. 3C), which
suggests that the epitope the mAbs recognizes is in the PD segment.
The second round of FACS (Fig. 3D-I)
determined that, compared with negative control (Fig. 3D), the mAb could not bind to
PD2, PD3 or PD5 (Fig. 3F, G and I, respectively), whereas it
could strongly bind to PD1 and PD4 (Fig. 3E and H). This revealed that the
epitope that the mAb binds to was in the region overlapping the
PD1 and PD4 segments, a specific fifteen
amino acid sequence (YQSEAHGLPLHLPGN).
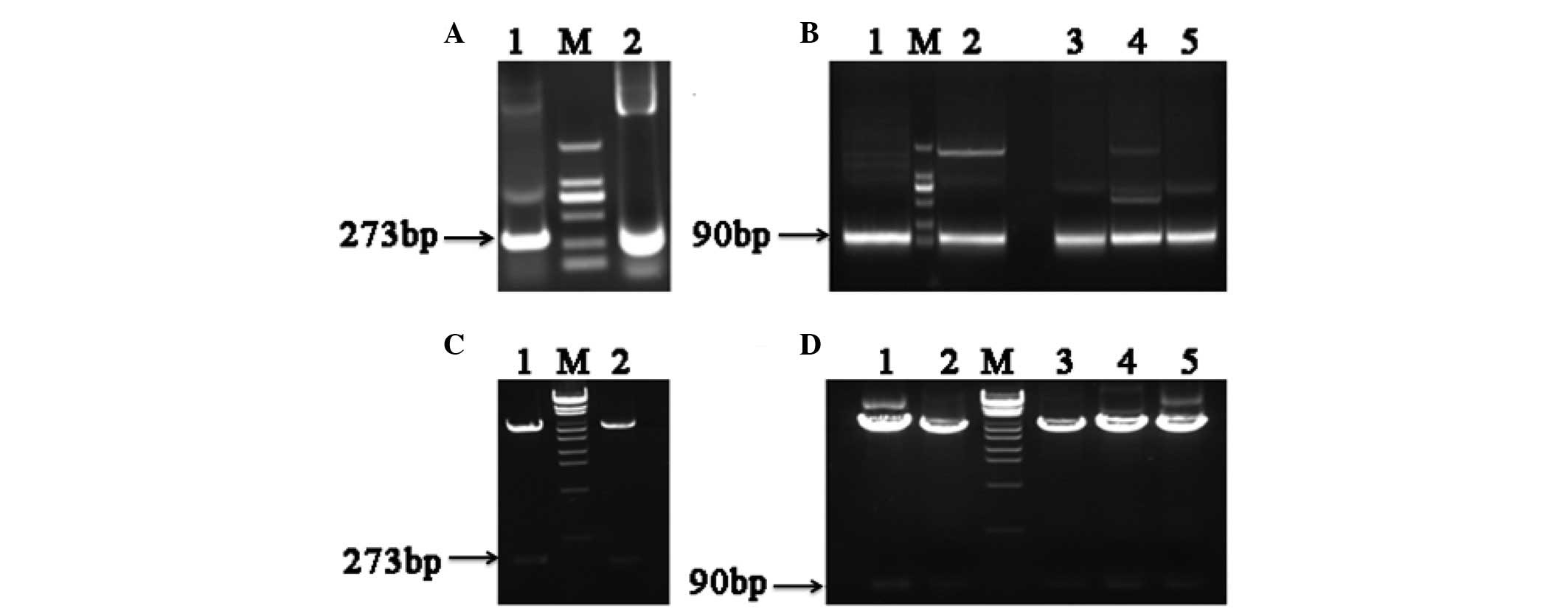 | Figure 2.Electrophoretic analysis of
polymerase chain reaction (PCR) human fibroblast growth factor-21
(hFGF-21) segment products and restriction enzyme analysis of
anchored periplasmic expression (APEx) bacterial display vectors.
(A) Electrophoretic analysis of PCR products. 1, polymerase chain
reaction upper sequence PCR product; M, DNA marker; 2, polymerase
chain reaction downstream sequence PCR product. (B) Electrophoretic
analysis of PCR products. 1, PD1 product; M, DNA marker;
2, PD2 product; 3, PD3 product; 4,
PD4 product; 5, PD5 product. (C) Restriction
enzyme analysis. 1, APEx-PU; M, λEcoT14 DNA marker; 2,
APEx-PD. (D) Restriction enzyme analysis. 1, APEx-PD1;
2, APEx-PD2; M, λEcoT14 DNA marker; 3,
APEx-PD3; 4, APEx-PD4; 5,
APEx-PD5. PU, 1–91 amino acids upstream of hFGF-21; PD,
92–181 amino acids downstream of hFGF-21. Bp, base pairs. |
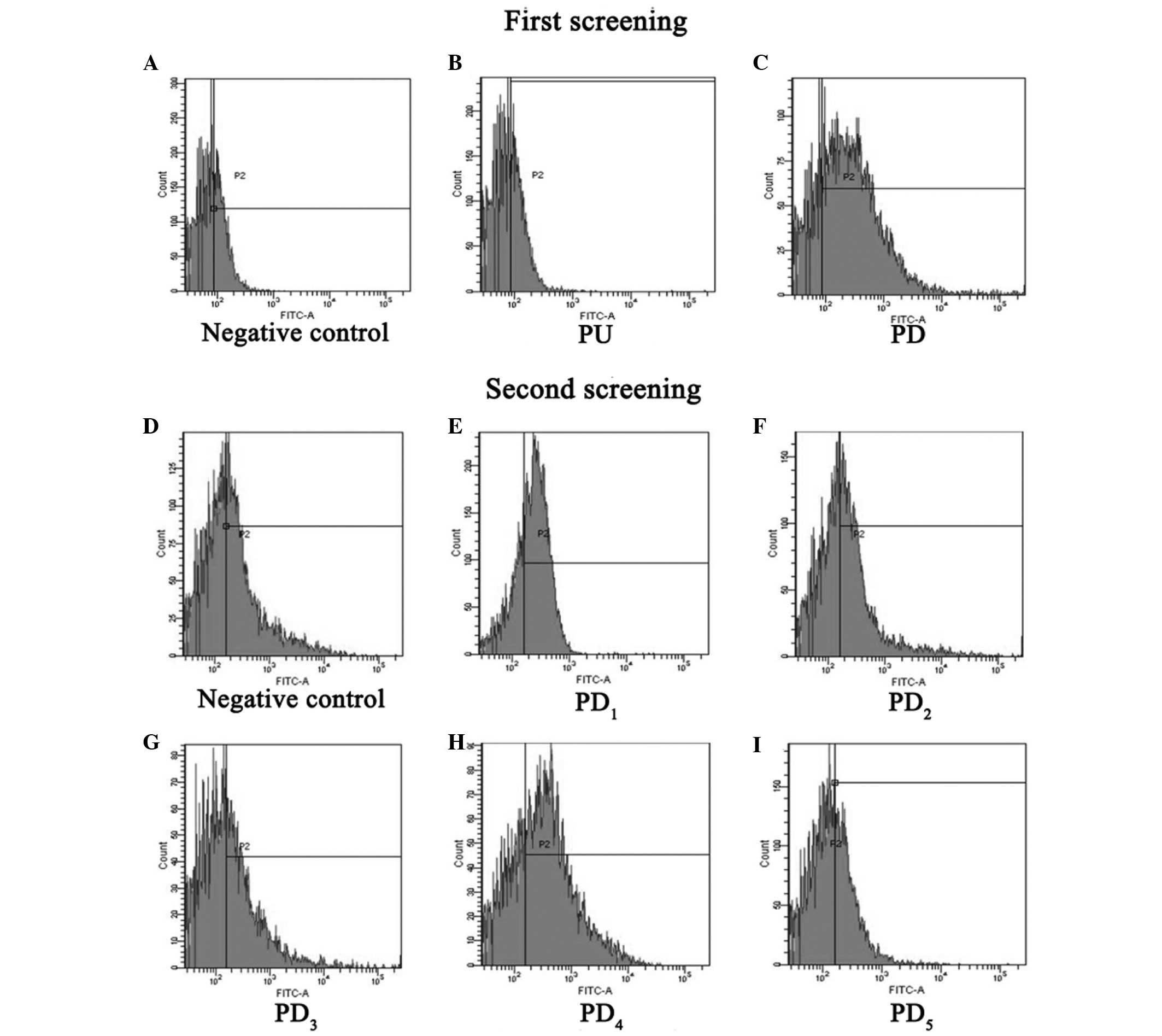 | Figure 3.Epitope screening by
fluorescence-activated cell sorting analysis. The first screening
included the a (A) negative control, (B) PU and (C) PD. The second
screening included a (D) negative control, (E) PD1, (F)
PD2, (G) PD3, (H) PD4 and (I)
PD5. PU, 1–91 amino acids upstream of hFGF-21; PD,
92–181 amino acids downstream of hFGF-21; FITC, fluorescein
isothiocyanate; PU, polymerase chain reaction upper sequence; PD,
polymerase chain reaction downstream sequence. |
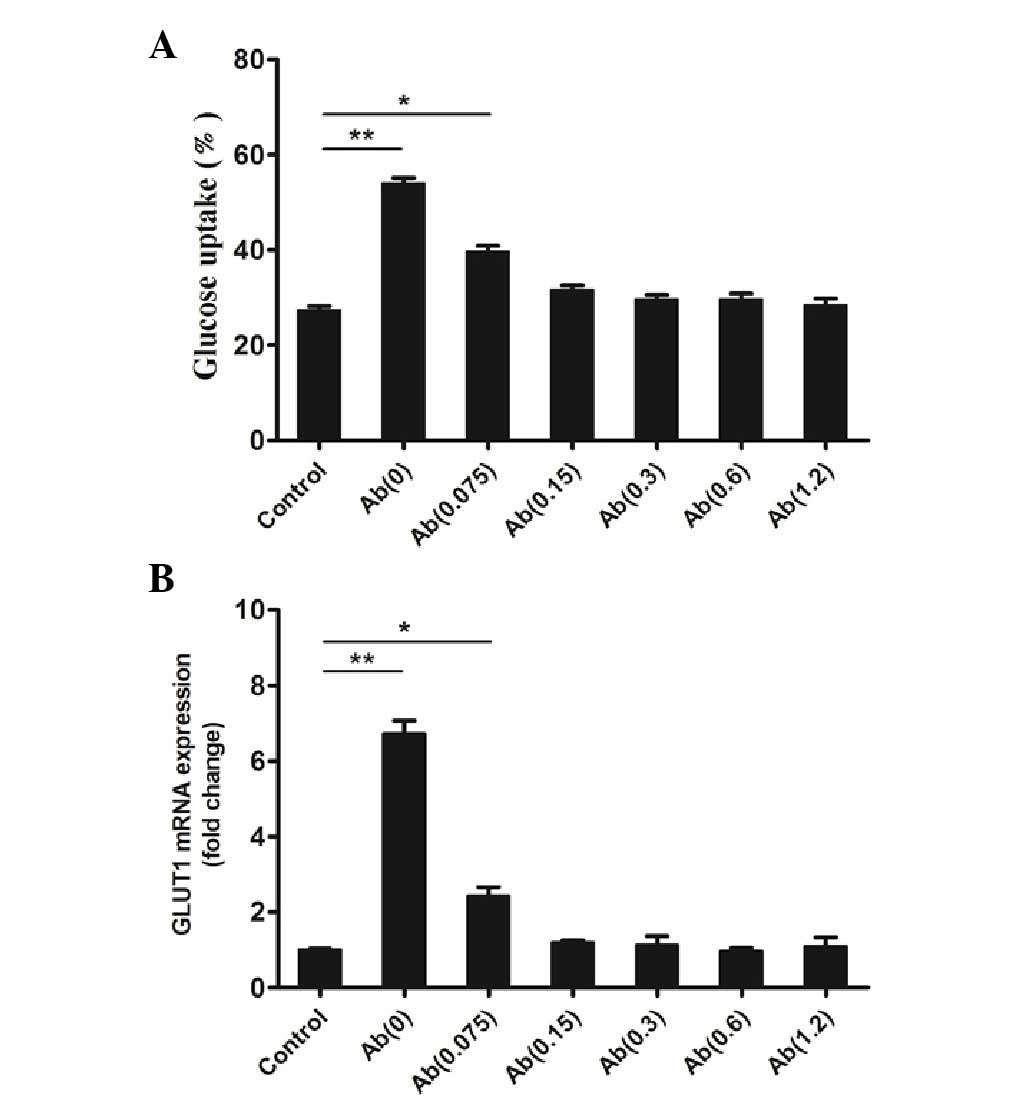
Glucose uptake assay
In the glucose uptake assay a control group (without
hFGF-21 and the mAb) and 6 groups stimulated with hFGF-21 (1,000
nmol/l) and different concentrations of the mAb were tested.
Results of the assays determined that, compared with the control,
the mAb (0.0 µg/ml) and hFGF-21 group had significantly increased
glucose uptake (P<0.01; Fig. 4A),
which is consistent with the results of a previous study (1). The mAb (0.075 µg/ml) and hFGF-21 group
also had a significantly increased glucose uptake compared with the
control (P<0.05; Fig. 4A).
However, the mAb (0.15, 0.3, 0.6 or 1.2 µg/ml) and hFGF-21 groups
did not have increased glucose uptake compared with the mAb(0)
group (Fig. 4A), which indicates
that the mAb reduces hFGF-21-dependent glucose uptake in
vitro.
GLUT1 mRNA expression
GLUT1 mRNA expression was quantified in a control
group (without hFGF-21 and the mAb) and in 6 groups stimulated with
hFGF-21 (1,000 nmol/l) and different concentrations of the mAb. The
results of qPCR analysis identified that, compared with the
control, the mAb (0.0 µg/ml) and hFGF-21 group had significantly
increased GLUT1 mRNA expression (P<0.01; Fig. 4B), which is consistent with the
results of a previous study (1). The
mAb (0.075 µg/ml) and hFGF-21 group also had significantly
increased GLUT1 mRNA expression (P<0.05; Fig. 4B). However, mAb (0.15, 0.3, 0.6 or
1.2 µg/ml) and hFGF-21 groups did not have increased GLUT1 mRNA
expression (Fig. 4B). This indicates
that the mAb reduces hFGF-21-dependent GLUT1 mRNA expression in
vitro compared with the Ab(0) group.
Discussion
An anti-hFGF-21 2D8 mAb was prepared and purified in
the present study. Bacterial display technology was used to screen
for the epitope recognized by the mAb. Harvey et al
(20) first reported the NlpA-based
bacterial display technology. Our APEx bacterial display vectors
were obtained from our laboratory previous studies (17–19).
Different hFGF-21 segments were anchored to the inner membrane of
the host cell, in order to allow screening for the epitope that
anti-hFGF-21 mAb (2D8) recognizes. Bacterial cells are the host of
choice for expression of recombinant proteins, due to their fast
expression foreign genes, rapid growth rate and ease of genetic
manipulation (20,22–26).
APEx-FACS enables real-time visualization of mAb-peptide binding,
which allowed identification of the epitope that the anti-hFGF-21
mAb recognized. The anti-hFGF-21 mAb-binding ability of different
hFGF-21 segments could be visualized via the FITC-label on the mAb
during FACS. In addition, the intensity of fluorescence during FACS
reflected the binding affinity. The segments PD1 and
PD4 showed high mAb-binding ability, therefore, the
anti-hFGF-21 mAb epitope was located in the region overlapping both
segments. Compared with traditional method, the APEx-FACS reduced
the time taken to screen for epitopes, reduced costs and was thus
more efficient (17). Numerous
previous studies have shown that hFGF-21 serves an important role
in metabolism, for example, hFGF-21 has been shown to activate the
coreceptor β-Klotho (27–29). The present study identified that the
epitope of the anti-hFGF-21 mAb was close to the binding sites of
the coreceptor β-Klotho and hFGF-21. Therefore, the results of the
current study suggest an interaction between hFGF-21 and the
coreceptor β-Klotho.
The results of the in vitro glucose uptake
bioactivity of the mAb indicated that the mAb decreased
hFGF-21-dependent glucose uptake compared with the Ab(0) in 3T3-L1
adipocytes. To determine a potential molecular mechanism for this
effect of the mAb, its effect on hFGF-21-dependent GLUT1 mRNA
expression in 3T3-L1 adipocytes was investigated. As expected,
compared with the Ab(0) hFGF-21-dependent GLUT1 upregulation was
reduced with increasing concentrations of the mAb, suggesting that
this is the mechanism through which the mAb decreased glucose
uptake. These assays provided evidence that the mAb had
hFGF-21-neutralizing activity.
Therapeutic antibodies, including those for cancer,
autoimmune diseases and viral infections, are widespread.
Therapeutic effect of the humanized mAb (Palivizumab) to treat
respiratory syncytial virus infection was confirmed a decade ago
(30). The anti-hFGF-21 mAb (clone
2D8) prepared and analyzed in the present study has potential as a
novel clinical treatment for hFGF-21-related diseases. Therefore,
the current study provides a solid foundation for future research
and development of the humanized anti-FGF-21 mAb (clone 2D8).
In conclusion, the present study prepared and
characterized an anti-hFGF-21 mAb (2D8), which will provide a novel
tool for future research investigating the role that hFGF-21 serves
in metabolism. The epitope of the mAb was identified as a specific
fifteen amino acid sequence (YQSEAHGLPLHLPGN). In addition,
compared with the Ab(0) the mAb was found to reduce
hFGF-21-dependent glucose uptake by decreasing hFGF-21
dependent-GLUT1 mRNA expression in 3T3-L1 adipocytes, which
demonstrated that the mAb had hFGF-21-neutralizing activity. This
study, to the best of our knowledge, is the first to identify the
epitope of anti-hFGF-21 mAb (2D8) and study its bioactivity in
vitro. The 2D8 anti-hFGF mAb described in the present study has
potential as a therapeutic agent and further research should be
conducted to investigate the this application.
Acknowledgements
The present study was supported by The National
Natural Science Foundation of China Youth Science Project Fund
(grant no. 31001084).
References
|
1
|
Kharitonenkov A, Shiyanova TL, Koester A,
Ford AM, Micanovic R, Galbreath EJ, Sandusky GE, Hammond LJ, Moyers
JS, Owens RA, et al: FGF-21 as a novel metabolic regulator. J Clin
Invest. 115:1627–1635. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hou YT, Li JN, Ren GP, Liu MY, Sun GP,
Wang WF and Li DS: Cloning, expression and glucose regulation
activity of human FGF-21. Yi Chuan. 32:583–587. 2010.(In Chinese).
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Adams AC and Kharitonenkov A: FGF21: The
center of a transcriptional nexus in metabolic regulation. Curr
Diabetes Rev. 8:285–293. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gälman C, Lundåsen T, Kharitonenkov A,
Bina HA, Eriksson M, Hafström I, Dahlin M, Amark P, Angelin B and
Rudling M: The circulating metabolic regulator FGF21 is induced by
prolonged fasting and PPARalpha activation in man. Cell Metab.
8:169–174. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Badman MK, Pissios P, Kennedy AR, Koukos
G, Flier JS and Maratos-Flier E: Hepatic fibroblast growth factor
21 is regulated by PPARalpha and is a key mediator of hepatic lipid
metabolism in ketotic states. Cell Metab. 5:426–437. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Inagaki T, Dutchak P, Zhao G, Ding X,
Gautron L, Parameswara V, Li Y, Goetz R, Mohammadi M, Esser V, et
al: Endocrine regulation of the fasting response by
PPARalpha-mediated induction of fibroblast growth factor 21. Cell
Metab. 5:415–425. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Moyers JS, Shiyanova TL, Mehrbod F, Dunbar
JD, Noblitt TW, Otto KA, Reifel-Miller A and Kharitonenkov A:
Molecular determinants of FGF-21 activity-synergy and cross-talk
with PPARgamma signaling. J Cell Physiol. 210:1–6. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xiao Y, Xu A, Law LS, Chen C, Li H, Li X,
Yang L, Liu S, Zhou Z and Lam KS: Distinct changes in serum
fibroblast growth factor 21 levels in different subtypes of
diabetes. J Clin Endocrinol Metab. 97:E54–E58. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen WW, Li L, Yang GY, Li K, Qi XY, Zhu
W, Tang Y, Liu H and Boden G: Circulating FGF-21 levels in normal
subjects and in newly diagnose patients with Type 2 diabetes
mellitus. Exp Clin Endocrinol Diabetes. 116:65–68. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mashili FL, Austin RL, Deshmukh AS, Fritz
T, Caidahl K, Bergdahl K, Zierath JR, Chibalin AV, Moller DE,
Kharitonenkov A and Krook A: Direct effects of FGF21 on glucose
uptake in human skeletal muscle: Implications for type 2 diabetes
and obesity. Diabetes Metab Res Rev. 27:286–297. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stein S, Bachmann A, Lössner U, Kratzsch
J, Blüher M, Stumvoll M and Fasshauer M: Serum levels of the
adipokine FGF21 depend on renal function. Diabetes care.
32:126–128. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dushay J, Chui PC, Gopalakrishnan GS,
Varela-Rey M, Crawley M, Fisher FM, Badman MK, Martinez-Chantar ML
and Maratos-Flier E: Increased fibroblast growth factor 21 in
obesity and nonalcoholic fatty liver disease. Gastroenterology.
139:456–463. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang X, Yeung DC, Karpisek M, Stejskal D,
Zhou ZG, Liu F, Wong RL, Chow WS, Tso AW, Lam KS and Xu A: Serum
FGF21 levels are increased in obesity and are independently
associated with the metabolic syndrome in humans. Diabetes.
57:1246–1253. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li L, Yang G, Ning H, Yang M, Liu H and
Chen W: Plasma FGF-21 levels in type 2 diabetic patients with
ketosis. Diabetes Res Clin Pract. 82:209–213. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ding LJ, Li JC, Fu R, Zhou YJ and Huo GC:
Preparation and characteristic identification of monoclonal
antibody against sulfamethazine. Journal of Northeast Agricultural
University (English edition). 13:145–148. 2006.
|
|
16
|
Jiang YY, Liu MY, Ren GP, Wang WF, Liu XM
and Li DS: Cloning, expression and purification of mouse fibroblast
growth factor-21 and its function in adipocyte glucose metabolism.
Sheng Wu Hua Xue Yu Sheng Wu Wu Li Jin Zhan Bian Ji Bu. 36:157–164.
2009.(In Chinese).
|
|
17
|
Guo M, Xu LM, Zhou B, Yin JC, Ren GP and
Li DS: A novel efficient method for B cell epitopes mapping base on
bacterial display. Zhongguo Mianyixue Zazhi. 30:366–372. 2014.(In
Chinese).
|
|
18
|
Xu WJ, Zhang YJ, Li QQ, Wu Q, Yu XF, Guo
XC, Yin JC, Ren GP and Li DS: Biopharmaceutical Lab, College of
Life Science, Northeast Agricultural University: Systematic mapping
the linear antigenic domains of the porcine circovirus 2b Cap
protein by bacterial display technology. Zhong Guo Yu Fang Shou Yi
Xue Bao Bian Ji Bu. 38:398–402. 2016.(In Chinese).
|
|
19
|
Xu LM, Yin CK, Ren GP, Tian H, Wang XQ,
Ding LJ and Li DS: Establishment of bacterial display technology
for fab antibody library screening. Xi Bao Yu Fen Zi Mian Yi Xue Za
Zhi. 27:1090–1093. 2011.(In Chinese). PubMed/NCBI
|
|
20
|
Harvey BR, Georgiou G, Hayhurst A, Jeong
KJ, Iverson BL and Rogers GK: Anchored periplasmic expression, a
versatile technology for the isolation of high-affinity antibodies
from Escherichia coli-expressed libraries. Proc Natl Acad Sci USA.
101:9193–9198. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wingfield PT: Overview of the purification
of recombinant proteins produced in Escherichia coli. Curr Protoc
Protein Sci Chapter. 6:Unit 6.1. 2003.
|
|
23
|
Georgiou G: Analysis of large libraries of
protein mutants using flow cytometry. Adv Protein Chem. 55:293–315.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Daugherty PS, Olsen MJ, Iverson BL and
Georgiou G: Development of an optimized expression system for the
screening of antibody libraries displayed on the Escherichia coli
surface. Protein Eng. 12:613–621. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Daugherty PS: Protein engineering with
bacterial display. Curr Opin Struct Biol. 17:474–480. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rice JJ, Schohn A, Bessette PH, Boulware
KT and Daugherty PS: Bacterial display using circularly permuted
outer membrane protein OmpX yields high affinity peptide ligands.
Protein Sci. 15:825–836. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kurosu H, Choi M, Ogawa Y, Dickson AS,
Goetz R, Eliseenkova AV, Mohammadi M, Rosenblatt KP, Kliewer SA and
Kuro-o M: Tissue-specific expression of betaKlotho and fibroblast
growth factor (FGF) receptor isoforms determines metabolic activity
of FGF19 and FGF21. J Biol Chem. 282:26687–26695. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ogawa Y, Kurosu H, Yamamoto M, Nandi A,
Rosenblatt KP, Goetz R, Eliseenkova AV, Mohammadi M and Kuro-o M:
BetaKlotho is required for metabolic activity of fibroblast growth
factor 21. Proc Natl Acad Sci USA. 104:7432–7437. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kharitonenkov A, Dunbar JD, Bina HA,
Bright S, Moyers JS, Zhang C, Ding L, Micanovic R, Mehrbod SF,
Knierman MD, et al: FGF-21/FGF-21 receptor interaction and
activation is determined by betaKlotho. J Cell Physiol. 215:1–7.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang D, Cummins C, Bayliss S, Sandercock J
and Burls A: Immunoprophylaxis against respiratory syncytial virus
(RSV) with palivizumab in children: A systematic review and
economic evaluation. Health Technol Assess. 12(iii): ix-x. 1–86.
2008.
|