Introduction
Coronary heart disease (CHD) is typically caused by
arteriosclerosis, which is characterized by the narrowing or
blockage of coronary arteries (1).
CHD is a complex disease, and a variety of genetic and epigenetic
factors have been associated with an increased risk of CHD
(2–4). Aberrant methylation of cytosine
residues in gene CpG islands is an important molecular mechanism
involved in the regulation of gene expression in response to the
environment (5). Global DNA
hypomethylation has been described in the atherosclerotic human
aorta (6). DNA methylation studies
on CHD have primarily focused on genes related to estrogen
receptors (ESR1 and ESR2), the immune system (FOXP3, PLA2G7 and
MMP-9), lipid metabolism (ABCA1, KLF2 and LRP1), oxidative stress
(GSTP1, BNIP3 and EC-COD), blood coagulation (TM and P2Y12) and
genes on chromosome 9p21 (BAX, BCL-2 and TIMP3) (7). In addition, low promoter methylation
was found to account for increased expression of the
15-lipoxygenase (ALOX15) gene (8).
Furthermore, coagulation factor VII (F7) promoter hypomethylation
has been correlated with higher plasma expression levels of
activated coagulation factor VII (FVIIa), and has been shown to
contribute towards an increased risk of CHD (9).
The catechol-O-methyltransferase (COMT) gene,
located on chromosome 22q 11.21, encodes a regulatory enzyme,
catecholamine and other catechols (10,11).
COMT is able to transfer a methyl group to degrade dopamine,
catecholamine, epinephrine and norepinephrine (12). In addition, COMT is associated with
numerous human disorders, such as schizophrenia and bipolar
disorder (13,14). Furthermore, a number of studies have
revealed an association between COMT and metabolic syndromes, such
as hypertension and diabetes, that are closely associated with CHD
(15,16). COMT gene variation serves an
important role in the increased risk of acute coronary events
(17).
Hypomethylation of the COMT promoter has been
detected in patients with schizophrenia and bipolar disorder
(13,18). However, it is unknown whether
methylation of the COMT promoter contributes towards the risk of
CHD. In light of previous findings, the aim of the current study
was to assess the association between COMT gene promoter
methylation and the risk of CHD.
Materials and methods
Sample selection
In total, 48 cases of CHD (24 males and 24 females)
and 48 gender-matched non-CHD controls from a Han Chinese
population were selected from Ningbo First Hospital of Ningbo
University (Ningbo, China). The mean ages of the CHD cases and
non-CHD controls were 64.0 years (range, 50.0–85.0 years) and 62.2
years (range, 52.0–71.0 years), respectively. Unrelated individuals
were examined by coronary angiography and diagnosed by experienced
cardiologists. The classification details and inclusion criteria
were as previously described (19–21).
Peripheral blood samples were collected between June 2013 and June
2014. The current case-control study was approved by the Ethics
Committee of Ningbo First Hospital. Written informed consent was
obtained from each of the participants.
DNA methylation assay
Genomic DNA was collected from peripheral blood
samples using a nucleic acid extraction automatic analyzer (Lab-Aid
820; Zeesan Biotech Co., Ltd., Xiamen, China). DNA was quantified
using a PicoGreen double-stranded DNA Quantification kit (Molecular
Probes; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according
to the manufacturer's protocol. Pyrosequencing using PyroMark Gold
Q24 Reagent (Qiagen, Hilden, Germany) was conducted to identify the
methylation level of the promoter region of the COMT gene. Sodium
bisulfite was used as a chemical modifier of genomic DNA to convert
unmethylated cytosine to uracil. Polymerase chain reaction
(Pyromark PCR kit; Qiagen) was performed using a mixture of primers
designed using PyroMark Assay Design software, version 2.0 (Qiagen)
yielding a fragment of COMT gene promoter (103 bp). The PCR
reaction mixtures contained 10 µl ZymoTaq™ Premix (Zymo Research
Corporation, Irvine, CA, USA), 2 µl DNA template, 1.5 µl each
primer and 5 µl DNase/RNase-free water. PCR amplification was
conducted under the following conditions: 95°C for 10 min, followed
by 45 cycles of 95°C for 20 sec, 57.5°C for 20 sec and 72°C for 30
sec The primer sequences used were: Forward,
5′-GGGTTTTTGGGGTAGTTAG-3′; reverse,
5′-biotin-TAACCAACCCTCTCACCT-3′; and 5′-TTTGGGGTAGTTAGG-3′ for the
sequencing primer. The methylation rate of each CG site was
quantified and analyzed using the PyroMark Assay Design
software.
Statistical analysis
Statistical tests were analyzed using SPSS version
16.0 (SPSS, Inc., Chicago, IL, USA). An independent sample t-test
was applied to identify COMT methylation differences between CHD
cases and controls. A non-parametric approach was used for data
that could not be normalized. P<0.05 was considered to indicate
a statistically significant difference. The figures were created
using GraphPad Prism version 6 software (GraphPad Software, Inc.,
La Jolla, CA, USA) or R software (version 3.1; https://www.r-project.org/).
Results
Correlation between the DNA
methylation levels of the three CpG sites
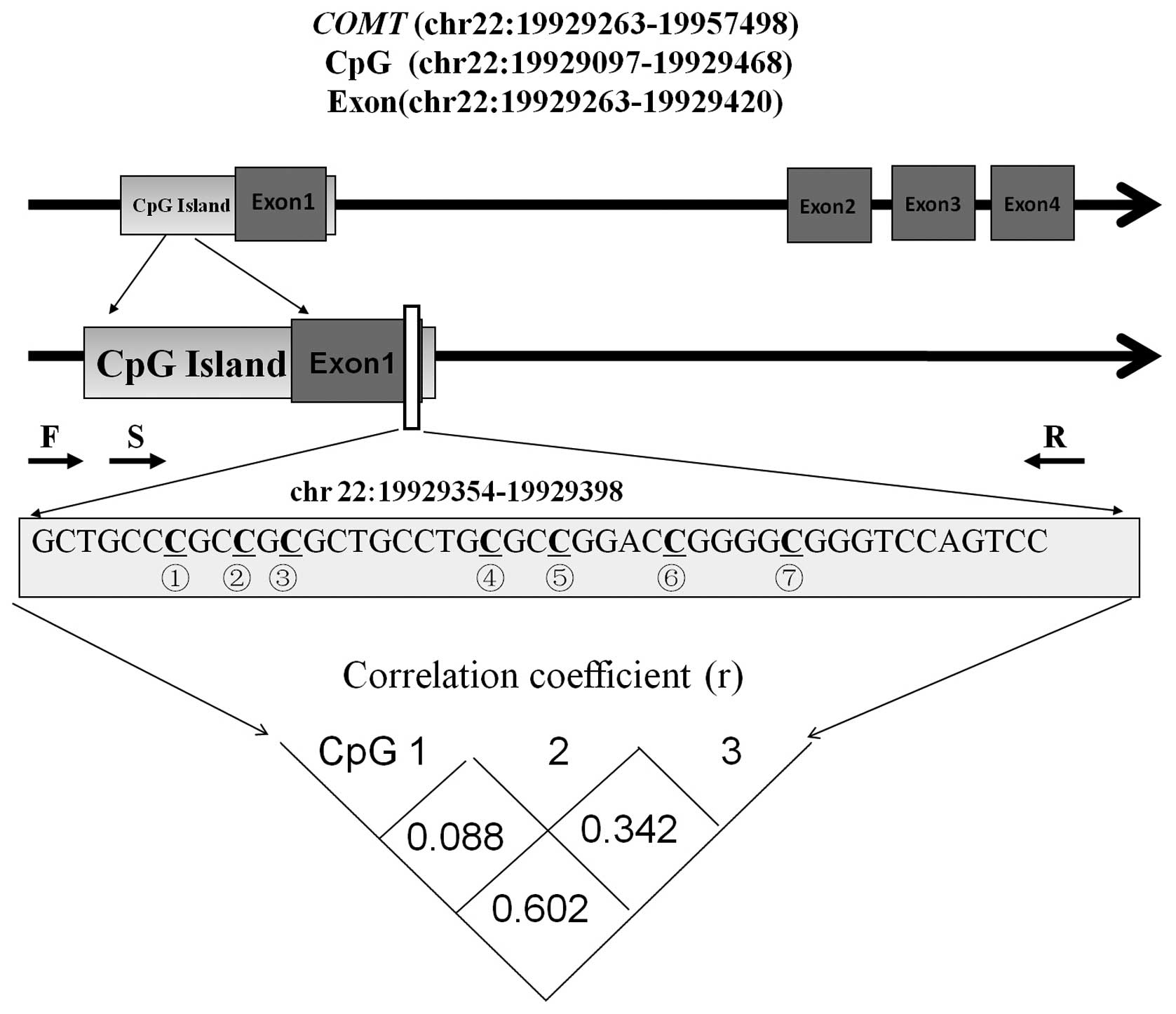
As shown in Fig. 1,
seven CpG sites were identified in the selected fragment
(chr22:19929354-19929398, Human GRCh37/hg19 Assembly) of the
promoter region of the COMT gene. However, due to sequencing
limitations, reliable methylation results were only available for
the three CpG sites preceding the fragment and, thus, these three
sites were chosen to represent promoter methylation of the COMT
gene. The results demonstrated that there was a moderate
correlation between the DNA methylation levels of the three CpG
sites (Fig. 1). Therefore, the three
CpG sites were tested separately in the subsequent association
analyses.
Comparison of the methylation levels
of the three CpG sites in CHD
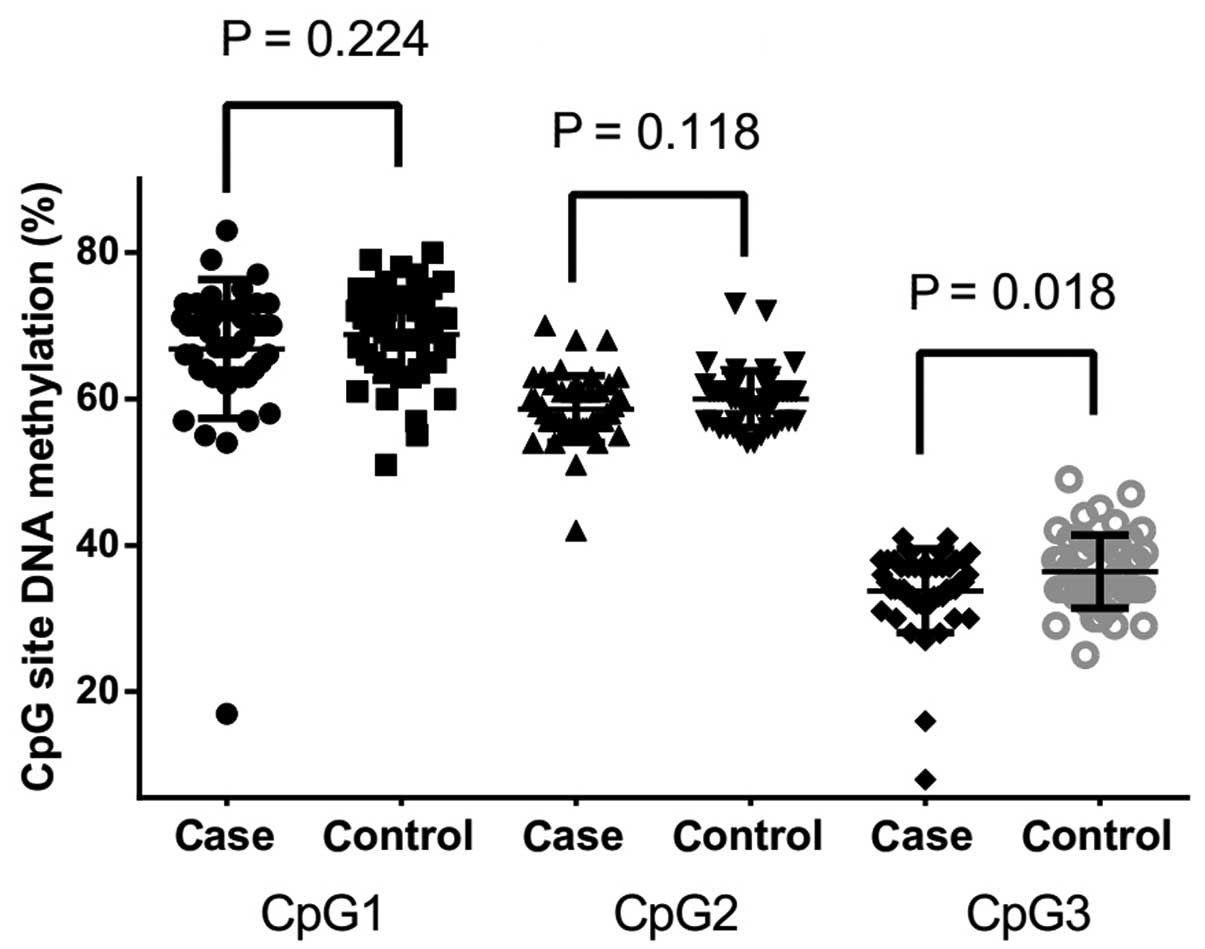
The follow-up case-control comparison showed that
there was a significantly lower level of CpG3 methylation in CHD
cases compared with non-CHD controls (Fig. 2; 33.77±5.71 vs. 36.42±5.00%;
P=0.018). However, no significant difference in the methylation
level was found between the cases and controls in CpG1 (Fig. 2; P=0.224) and CpG2 (Fig. 2, P=0.118). Further subgroup analysis
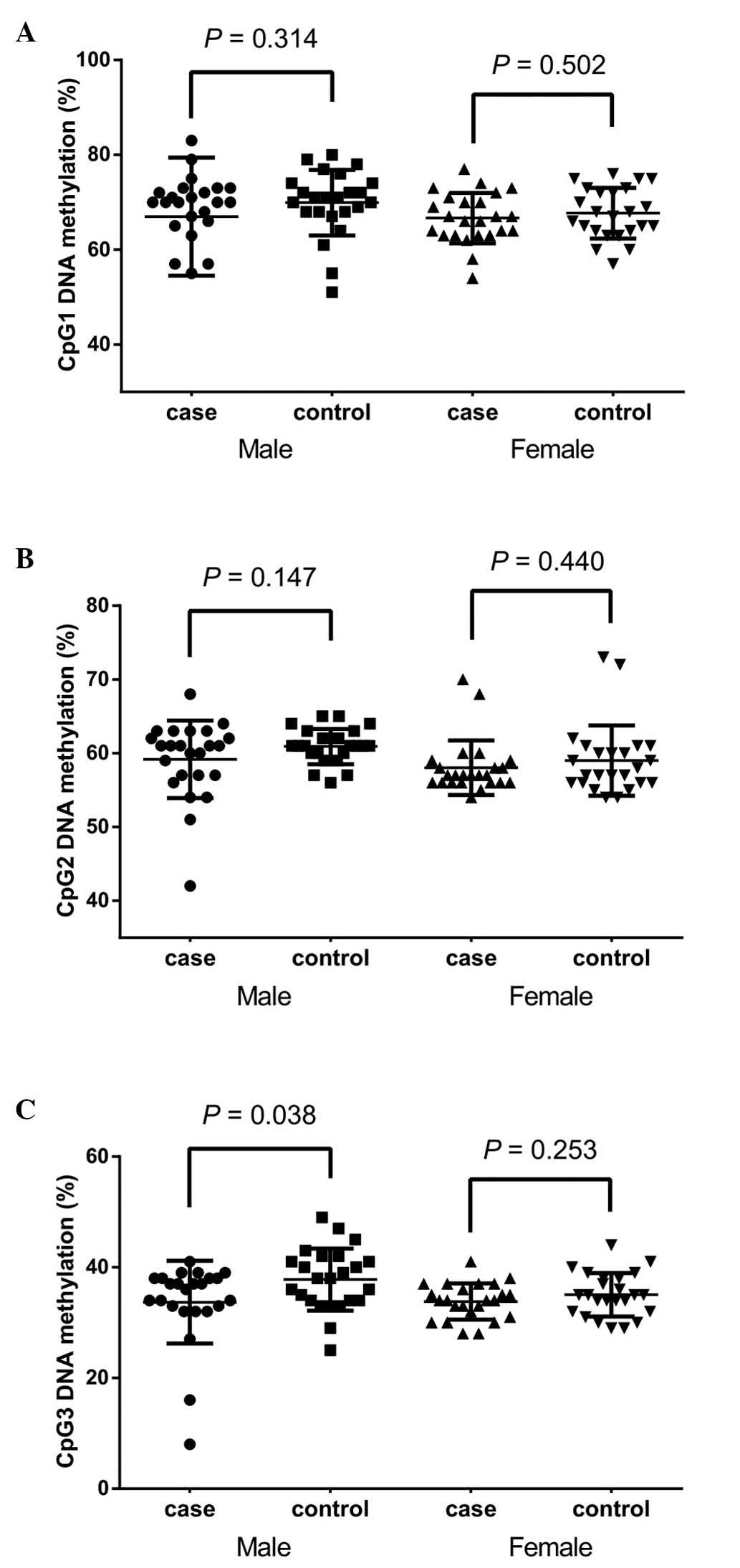
by gender (Fig. 3) suggested that
the significant association between CpG3 methylation was retained
in males (Fig. 3C; 33.71±7.49 vs.
37.79±5.61%; P=0.038) but not in females (Fig. 3C; P=0.253) at the CpG3 site.
Correlation between COMT and the three
CpG sites
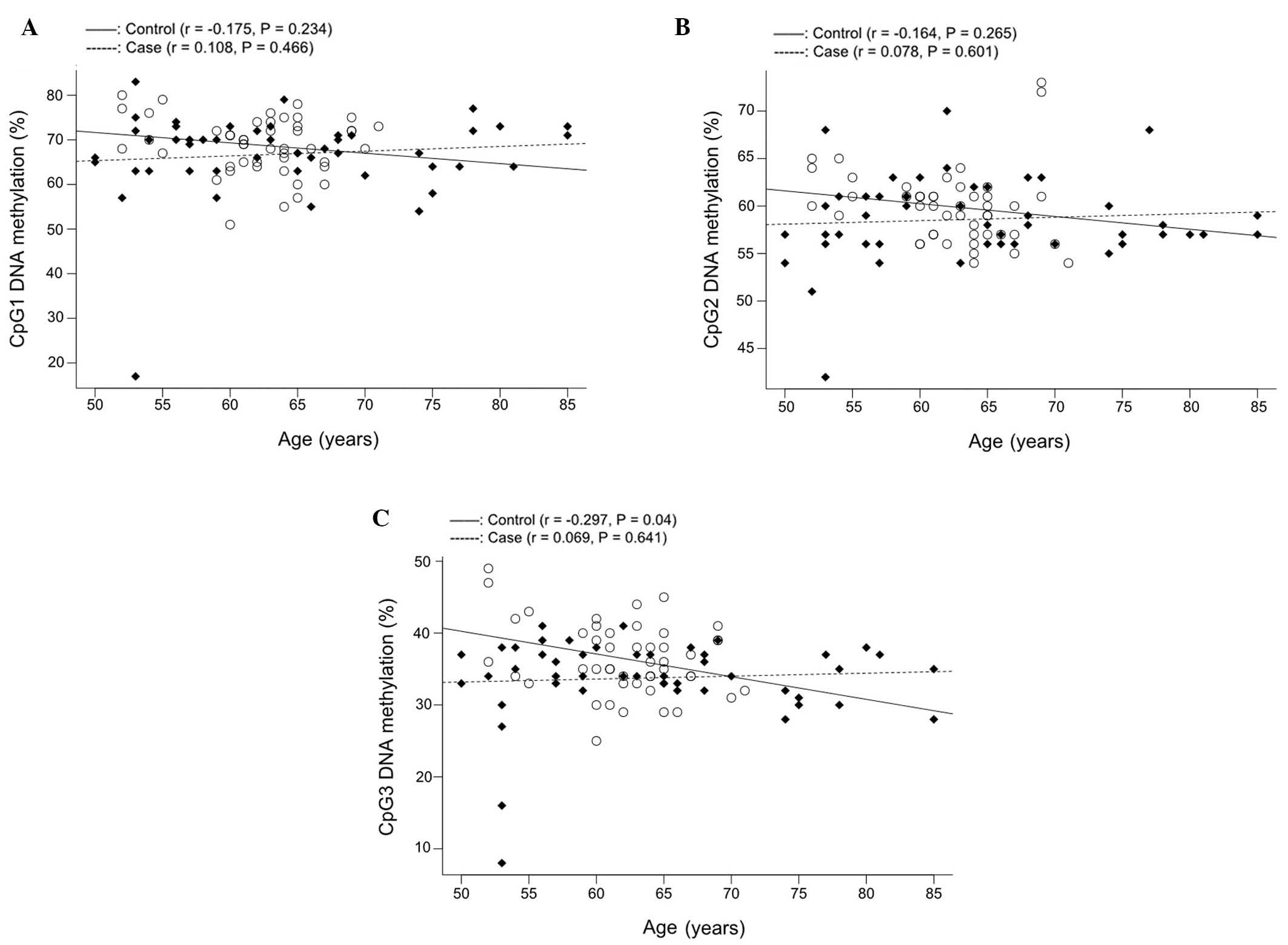
Further correlation tests (Fig. 4) demonstrated that COMT CpG3
methylation was associated with age in controls (Fig. 4C; r=−0.297, P=0.040) but not in CHD
cases. However, gender-stratified analysis in the control group did
not identify any significant association between males (P=0.072,
data not shown) or females (P=0.300, data not shown). Therefore,
additional studies are required to confirm this inverse correlation
in the non-CHD controls.
Discussion
Findings of the current study demonstrated that CHD
cases have lower levels of methylated COMT CpG3 compared with the
controls. Further breakdown analysis by gender showed that CpG3
methylation was significantly associated with CHD in males
(P=0.038) but not in females (P=0.253), suggesting a gender
disparity in the association between COMT methylation and CHD.
Additionally, the COMT CpG3 methylation level was inversely
associated with age in the controls but not in cases with CHD.
COMT encodes a methyltransferase that demethylates
s-adenosylmethionine to S-adenosyl-L-homocysteine, the immediate
precursor of homocysteine (22).
Therefore, COMT, as a homocysteine metabolism-mediated gene, has
been hypothesized to serve a vital role in the increased risk of
CHD (17). A low activity COMT
genotype has a protective effect against cardiovascular diseases
(23). The results in the present
study show significant promoter hypomethylation of COMT among CHD
cases, indicating a potential biomarker for predicting the
occurrence and development of CHD.
Gender disparity exists in the pathogenesis of CHD
(24). The current study identified
a significant difference in the association between CpG3
methylation and CHD with regards to males and females. However, as
there is no evidence suggesting that COMT serves an important role
in the metabolism of estrogen, further studies are required to
determine whether COMT interacts with other proteins associated
with the metabolism of estrogen (25).
Altered DNA methylation patterns are one of the
molecular mechanisms that underlie the phenotypic changes
associated with human aging (26).
In the present study, it was observed that COMT CpG3 methylation
levels was reduced with increasing age in controls. It can,
therefore, be hypothesized that COMT hypermethylation serves an
important protective role against cardiovascular diseases, although
the exact mechanisms underlying this role need to be investigated
in future studies.
The current study had a number of limitations.
Firstly, the sample size in the present study was relatively small.
Larger sample sizes and the inclusion of other ethnic populations
are required in order to confirm the findings. Secondly, COMT
methylation was measured in the DNA of the peripheral blood, which
included numerous types of cells, such as granulocytes and
lymphocytes, which potentially affect the specificity of the
methylation assay, as COMT methylation levels may vary between cell
types. Thirdly, only three CpG sites from a fragment of the gene
promoter region were selected to represent the entire COMT
promoter. Although the cases with CHD were gender matched with the
controls, the potential effects of other unknown environmental
factors on the findings regarding COMT methylation cannot be
excluded.
In conclusion, it was observed that COMT promoter
hypomethylation is associated with CHD in males. The results
contribute towards a better understanding of the role of COMT
methylation in the pathophysiology of CHD.
Acknowledgements
The present study was supported by the grants from
the National Natural Science Foundation of China (grant nos.
31100919 and 81371469), the Natural Science Foundation of Zhejiang
Province (grant no. LR13H020003) and the K.C. Wong Magna Fund from
Ningbo University.
References
|
1
|
Jia X, Li W, Miao Z, Feng C, Liu Z, He Y,
Lv J, Du Y, Hou M, He W, et al: Identification of modules related
to programmed cell death in CHD based on EHEN. BioMed Res Int.
2014:4753792014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ding H, yan F, Zhou LL, Ji XH, Gu XN, Tang
ZW and Chen RH: Association between previously identified loci
affecting telomere length and coronary heart disease (CHD) in Han
Chinese population. Clin Interv Aging. 9:857–861. 2014.PubMed/NCBI
|
|
3
|
Evans A, Van Baal GCM, McCarron P, DeLange
M, Soerensen TI, De Geus EJ, Kyvik K, Pedersen NL, Spector TD,
Andrew T, et al: The genetics of coronary heart disease: The
contribution of twin studies. Twin Res. 6:432–441. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kelpis TG, Anastasiadis K, Nimatoudis I,
Kelpi MG, Hadjimiltiades S and Papakonstantinou C: Prevalence of
‘distressed’ personality in patients with coronary artery disease
and its correlation with morbidity after coronary surgery. Hellenic
J Cardiol. 54:362–367. 2013.PubMed/NCBI
|
|
5
|
Tang L, Ye H, Hong Q, Wang L, Wang Q, Wang
H, Xu L, Bu S, Zhang L, Cheng J, et al: Elevated CpG island
methylation of GCK gene predicts the risk of type 2 diabetes in
Chinese males. Gene. 547:329–333. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yamada Y, Nishida T, Horibe H, Oguri M,
Kato K and Sawabe M: Identification of hypo- and hypermethylated
genes related to atherosclerosis by a genome-wide analysis of DNA
methylation. Int J Mol Med. 33:1355–1363. 2014.PubMed/NCBI
|
|
7
|
Ye H, Hong Q, Tang L, Zhou A, Jiang D, Li
Y, Dai D and Duan S: DNA methylation in coronary heart disease. Xi
Bao Sheng Wu Xue Za Zhi. 36:1422–1429. 2014.(In Chinese).
|
|
8
|
Hiltunen MO, Tuomisto TT, Niemi M, Bräsen
JH, Rissanen TT, Törönen P, Vajanto I and Ylä-Herttuala S: Changes
in gene expression in atherosclerotic plaques analyzed using DNA
array. Atherosclerosis. 165:23–32. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Friso S, Lotto V, Choi SW, Girelli D,
Pinotti M, Guarini P, Udali S, Pattini P, Pizzolo F, Martinelli N,
et al: Promoter methylation in coagulation F7 gene influences
plasma FVII concentrations and relates to coronary artery disease.
J Med Genet. 49:192–199. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Männistö PT and Kaakkola S:
Catechol-O-methyltransferase (COMT): Biochemistry, molecular
biology, pharmacology and clinical efficacy of the new selective
COMT inhibitors. Pharmacol Rev. 51:593–628. 1999.PubMed/NCBI
|
|
11
|
Wardle MC, de Wit H, Penton-Voak I, Lewis
G and Munafò MR: Lack of association between COMT and working
memory in a population-based cohort of healthy young adults.
Neuropsychopharmacolgy. 38:1253–1263. 2013. View Article : Google Scholar
|
|
12
|
Baud P, Courtet P, Perroud N, Jollant F,
Buresi C and Malafosse A: Catechol-O-methyltransferase polymorphism
(COMT) in suicide attempters: A possible gender effect on anger
traits. Am J Med Genet B Neuropsychiatr Genet 144B. 1042–1047.
2007. View Article : Google Scholar
|
|
13
|
Abdolmaleky HM, Cheng KH, Faraone SV,
Wilcox M, Glatt SJ, Gao F, Smith CL, Shafa R, Aeali B, Carnevale J,
et al: Hypomethylation of MB-COMT promoter is a major risk factor
for schizophrenia and bipolar disorder. Hum Mol Genet.
15:3132–3145. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bosia M, Pigoni A, Pirovano A, Lorenzi C,
Spangaro M, Buonocore M, Bechi M, Cocchi F, Guglielmino C, Bramanti
P, et al: COMT and STH polymorphisms interaction on cognition in
schizophrenia. Neurol Sci. 36:215–220. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kring SI, Werge T, Holst C, Toubro S,
Astrup A, Hansen T, Pedersen O and Sørensen TI: Polymorphisms of
serotonin receptor 2A and 2C genes and COMT in relation to obesity
and type 2 diabetes. PloS One. 4:e66962009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hagen K, Pettersen E, Stovner LJ, Skorpen
F, Holmen J and Zwart JA: High systolic blood pressure is
associated with Val/Val genotype in the
catechol-O-methyltransferase gene. The nord-trøndelag health study
(HUNT). Am J Hypertens. 20:21–26. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Voutilainen S, Tuomainen TP, Korhonen M,
Mursu J, Virtanen JK, Happonen P, Alfthan G, Erlund I, North KE,
Mosher MJ, et al: Functional COMT Val158Met polymorphism, risk of
acute coronary events and serum homocysteine: The kuopio ischaemic
heart disease risk factor study. PloS One. 2:e1812007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nohesara S, Ghadirivasfi M, Mostafavi S,
Eskandari MR, Ahmadkhaniha H, Thiagalingam S and Abdolmaleky HM:
DNA hypomethylation of MB-COMT promoter in the DNA derived from
saliva in schizophrenia and bipolar disorder. J Psychiatr Res.
45:1432–1438. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang D, Zheng D, Wang L, Huang Y, Liu H,
Xu L, Liao Q, Liu P, Shi X, Wang Z, et al: Elevated PLA2G7 gene
promoter methylation as a gender-specific marker of aging increases
the risk of coronary heart disease in females. PloS One.
8:e597522013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou J, Huang Y, Huang RS, Wang F, Xu L,
Le Y, Yang X, Xu W, Huang X, Lian J and Duan S: A case-control
study provides evidence of association for a common SNP rs974819 in
PDGFD to coronary heart disease and suggests a sex-dependent
effect. Thromb Res. 130:602–606. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ye H, Zhao Q, Huang Y, Wang L, Liu H, Wang
C, Dai D, Xu L, Ye M and Duan S: Meta-analysis of low density
lipoprotein receptor (LDLR) rs2228671 polymorphism and coronary
heart disease. BioMed Res Int. 2014:5649402014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Müller T: Catechol-O-methyltransferase
inhibitors in Parkinson's disease. Drugs. 75:157–174. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Eriksson AL, Skrtic S, Niklason A, Hultén
LM, Wiklund O, Hedner T and Ohlsson C: Association between the low
activity genotype of catechol-O-methyltransferase and myocardial
infarction in a hypertensive population. Eur Heart J. 25:386–391.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hyvärinen M, Qiao Q, Tuomilehto J,
Söderberg S, Eliasson M and Stehouwer CDA: The difference between
acute coronary heart disease and ischaemic stroke risk with regard
to gender and age in Finnish and Swedish populations. Int J Stroke.
5:152–156. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Martorell L, Costas J, Valero J,
Gutierrez-Zotes A, Phillips C, Torres M, Brunet A, Garrido G,
Carracedo A, Guillamat R, et al: Analyses of variants located in
estrogen metabolism genes (ESR1, ESR2, COMT and APOE) and
schizophrenia. Schizophr Res. 100:308–315. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Raddatz G, Hagemann S, Aran D, Söhle J,
Kulkarni PP, Kaderali L, Hellman A, Winnefeld M and Lyko F: Aging
is associated with highly defined epigenetic changes in the human
epidermis. Epigenetics Chromatin. 6:362013. View Article : Google Scholar : PubMed/NCBI
|