Introduction
Renal cell carcinoma (RCC) has the highest mortality
rate of all urological malignancies worldwide and accounts for
~65,000 new cancer cases per year in the United States alone
(1). Clear cell (cc) RCC is the most
common subtype and accounts for 70–80% of all RCC cases (2,3).
Metastasis and progression are the most common events in ccRCC and
~33% of affected patients are in the terminal stage at the time of
diagnosis (4,5). Patients with metastatic ccRCC have a
poor prognosis and the number of therapeutic strategies available
is limited (6–8). Therefore, further research into the
underlying molecular mechanisms of ccRCC metastasis to identify
novel diagnostic biomarkers for ccRCC in the early stages is
urgently required.
Long non-coding RNAs (lncRNAs) are a newly
identified class of non-coding RNAs, with transcripts of >200-bp
nucleotides that have no protein-coding function (9). It has been demonstrated that lncRNAs
may serve a crucial role in various cellular biological processes
and human disease (10,11). As microRNAs, lncRNAs may serve a key
role in regulating human cancer cell growth, invasion and apoptosis
(12,13). Li et al (14) demonstrated that the upregulation of
lncRNA urothelial cancer associated (UCA) 1 was correlated with
advanced clinical stage and a poor prognosis in patients with
esophageal squamous cell carcinoma. In addition, knockdown of UCA1
was observed to inhibit cell growth, migration and invasion
(14). The lncRNA HOXA cluster
antisense RNA 2 promotes cancer cell proliferation by
epigenetically silencing P21/Polo-like kinase 3/DNA damage
inducible transcript 3 expression in gastric carcinoma (15). Furthermore, the lncRNA taurine
upregulated gene 1 is highly expressed in hepatocellular carcinoma
and promotes cell proliferation and apoptosis by epigenetically
silencing Krüppel-like factor 2 (16).
LOC389332 is a 723-bp intragenic lncRNA transcribed
from chromosome 5 in the human genome (17). Analysis of previous lncRNA expression
signatures by microarray showed that LOC389332 was significantly
downregulated in ccRCC (18,19); however, its biological functions in
ccRCC have remained elusive. The present study was performed to
verify the expression pattern of LOC389332 and evaluate the
clinical significance of lncRNA LOC389332 in ccRCC. In addition,
the impact of LOC389332 on ccRCC cell proliferation and migration
was assessed in vitro. The present study aimed to identify
whether LOC389332 downregulation was associated with poor prognosis
and tumor progression in ccRCC, therefore determining whether
LOC389332 may be developed as a novel prognostic biomarker and
therapeutic target for ccRCC treatment.
Materials and methods
Patients and specimens
All 30 samples of ccRCC tissues and paired adjacent
non-tumor tissues used in the present study were obtained from 30
patients who had undergone radical nephrectomy at the Department of
Urology of Renmin Hospital, Hubei University of Medicine (Hubei,
China) between January 2009 and February 2011. Patients were
selected on the basis of the following inclusion and exclusion
criteria: i) Definite pathological diagnosis of ccRCC according to
the World Health Organization criteria (20); ii) suitable formalin-fixed,
paraffin-embedded tissue specimens; iii) complete
clinicopathological data. None of the patients received
radiotherapy or chemotherapy prior to radical nephrectomy.
Following surgical resection, specimens were immediately immersed
in RNAlater® (Qiagen GmbH, Hilden, Germany) for 30 min
and then transferred into liquid nitrogen for cryopreservation
until use. The clinicopathological features of the patients are
presented in Table I. Follow-up of
patients was completed in May 2015 and the median observation time
was 30 months. All specimens were collected on the basis of their
availability for research and following a protocol approved by the
Medical Ethics committee of the Renmin Hospital, Hubei University
of Medicine (Hubei, China). Written consent was obtained from all
patients participating in the study.
 | Table I.Association between LOC389332
expression and clinicopathological parameters of patients with
ccRCC. |
Table I.
Association between LOC389332
expression and clinicopathological parameters of patients with
ccRCC.
|
|
| LOC389332
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
parameter | Number of cases | Low (n) | High (n) | P-value |
|---|
| Gender |
|
|
| 1.000 |
| Male | 15 | 11 | 4 |
|
|
Female | 15 | 10 | 5 |
|
| Age, years |
|
|
| 0.418 |
| ≤60 | 18 | 14 | 4 |
|
|
>60 | 12 | 7 | 5 |
|
| Tumor size, cm |
|
|
| 0.100 |
| ≤4 | 19 | 11 | 8 |
|
|
>4 | 11 | 10 | 1 |
|
| Fuhrman grade |
|
|
| 0.001 |
|
T1-T2 | 7 | 1 | 6 |
|
|
T3-T4 | 23 | 20 | 3 |
|
| AJCC stage |
|
|
| 0.001 |
| I–II | 9 | 2 | 7 |
|
|
III–IV | 21 | 19 | 2 |
|
| Lymph node
metastasis |
|
|
| <0.001 |
|
Absent | 10 | 1 | 9 |
|
|
Present | 20 | 20 | 0 |
Cell culture
Human ccRCC cell lines (786-O, 769-P, CaKi-1 and
RLC-310) and a normal immortalized human proximal tubule epithelial
cell line (HK-2) were obtained from the American Type Culture
Collection (Manassas, VA, USA). All cells were cultured in
RPMI-1640 medium (Thermo Fisher Scientific, Inc., Waltham, MA,
USA), supplemented with 10% fetal bovine serum (Invitrogen; Thermo
Fisher Scientific, Inc.) in a 100% humidified atmosphere of 5%
CO2/95% air at 37°C.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from 30 cases of fresh ccRCC tissues and
cell lines were extracted using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). RNA was detected and
quantified using a NanoDrop™ 2000c spectrophotometer (Thermo Fisher
Scientific, Inc., Wilmington, DE, USA). Subsequently, RNA samples
were reverse-transcribed into complementary first-strand DNA using
a High-Capacity RNA-to-cDNA kit (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. The qPCR reaction was
then performed on the LightCycler® 480 Real-Time PCR
system (Roche Applied Science, Penzberg, Germany) using the
SYBR® Select Master Mix (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. The reaction
conditions for PCR were as follows: 95°C for 20 min, followed by 40
cycles of 95°C for 10 sec, 55°C for 30 sec and 72°C for 30 sec. The
specific primers for human LOC389332 were as follows: Forward,
5′-GCGCTCGTCGTCCTCTTCATCG-3′ and reverse,
5′-CGTGGCTCAGTCCCAAGCTACACC-3′. The GAPDH gene was used as an
internal control, and the primers for GAPDH were: Forward,
5′-GGCACCACACCTTCTACAATGAG-3′ and reverse,
5′-GGATAGCACAGCCTGGATAGCA-3′. All primers were obtained from
Invitrogen (Carlsbad, CA, USA). The relative expression levels of
LOC389332 in ccRCC tissues and cell lines were analyzed using the
2−ΔΔCq method (21).
Cell transfection
As reduction in the expression of LOC389332 in ccRCC
tissues and cells was observed, a gain-of-function study was
performed by inducing the overexpression of LOC389332 to identify
its function in the 786-O and 769-P cell lines. An LOC389332
overexpression plasmid (pcDNA3.1-LOC389332) was purchased from
Thermo Fisher Scientific, Inc. 786-O and 769-P cells were treated
with the indicated amounts of pcDNA3.1-LOC389332 plasmid using
Lipofectamine® 2000 reagent (Thermo Fisher Scientific,
Inc.). The empty vector pcDNA3.1- was used as a negative
control.
Cell migration assay
A scratch wound assay was applied to evaluate the
ability of 786-O and 769-P cells to migrate in vitro. Cells
(~2×106/well) were seeded into 6-well plates and grown
to 70% confluence ~24 h prior to infection. Following 6 h of
transfection, similar-sized wounds were generated to the cell
monolayers using sterile 100-µl pipette tips. Following three
washes with phosphate-buffered saline to remove cell debris, cells
were cultured in the incubator at 37°C. In each sample, images of
the same area were acquired with a Leica DM LB2 microscope digital
camera system (Leica Microsystems GmbH, Wetzlar, Germany) at 0 and
12 h after the wounds were made to determine the amount of wound
closure. The data was calculated using the software program
MIAS-2000 (Leica Microsystems GmbH). This experiment was performed
in triplicate and repeated at least three times.
Cell proliferation assay
786-O and 769-P cell proliferation was assessed
using an MTT assay. Transfected cells were seeded onto each well of
96-well plates at a density of 5×103 cells per well.
Following 0, 24, 48, 72 and 96 h of transfection, 15 µl of MTT (5
mg/ml; Sigma-Aldrich, St. Louis, MO, USA) was added to each well
and plates were incubated for 4 h at room temperature.
Subsequently, 130 µl dimethyl sulfoxide was added to each well and
wells were shaken for 10 min at 37°C to solubilize the formazan
crystals. The absorbance of 786-O and 769-P cells was measured at
490 nm using a Bio-Rad model 680 microplate reader (Bio-Rad
Laboratories Inc., Hercules, CA, USA). Results from the MTT assay
are presented as the mean of at least three independent
experiments.
Statistical analysis
Statistical analysis was performed with SPSS 16.0
(SPSS, Inc., Chicago, IL, USA). Values are expressed as the mean ±
standard deviation. To compare LOC389332 expression levels in ccRCC
tissues vs. matched tumor normal tissues, a paired Student's
t-test. was used. The χ2 test was applied to compare the
levels of LOC389332 expression with various clinicopathological
parameters. The results of the MTT assay were analyzed using
one-way analysis of variance. Overall survival curves were
generated using the Kaplan-Meier method. P<0.05 was considered
to indicate a statistically significant difference.
Results
Expression of LOC389332 is
downregulated in ccRCC tissues and cells
Previous lncRNA expression profiling of ccRCC
indicated that LOC389332 was significantly downregulated in RCC
tissues (18,19). To confirm that LOC389332 expression
is decreased in ccRCC, RT-qPCR was performed to quantify LOC389332
expression in the 30 ccRCC tissues and ccRCC cell lines. The
results demonstrated that LOC389332 expression was significantly
lower in ccRCC tissues compared with matched adjacent normal
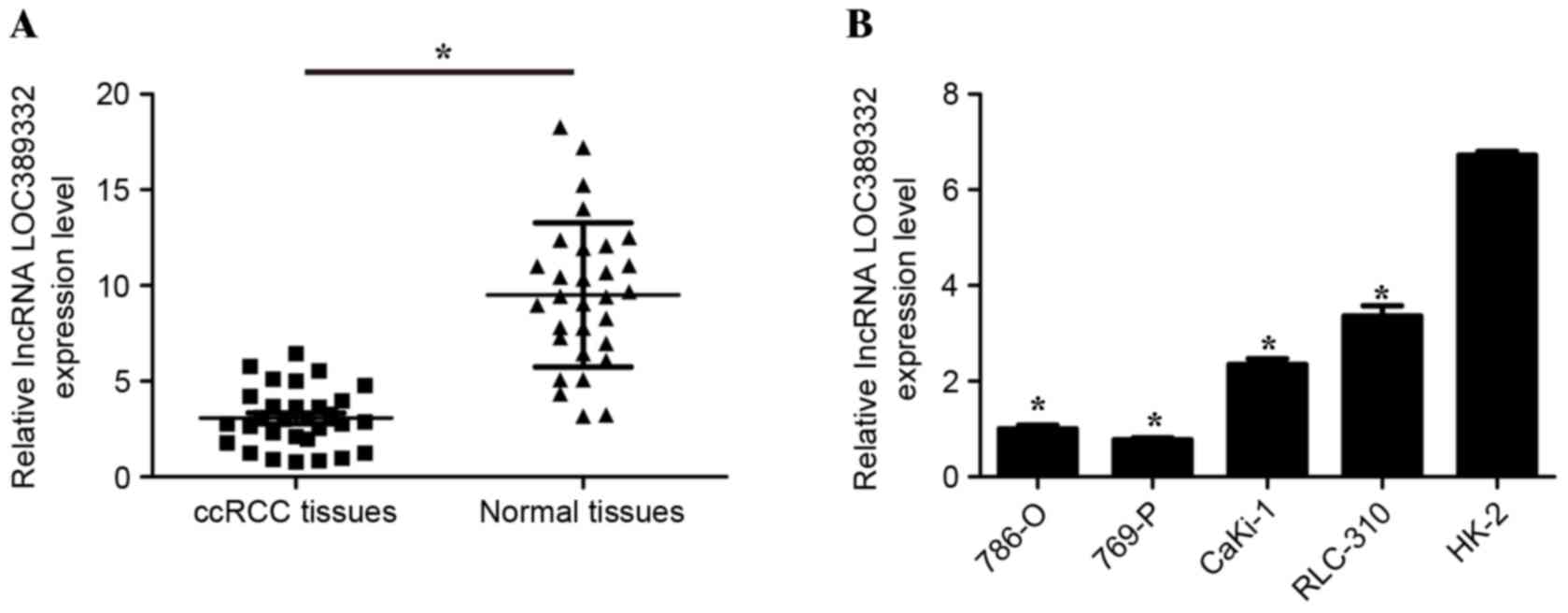
tissues (P<0.05; Fig. 1A). In
addition, LOC389332 expression was significantly downregulated in
786-O, 769-P, CaKi-1 and RLC-310 cell lines, compared with that in
the normal HK-2 cell line (P<0.05; Fig. 1B). These results suggested that
LOC389332 expression is suppressed in ccRCC.
Association between LOC389332
expression and clinicopathological parameters in ccRCC
To evaluate the association between LOC389332
expression and the patients' clinicopathological features, the 30
ccRCC samples were divided into low (n=21) and high (n=9) LOC389332
expression groups based on the median value of relative LOC389332
expression. As presented in Table I,
downregulation of LOC389332 expression was correlated with tumor
American Joint Commission on Cancer (AJCC) stage (22) (P=0.001), Fuhrman grade (23) (P=0.001) and lymph node metastasis
(P<0.001). However, no correlation was detected between
LOC389332 expression and patient gender (P=1.000), age (P=0.418) or
tumor size (P=0.100). These results demonstrated that decreased
LOC389332 expression was associated with ccRCC progression and
development.
Downregulation of LOC389332 predicts
poor prognosis in ccRCC patients
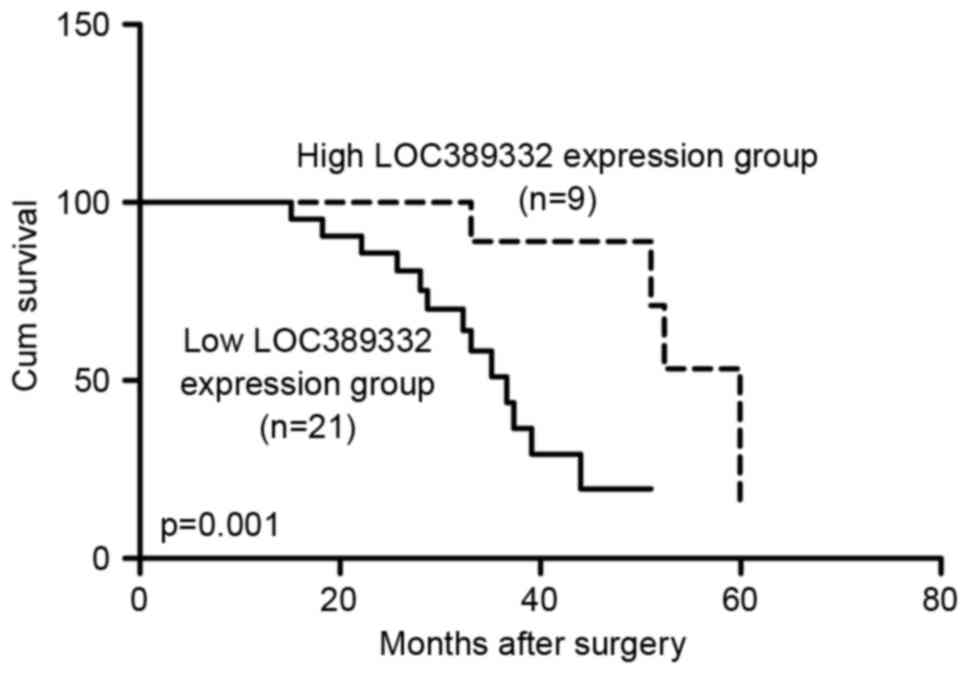
Kaplan-Meier analysis revealed that the overall
survival rate of the low LOC389332 expression group was
significantly lower than the overall survival of the high LOC389332
expression group (P=0.001; Fig. 2).
The 3-year survival rate for ccRCC patients with low LOC389332
expression was 54% compared with 83% in patients with high
LOC389332 expression (P<0.05).
Downregulation of LOC389332 inhibits
growth of ccRCC cells in vitro
To identify the biological functions of LOC389332 on
ccRCC cells in vitro, transfection of pcDNA3.1-LOC389332
overexpression plasmid were applied to restore LOC389332 expression
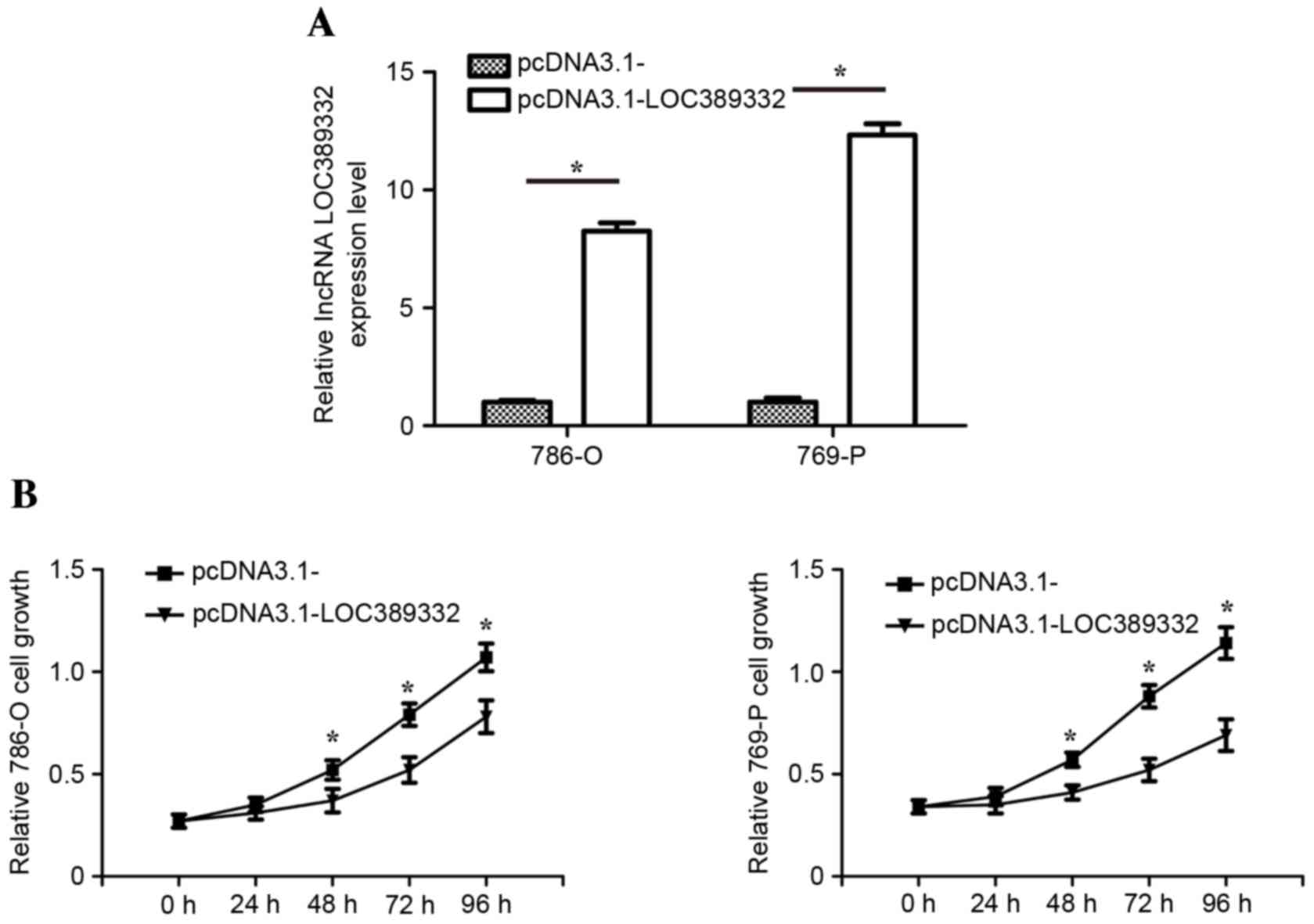
in 786-O and 769-P cells. RT-qPCR analysis revealed that LOC389332
expression in 786-O and 769-P cells transfected with the
pcDNA3.1-LOC389332 plasmid was significantly upregulated compared
with that in the pcDNA3.1-empty vector group (Fig. 3A). The MTT assay revealed that the
proliferation of the 786-O and 769-P cell lines was significantly
decreased in the pcDNA3.1-LOC389332 groups compared with that in
the empty vector-transfected groups (Fig. 3B).
Downregulation of LOC389332 inhibits
migration of ccRCC cells in vitro
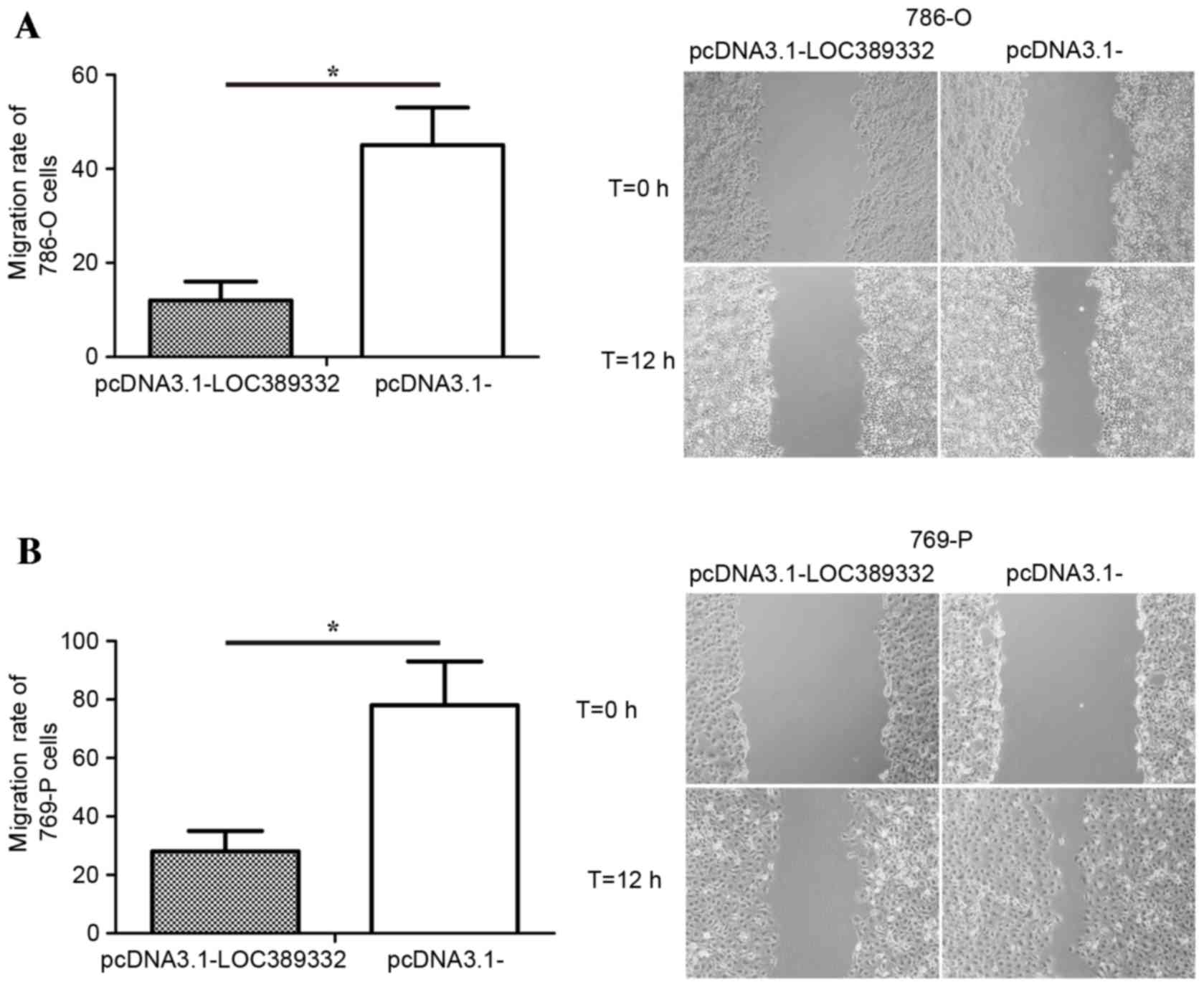
The effect of LOC389332 on ccRCC cell migration was
observed using a scratch wound assay. Compared with the
pcDNA3.1-empty vector group, overexpression of LOC389332
significantly suppressed the migration capacity of 786-O and 769-P
cells (P=0.002 in each; Fig. 4),
suggesting that ectopic overexpression of LOC389332 impairs the
migratory capacity of ccRCC cells.
Discussion
At present, ccRCC has one of the highest mortality
rates of all types of kidney disease (24). In spite of improvements in
therapeutic strategies for cancer, major challenges still exist
regarding the management of patients with ccRCC. Therefore,
identifying novel molecular targets to diagnose and treat ccRCC is
important to improve the prognosis and clinical outcomes of
patients affected.
Over the past few decades, gene expression
microarray analysis has been regarded as a useful and promising
approach to identify the molecular signatures of ccRCC (25,26). A
number of novel lncRNAs have been identified using lncRNA
expression profiling and certain altered lncRNA expression patterns
have been found to be associated with the genesis and progression
of various types of cancer (27,28). It
was demonstrated that high expression of the lncRNA metastasis
associated lung adenocarcinoma transcript 1 is correlated with
tumor progression and poor prognosis in patients with ccRCC
(29). Furthermore, upregulation of
lncRNA Sprouty RTK signaling antagonist 4-intronic transcript 1
predicts poor prognosis in patients with ccRCC (30). Downregulation of lncRNA growth arrest
specific 5 suppresses tumor growth in ccRCC (31). Furthermore, it was demonstrated that
decreased expression of neuroblastoma associated transcript 1 is
associated with poor prognosis in patients with ccRCC (32). However, in addition to these few
abovementioned lncRNAs, further ones remain to be identified in
ccRCC.
Previous studies using lncRNA expression profiling
have indicated that lncRNA LOC389332 is significantly downregulated
in ccRCC tissue (18,19). However, despite being the most common
urological cancer, it remains unknown whether aberrant LOC389332
expression is associated with ccRCC carcinogenesis. The present
study investigated the clinical significance of LOC389332 in
patients with ccRCC. The RT-qPCR results showed that LOC389332
expression was markedly downregulated in ccRCC tissues and cell
lines compared with that in adjacent non-tumor tissues and the
normal human proximal tubule epithelial cell line HK-2. In
addition, it was demonstrated that LOC389332 expression was
strongly associated with the AJCC tumor stage, Fuhrman grade and
lymph node metastasis. However, LOC389332 expression was not
associated with patient age, gender and tumor size. These results
suggested that downregulation of LOC389332 serves an important role
in ccRCC progression and development.
The clinicopathological features of patients with
ccRCC suggest that LOC389332 may affect the growth and metastasis
of ccRCC cells. Therefore, MTT and scratch wound assays were
performed to evaluate the biological function of LOC389332 in ccRCC
cells. The results demonstrated that ectopic overexpression of
LOC389332 significantly inhibits the growth and migratory capacity
of 786-O and 769-P cells compared with that of empty
vector-transfected cells. This suggested that restoration of
LOC389332 expression may inhibit ccRCC development and progression
and that high expression of LOC389332 may inhibit malignant
phenotypes of ccRCC.
In conclusion, to the best of our knowledge, the
present study was the first to identify that the novel lncRNA
LOC389332 is downregulated in ccRCC tissues, which is associated
with advanced tumor progression. Furthermore, the results suggested
that LOC389332 affects ccRCC cell proliferation and migration.
Therefore, LOC389332 may be developed as a novel and effective
diagnostic and prognostic marker, and ectopic overexpression of
LOC389332 may represent a therapeutic strategy for ccRCC.
Acknowledgements
The present study was supported by the Scientific
and Technological Project of Shiyan City of Hubei Province (grant
no. ZD2012020).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Martel CL and Lara PN: Renal cell
carcinoma: Current status and future directions. Crit Rev Oncol
Hematol. 45:177–190. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rini BI, Rathmell WK and Godley P: Renal
cell carcinoma. Curr Opin Oncol. 20:300–306. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rini BI, Campbell SC and Escudier B: Renal
cell carcinoma. Lancet. 373:1119–1132. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Russo P: Renal cell carcinoma:
Presentation, staging, and surgical treatment. Semin Oncol.
27:160–176. 2000.PubMed/NCBI
|
|
6
|
Bhatt JR and Finelli A: Landmarks in the
diagnosis and treatment of renal cell carcinoma. Nat Rev Urol.
11:517–525. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Motzer RJ, Bander NH and Nanus DM:
Renal-cell carcinoma. N Engl J Med. 335:865–875. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cho IC and Chung J: Current status of
targeted therapy for advanced renal cell carcinoma. Korean J Urol.
53:217–228. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wapinski O and Chang HY: Long noncoding
RNAs and human disease. Trends Cell Biol. 21:354–361. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li JY, Ma X and Zhang CB: Overexpression
of long non-coding RNA UCA1 predicts a poor prognosis in patients
with esophageal squamous cell carcinoma. Int J Clin Exp Pathol.
7:7938–7944. 2014.PubMed/NCBI
|
|
15
|
Xie M, Sun M, Zhu YN, Xia R, Liu YW, Ding
J, Ma HW, He XZ, Zhang ZH, Liu ZJ, et al: Long noncoding RNA
HOXA-AS2 promotes gastric cancer proliferation by epigenetically
silencing P21/PLK3/DDIT3 expression. Oncotarget. 6:33587–33601.
2015.PubMed/NCBI
|
|
16
|
Huang MD, Chen WM, Qi FZ, Sun M, Xu TP, Ma
P and Shu YQ: Long non-coding RNA TUG1 is up-regulated in
hepatocellular carcinoma and promotes cell growth and apoptosis by
epigenetically silencing of KLF2. Mol Cancer. 14:1652015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schmutz J, Martin J, Terry A, Couronne O,
Grimwood J, Lowry S, Gordon LA, Scott D, Xie G, Huang W, et al: The
DNA sequence and comparative analysis of human chromosome 5.
Nature. 431:268–274. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu G, Yao W, Wang J, Ma X, Xiao W, Li H,
Xia D, Yang Y, Deng K, Xiao H, et al: LncRNAs expression signatures
of renal clear cell carcinoma revealed by microarray. PLoS One.
7:e423772012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qin C, Han Z, Qian J, Bao M, Li P, Ju X,
Zhang S, Zhang L, Li S, Cao Q, et al: Expression pattern of long
non-coding RNAs in renal cell carcinoma revealed by microarray.
PLoS One. 9:e993722014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lopez-Beltran A, Scarpelli M, Montironi R
and Kirkali Z: 2004 WHO classification of the renal tumors of the
adults. Eur Urol. 49:798–805. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: AJCC cancer staging manual. 7th. Springer
Verlag; New York, NY: pp. 479–489. 2010
|
|
23
|
Zimmermann U, Woenckhaus C, Pietschmann S,
Junker H, Maile S, Schultz K, Protzel C and Giebel J: Expression of
annexin II in conventional renal cell carcinoma is correlated with
Fuhrman grade and clinical outcome. Virchows Arch. 445:368–374.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chow WH and Devesa SS: Contemporary
epidemiology of renal cell cancer. Cancer J. 14:288–301. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
van Spronsen DJ, de Weijer KJ, Mulders PF
and De Mulder PH: Novel treatment strategies in clear-cell
metastatic renal cell carcinoma. Anticancer Drugs. 16:709–717.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Takahashi M, Teh BT and Kanayama HO:
Elucidation of the molecular signatures of renal cell carcinoma by
gene expression profiling. J Med Invest. 53:9–19. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Maruyama R and Suzuki H: Long noncoding
RNA involvement in cancer. BMB Rep. 45:604–611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lisitsyn NA, Chernyi AA, Karpov VL and
Beresten SF: A role of long noncoding RNAs in carcinogenesis. Mol
Biol (Mosk). 49:561–570. 2015.(In Russian). View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang HM, Yang FQ, Chen SJ, Che J and
Zheng JH: Upregulation of long non-coding RNA MALAT1 correlates
with tumor progression and poor prognosis in clear cell renal cell
carcinoma. Tumour Biol. 36:2947–2955. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang HM, Yang FQ, Yan Y, Che JP and Zheng
JH: High expression of long non-coding RNA SPRY4-IT1 predicts poor
prognosis of clear cell renal cell carcinoma. Int J Clin Exp
Pathol. 7:5801–5809. 2014.PubMed/NCBI
|
|
31
|
Qiao HP, Gao WS, Huo JX and Yang ZS: Long
non-coding RNA GAS5 functions as a tumor suppressor in renal cell
carcinoma. Asian Pac J Cancer Prev. 14:1077–1082. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xue S, Li QW, Che JP, Guo Y, Yang FQ and
Zheng JH: Decreased expression of long non-coding RNA NBAT-1 is
associated with poor prognosis in patients with clear cell renal
cell carcinoma. Int J Clin Exp Pathol. 8:3765–3774. 2015.PubMed/NCBI
|