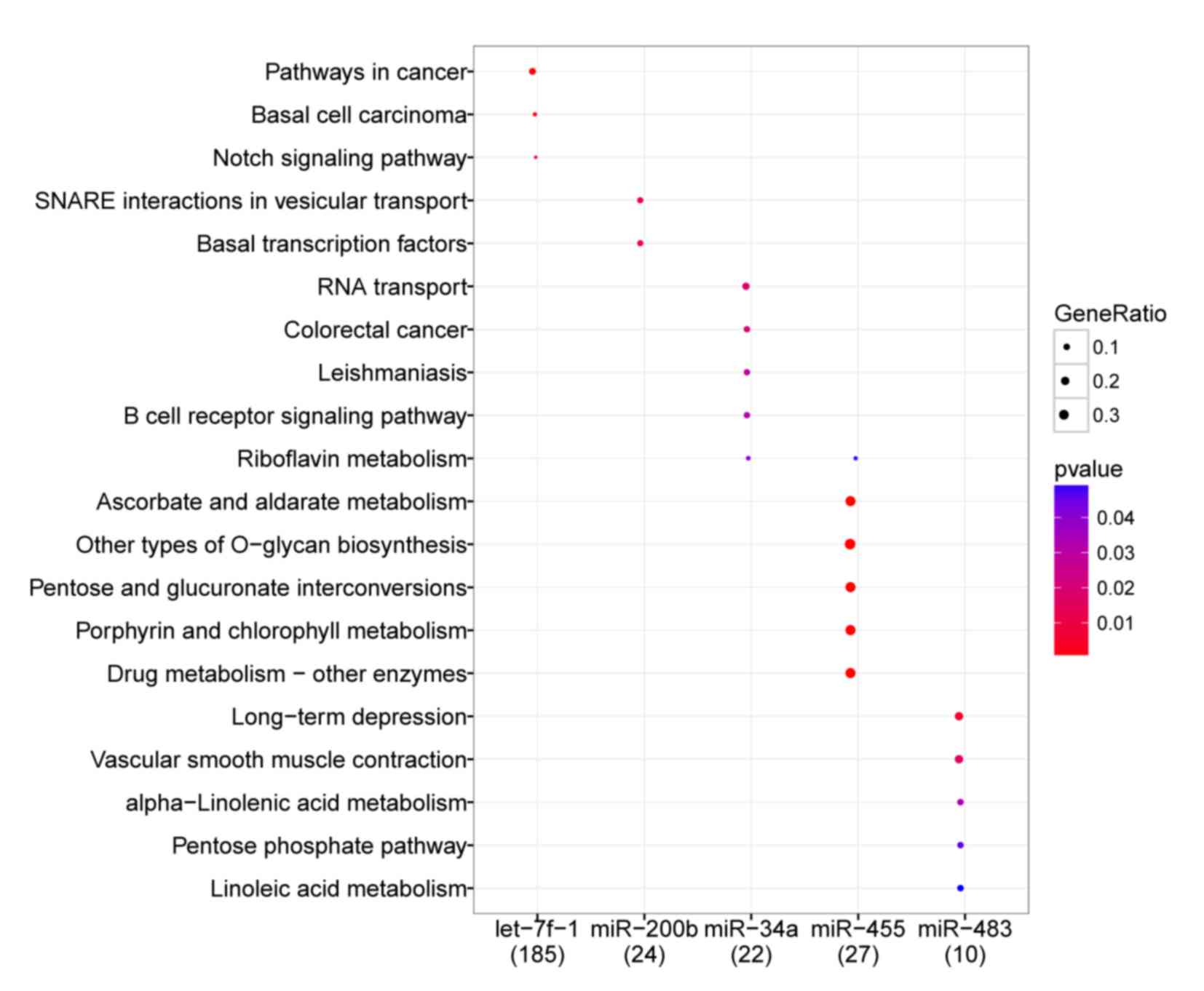
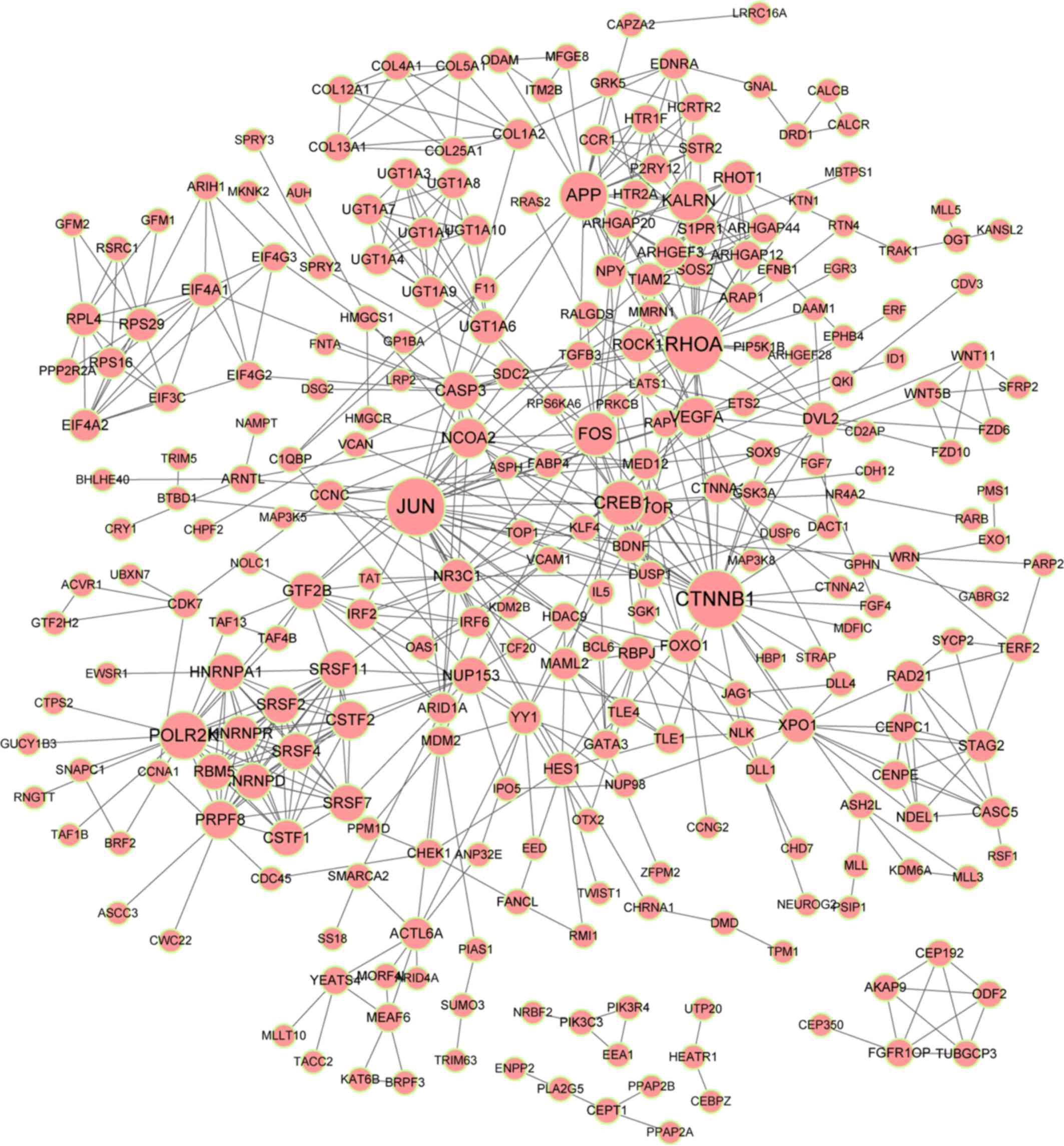
|
1
|
Organization WH: The top 10 causes of
death. July. 2013, simplewhoint/mediacentre/factsheets/fs310/en2014
|
|
2
|
Wunderink RG and Waterer GW: Clinical
practice. Community-acquired pneumonia. N Engl J Med. 370:543–551.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Said MA, Johnson HL, Nonyane BA,
Deloria-Knoll M, O'Brien KL; AGEDD Adult Pneumococcal Burden Study
Team, ; Andreo F, Beovic B, Blanco S, Boersma WG, et al: Estimating
the burden of pneumococcal pneumonia among adults: A systematic
review and meta-analysis of diagnostic techniques. PLoS One.
8:e602732013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rakha MA, Abdelmoneim AN, Farhoud S,
Pièche S, Cousens S, Daelmans B and Bahl R: Does implementation of
the IMCI strategy have an impact on child mortality? A
retrospective analysis of routine data from Egypt. BMJ Open.
3:pii.e0018522013. View Article : Google Scholar
|
|
5
|
Floyd J, Wu L, Hay Burgess D, Izadnegahdar
R, Mukanga D and Ghani AC: Evaluating the impact of pulse oximetry
on childhood pneumonia mortality in resource-poor settings. Nature.
528:S53–S59. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Choi SH, Hong SB, Ko GB, Lee Y, Park HJ,
Park SY, Moon SM, Cho OH, Park KH, Chong YP, et al: Viral infection
in patients with severe pneumonia requiring intensive care unit
admission. Am J Respir Crit Care Med. 186:325–332. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Berkley JA, Munywoki P, Ngama M, Kazungu
S, Abwao J, Bett A, Lassauniére R, Kresfelder T, Cane PA, Venter M,
et al: Viral etiology of severe pneumonia among Kenyan infants and
children. JAMA. 303:2051–2057. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sapru A, Hansen H, Ajayi T, Brown R,
Garcia O, Zhuo H, Wiemels J, Matthay MA and Wiener-Kronish J: 4G/5G
polymorphism of plasminogen activator inhibitor-1 gene is
associated with mortality in intensive care unit patients with
severe pneumonia. Anesthesiology. 110:1086–1091. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
España PP, Capelastegui A, Bilbao A, Diez
R, Izquierdo F, de Goicoetxea MJ Lopez, Gamazo J, Medel F, Salgado
J, Gorostiaga I, et al: Utility of two biomarkers for directing
care among patients with non-severe community-acquired pneumonia.
Eur J Clin Microbiol Infect Dis. 31:3397–3405. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wong KY, Huang X and Chim CS: DNA
methylation of microRNA genes in multiple myeloma. Carcinogenesis.
33:1629–1638. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Abd-El-Fattah AA, Sadik NA, Shaker OG and
Aboulftouh ML: Differential microRNAs expression in serum of
patients with lung cancer, pulmonary tuberculosis, and pneumonia.
Cell Biochem Biophys. 67:875–884. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ramirez G, Uribe-Boll J, Cruz A, Jimenez
L, Banales J, Romero S, Hidalgo A, Bautista E, Merino E and Moran
J: Circulating microRNA profiles in patients with severe pneumonia
associated to the A/H1N1 virus. Am J Respir Crit Care Med.
189:A26942014.
|
|
13
|
Ren S, Peng Z, Mao JH, Yu Y, Yin C, Gao X,
Cui Z, Zhang J, Yi K, Xu W, et al: RNA-seq analysis of prostate
cancer in the Chinese population identifies recurrent gene fusions,
cancer-associated long noncoding RNAs and aberrant alternative
splicings. Cell Res. 22:806–821. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang Z, Gerstein M and Snyder M: RNA-Seq:
A revolutionary tool for transcriptomics. Nat Rev Genet. 10:57–63.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Weiwu D: Guidelines for the diagnosis and
treatment of community acquired pneumonia. Chinese J Tuberculosis
Respiratory Dis. 29:651–655. 2001.
|
|
16
|
Griffiths-Jones S, Saini HK, van Dongen S
and Enright AJ: miRBase: Tools for microRNA genomics. Nucleic Acids
Res. 36:D154–D158. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Smyth GK: Limma: Linear models for
microarray dataBioinformatics and Computational Biology Solutions
Using R and Bioconductor. Springer; New York, NY: pp. 397–420.
2005, View Article : Google Scholar
|
|
18
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J Royal Statistical Society Series B
(Methodological). 57:289–300. 1995.
|
|
19
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human microRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Palmieri D, Capponi S, Geroldi A, Mura M,
Mandich P and Palombo D: TNFα induces the expression of genes
associated with endothelial dysfunction through p38MAPK-mediated
down-regulation of miR-149. Biochem Biophys Res Commun.
443:246–251. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Harris MA, Clark J, Ireland A, Lomax J,
Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C,
et al: The gene ontology (GO) database and informatics resource.
Nucleic Acids Res. 32(Database Issue): D258–D261. 2004.PubMed/NCBI
|
|
22
|
Kanehisa M, Goto S, Sato Y, Furumichi M
and Tanabe M: KEGG for integration and interpretation of
large-scale molecular data sets. Nucleic Acids Rese. 40(Database
Issue): D109–D114. 2012. View Article : Google Scholar
|
|
23
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tan KS, Choi H, Jiang X, Yin L, Seet JE,
Patzel V, Engelward BP and Chow VT: Micro-RNAs in regenerating
lungs: An integrative systems biology analysis of murine influenza
pneumonia. BMC Genomics. 15:5872014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao B, Han H, Chen J, Zhang Z, Li S, Fang
F, Zheng Q, Ma Y, Zhang J, Wu N and Yang Y: MicroRNA let-7c
inhibits migration and invasion of human non-small cell lung cancer
by targeting ITGB3 and MAP4K3. Cancer Lett. 342:43–51. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang X, Cao L, Wang Y, Wang X, Liu N and
You Y: Regulation of let-7 and its target oncogenes (Review). Oncol
Lett. 3:955–960. 2012.PubMed/NCBI
|
|
30
|
Liu XS, Chopp M, Zhang RL, Tao T, Wang XL,
Kassis H, Hozeska-Solgot A, Zhang L, Chen C and Zhang ZG: MicroRNA
profiling in subventricular zone after stroke: MiR-124a regulates
proliferation of neural progenitor cells through Notch signaling
pathway. PLoS One. 6:e234612011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pillé JY, Denoyelle C, Varet J, Bertrand
JR, Soria J, Opolon P, Lu H, Pritchard LL, Vannier JP, Malvy C, et
al: Anti-RhoA and Anti-RhoC siRNAs inhibit the proliferation and
invasiveness of MDA-MB-231 breast cancer cells in vitroin vivo. Mol
Ther. 11:267–274. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chan CH, Lee SW, Li CF, Wang J, Yang WL,
Wu CY, Wu J, Nakayama KI, Kang HY, Huang HY, et al: Deciphering the
transcriptional complex critical for RhoA gene expression and
cancer metastasis. Nat Cell Biol. 12:457–467. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
DeLeo FR and Musser JM: Axis of
coinfection evil. J Infect Dis. 201:488–490. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Soong G, Martin FJ, Chun J, Cohen TS, Ahn
DS and Prince A: Staphylococcus aureus protein A mediates invasion
across airway epithelial cells through activation of RhoA GTPase
signaling and proteolytic activity. J Biol Chem. 286:35891–35898.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ganter MT, Roux J, Su G, Lynch SV,
Deutschman CS, Weiss YG, Christiaans SC, Myazawa B, Kipnis E and
Wiener-Kronish JP: Role of small GTPases and alphavbeta5 integrin
in Pseudomonas aeruginosa-induced increase in lung endothelial
permeability. Am J Respir Cell Mol Biol. 40:108–118. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lin Y, Li Z, Ozsolak F, Kim SW,
Arango-Argoty G, Liu TT, Tenenbaum SA, Bailey T, Monaghan AP, Milos
PM and John B: An in-depth map of polyadenylation sites in cancer.
Nucleic Acids Res. 40:8460–8471. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Misquitta-Ali CM, Cheng E, O'Hanlon D, Liu
N, McGlade CJ, Tsao MS and Blencowe BJ: Global profiling and
molecular characterization of alternative splicing events
misregulated in lung cancer. Mol Cell Biol. 31:138–150. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Krause K, Karger S, Sheu SY, Aigner T,
Kursawe R, Gimm O, Schmid KW, Dralle H and Fuhrer D: Evidence for a
role of the amyloid precursor protein in thyroid carcinogenesis. J
Endocrinol. 198:291–299. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Takayama K, Tsutsumi S, Suzuki T,
Horie-Inoue K, Ikeda K, Kaneshiro K, Fujimura T, Kumagai J, Urano
T, Sakaki Y, et al: Amyloid precursor protein is a primary androgen
target gene that promotes prostate cancer growth. Cancer Res.
69:137–142. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Polakis P: Casein kinase 1 A Wnt'er of
disconnect. Curr Biol. 12:R499–R501. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Woenckhaus M, Merk J, Stoehr R, Schaeper
F, Gaumann A, Wiebe K, Hartmann A, Hofstaedter F and Dietmaier W:
Prognostic value of FHIT, CTNNB1, and MUC1 expression in non-small
cell lung cancer. Hum Pathol. 39:126–136. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shen J, Liao J, Guarnera MA, Fang H, Cai
L, Stass SA and Jiang F: Analysis of MicroRNAs in sputum to improve
computed tomography for lung cancer diagnosis. J Thorac. 9:33–40.
2014. View Article : Google Scholar
|
|
43
|
Tan KS, Choi H, Jiang X, Yin L, Ju ES,
Patzel V, Engelward BP and Chow VT: Micro-RNAs in regenerating
lungs: An integrative systems biology analysis of murine influenza
pneumonia. BMC Genomics. 15:1–13. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dewan AT, Egan KB, Hellenbrand K,
Sorrentino K, Pizzoferrato N, Walsh KM and Bracken MB: Whole-exome
sequencing of a pedigree segregating asthma. BMC Medical Genetics.
13:1–9. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li YJ, Ping C, Tang J and Zhang W:
MicroRNA-455 suppresses non-small cell lung cancer through
targeting ZEB1. Cell Biol Int. 40:621–628. 2016. View Article : Google Scholar : PubMed/NCBI
|