Introduction
Non-small cell lung cancer (NSCLC) is the leading
cause of cancer-associated mortality in both men and women
worldwide (1). In China, there has
been a drastic increase in lung cancer-associated mortality since
1970 (2). At present, treatments for
NSCLC are marginally effective (3)
and the primary target for treatment of NSCLCs is the epidermal
growth factor receptor (EGFR) pathway (4). Other than genetic factors, patient
characteristics including gender, race and lifestyle influence the
effectiveness of the treatment for NSCLC (5). The most prominent genetic factor
associated with the responsiveness to NSCLC treatment is a mutation
in EGFR (6,7). Mutations in EGFR mediate the
carcinogenic effect by manipulating the apoptotic mechanism
(8). Tyrosine kinase inhibitors
(TKIs) are the first line of treatment of cases featuring mutated
EGFR (9). The initial response to
TKIs is limited by acquired resistance and the T790M mutation in
EGFR has previously been demonstrated to have this acquired
resistance (10). A previous study
revealed that the role of the T790M mutation is associated with an
increase in the affinity of adenosine triphosphate (ATP) to mutated
EGFR (11). Naturally derived
compounds are being developed to treat NSCLC featuring a T790M
mutation (12). Furthermore,
computer-aided drug design has become a prominent tool in drug
discovery (13). In the present
study, an in-house Traditional Chinese Medicine (TCM) database was
used to investigate novel drugs for EGFR treatment of NSCLC
featuring this T790M mutation. In silico virtual screening
tools, absorption, distribution, metabolism and excretion
(ADME)/Tox analysis, and automated docking were all used to
identify an effective single TCM compound.
Materials and methods
Protein preparation
The crystallographic structure of the kinase domain
of the EGFR protein was retrieved from the Worldwide Protein Data
Bank (PDB; wwpdb.org; ID:1XKK) (14). The structure was cleared of water and
other ions and a T790M mutation was added using Discovery Studio
Visualizer (Release 3.5.′; Accelrys, Inc., San Diego, CA, USA). The
mutated structure was subjected to energy minimization using SPDB
viewer version 4.1 (Swiss institute of Bioinformatics, Lausanne,
Switzerland) following a previously documented protocol (15). GROMOS force field was used for the
energy minimization.
Virtual screening
An initial dataset of 3,000 in-house selected TCM
compounds, identified as exhibiting high activity, was used for the
present study. This dataset was subjected to analysis with
AutoDockVina version 1.1.2 (Scripps Research Institute, La Jolla,
CA, USA) (16) platform using a
Pymol interface (version 1.4.1; DeLano Scientific, Portland, OR,
USA) (17). A grid of 40 Å was
created around the ATP binding site. The number of compounds was
limited on the basis of Gibbs free energy (ΔG). Only those
compounds with an ΔG <-10 kcal/mol were selected for further
ADME/Tox analysis.
PreADME/Tox
An online sever (preadmet.bmdrc.org/) was used to evaluate the ADME of
25TCM compounds. The 4 TCM compounds, nardosinon, artesunate,
daidzin and emetine were selected based on their ADME properties.
The toxicity testing provided the mutagenicity and carcinogenicity
properties of the selected compounds and only those compounds with
no mutagenic and carcinogenic properties were selected for ADME
evaluation. The predictive in silico ADME values and
drug-likeliness values were used to further shortlist the
compounds. Properties including molecular weight, the octanol/water
partition coefficient (logP), number of hydrogen bond donors (HBD),
number of hydrogen bond acceptors (HBA) and total polar surface
area (tpsa) were taken into account.
Automated docking
The 4 most appropriate compounds, as determined by
their ADME properties, were subjected to automated docking by
AutoDock version 4.2 (Scripps Research Institute) (18). The Lamarckian algorithm of the tool
was used to evaluate the optimal TCM compound for binding to the
ATP binding pocket of EGFR. A total of 4 independent docking
experiments were performed for the limited TCM compounds with
maximum evaluation criteria based on number of runs for generating
the suitable pose. The optimum generated pose was studied using
LIGPLOT+ software version 4.5.3 (European bioinformatics
institute, Hinxton, UK).
Drug treatment and cell proliferation
assay
The human lung adenocarcinoma cell lines PC9GR4 and
H2347 were purchased from the American type culture collection
(ATCC, Manassas, VA, USA) and used in the present study. Cells were
seeded at a concentration of 1.5×104 cells/ml in a
24-well plate and cultured at 37°C in an incubator containing 5%
CO2 for 24 h in Dulbecco's modified Eagle's medium
supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). Concentrations of nardosinon
and gefitinib (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany)
ranging from 0.001 to 10 µM dissolved in dimethylsulfoxide (DMSO),
were subsequently added. The control group was treated with 0.1%
DMSO alone (vehicle control). Each experiment was performed five
times independently at each concentration. Cells were incubated at
37°C in a humidified atmosphere containing 5% CO2 for 24
h. The medium was subsequently removed and 0.1 mg/ml MTT solution
(Sigma-Aldrich; Merck Millipore) was added to the cells followed by
incubation for 4 h at 37.8°C in the dark. For control, 0.1 MTT
solution was added to a plate containing no cells. The supernatant
was subsequently removed and an equal volume of DMSO was added to
dissolve the formazan crystals. The absorbance was measured at 565
nm against background absorption at 650 nm using an EPOCH
Microplate Reader (BioTek Instruments, Inc., Winooski, VT,
USA).
Statistical analysis
One-way analysis of variance was used to calculate
significance. P-values between 0.001 and 0.01 were considered to
indicate a significant difference. All statistical tests were
performed using SPSS 18.0 for Windows (SPSS, Inc., Chicago, IL,
USA).
Results and Discussion
Protein preparation and virtual
screening
The T790M mutant form of EGFR kinase domain was
prepared from crystallographic structure (PDB ID: 1XKK). The
structure was subjected to energy minimization using GROMOS force
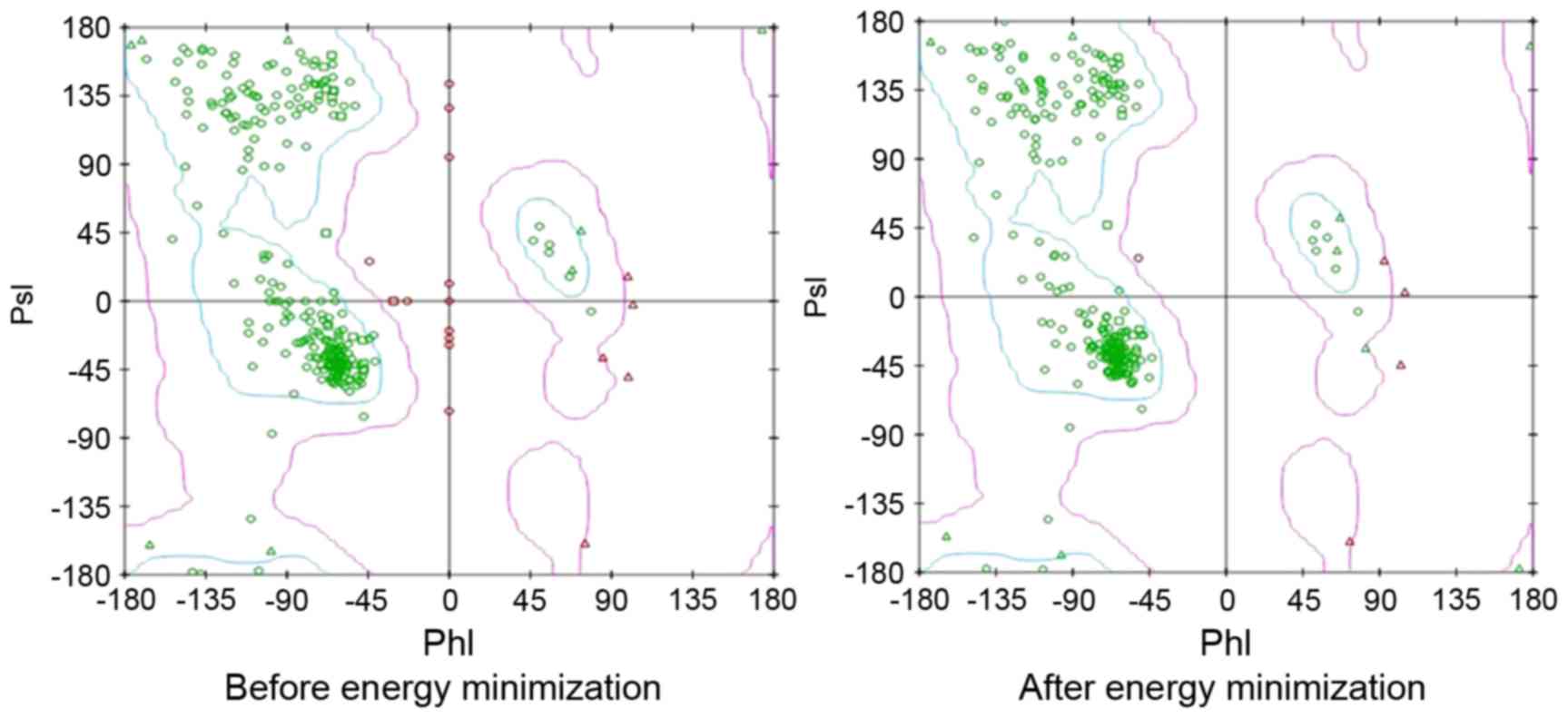
field with SPDB Viewer, and PsI-PhI distributions prior to and
following the energy minimization are depicted in Fig. 1. The purpose of virtual screening was
to limit the number of compounds without affecting the success rate
of compounds reaching further stages of drug development. A total
of 25 TCM compounds met the cut-off criteria (∆G <-10 kcal/mol)
set in virtual screening.
Predictive in silico ADME
The subset of 25 TCM compounds was further screened
on the basis of their drug-likeliness according to Lipinski's rule
of five (19). The online ADME/Tox
tool was used to check Lipinski's parameters and provided an
insight into the mutagenic and oncogenic potential of the compounds
tested, parameters that are an important basis for modern drug
design. A total of 4 compounds were shortlisted and used for
automated docking analysis. The following drug-likeliness
parameters were used for the shortlisted compounds: Total polar
surface area, molecular weight, calculated octanol/water partition
coefficient, number of hydrogen bond donors and number of hydrogen
bond acceptors (Table I).
 | Table I.Druglikeness and toxicity
prediction. |
Table I.
Druglikeness and toxicity
prediction.
| TCM No. | MW | logP | HBD | HBA | tpsa | Mutagenicity | Carcinogenicity |
|---|
| 00812 | 384.178 | 3.10 | 1 | 8 | 101 | NO | NO |
| 01273 | 416.111 | 0.46 | 5 | 9 | 146 | NO | NO |
| 01578 | 480.229 | 4.49 | 1 | 6 | 52.2 | NO | NO |
| 02108 | 250.333 | 2.96 | 1 | 3 | 35.5 | NO | NO |
Molecular docking analysis
The shortlisted TCM compounds were docked using the
AutoDock 4.2 tool into the optimized adenosine triphosphate (ATP)
binding site of the mutated EGFR protein and the results were
analyzed using Pymol and LIGPLOT+ software. The results
generated by the in silico analysis are presented in
Table II. All 4 shortlisted TCM
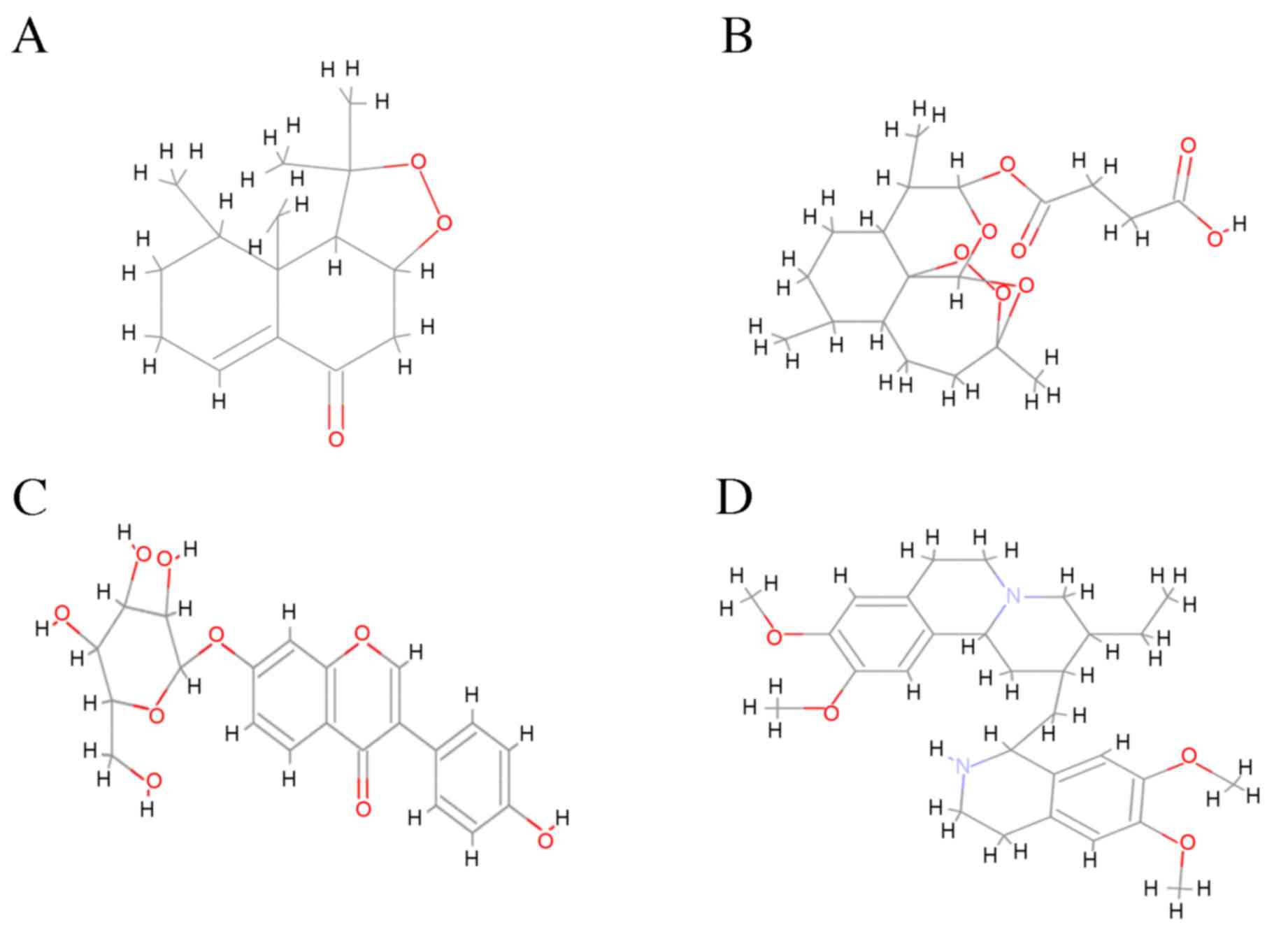
compounds (Fig. 2) interacted with
the ATP binding site via non-covalent interactions (hydrogen
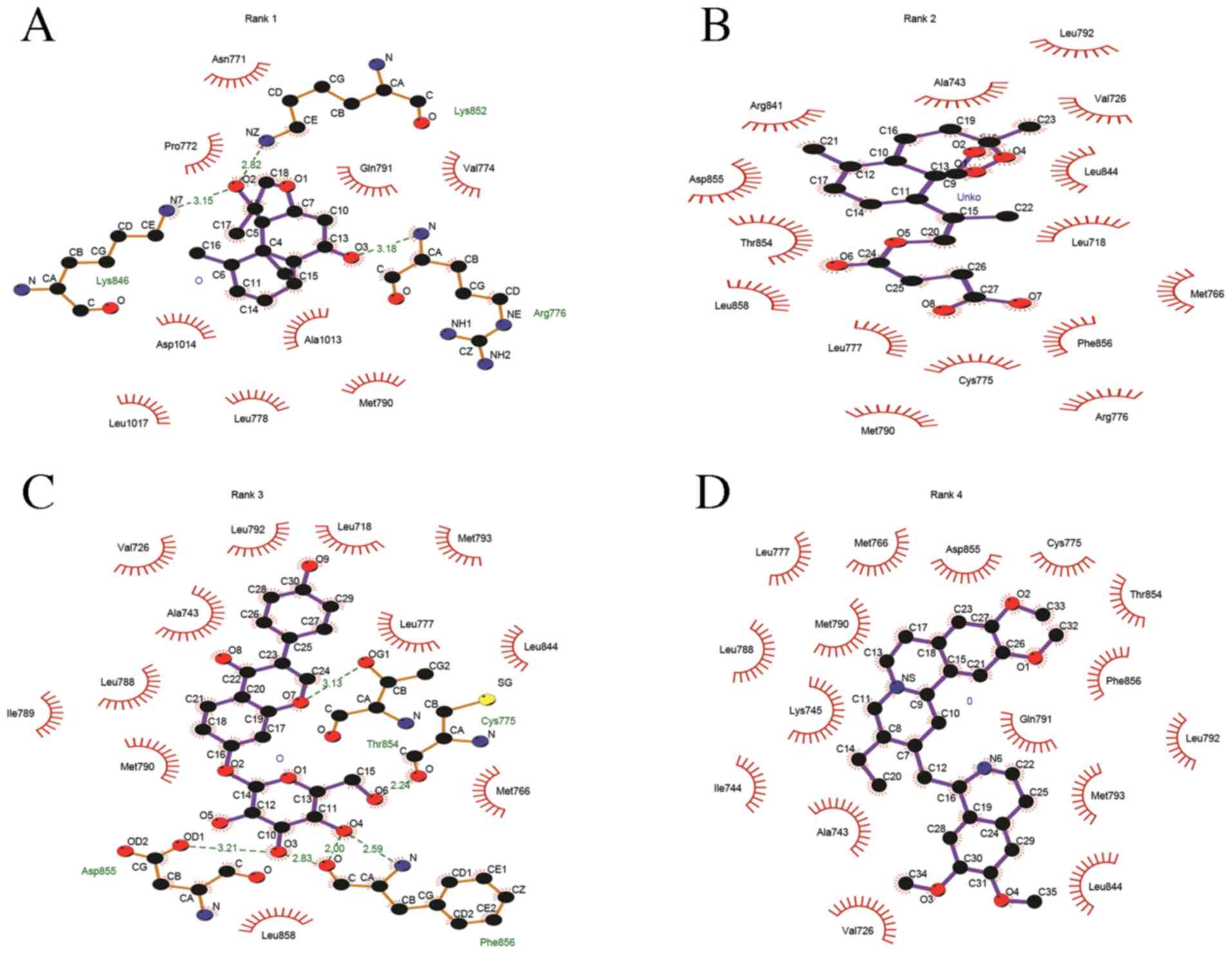
bonding). The two dimensional representation of the TCM-EGFR
interactions (hydrophobic or non covalent) are presented in
Fig. 3. The compounds were ranked
based on the spontaneity of their interaction with EGFR. Nardosinon
was determined to have a ΔG of −6.38 kcal/mol and form 3 hydrogen
bonds with the LYS852, ARG776 and LYS846 of the EGFR ATP binding
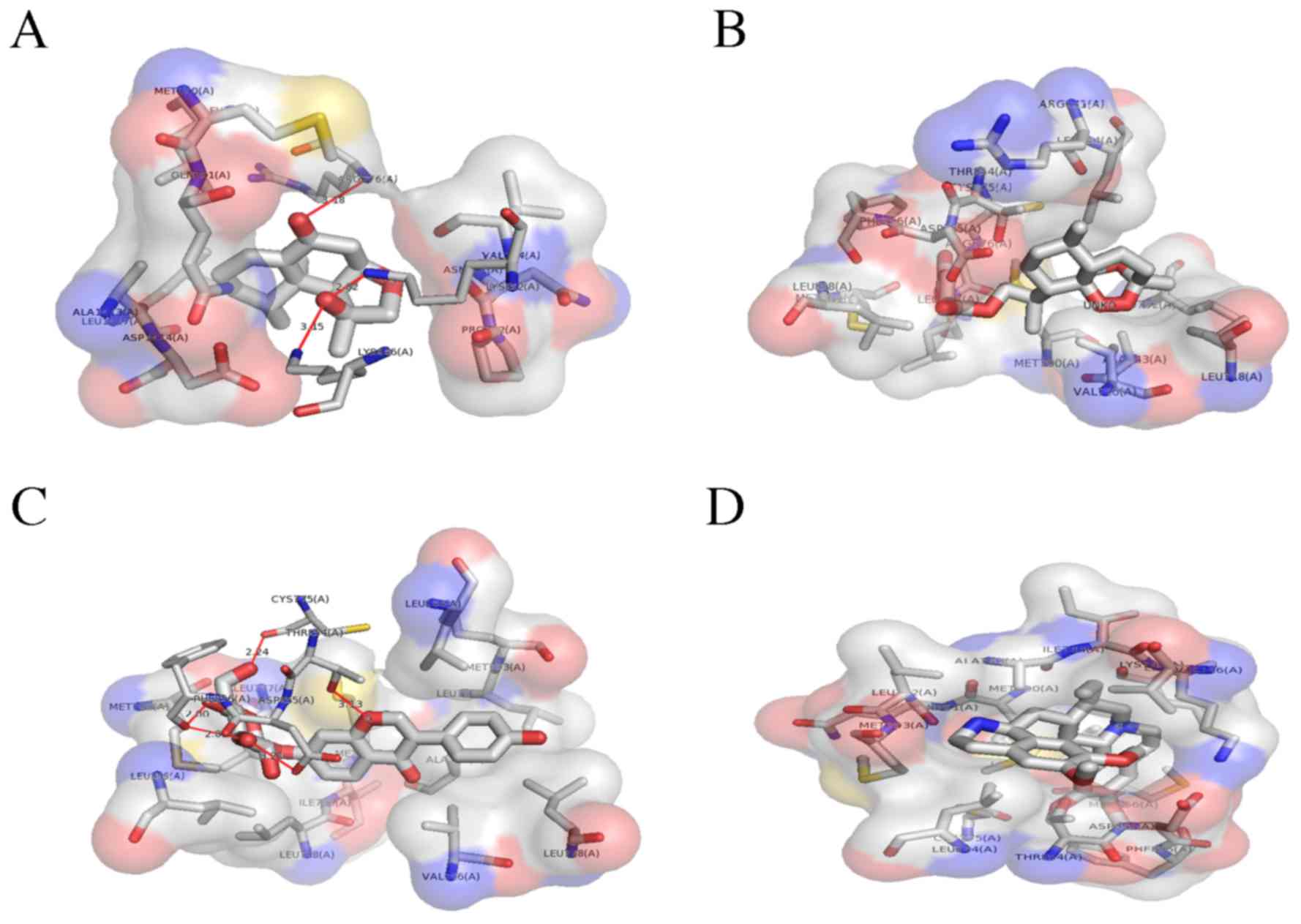
pocket (Fig. 4A). The hydrophobic
interaction pocket EGFR with nardosinon is comprised of the
following amino acids: ASN771, GLN791, VAL774, MET790, LEU778,
ALA1013, ASP1014, LEU1017 and PRO772. Oxygen at the second and
third positions of the ligand interacts with LYS 852, LYS 846 and
ARG776. The strongest interaction was between N on LYS852 and
O2 on nardosinon, with a distance of 2.82 Å.
 | Table II.Auto Dock analysis of four lead
natural products. |
Table II.
Auto Dock analysis of four lead
natural products.
| Name | TCM No. | ΔG, kcal/mol | Ligand binding
pocket | Hydrogen bonds |
|---|
| Lead 1 | 02108 | −6.38 | ASN771, GLN791,
VAL774, LYS852a, | LYS852:N-:
TCM:O2 (2.82 Å) |
|
|
|
| ARG776a, MET790, LEU778, ALA1013, | RG776:N-:
TCM:O3 (3.18 Å) |
|
|
|
| ASP1014, LEU1017,
LYS846a, PRO772 | LYS846:N-:
TCM:O2 (3.15 Å) |
| Lead 2 | 00812 | −5.82 | ALA743, LEU792,
VAL726, LEU844, | NIL |
|
|
|
| LEU718, MET766,
PHE856, CYS775, LEU777, |
|
|
|
|
| LEU858, THR854,
ASP855, ARG841 |
|
| Lead 3 | 01273 | −1.32 | ALA743, VAL726,
LEU792, LEU718, |
THR854:OG1-:TCM:O7
(3.13 Å) |
|
|
|
| MET793, LEU777,
LEU844, THR854a, |
CYS775:O-:TCM:O6 (2.24 Å) |
|
|
|
| CYS755a, MET766, PHE856a, LEU858, |
PHE856:N-:TCM:O4 (2.59 Å) |
|
|
|
| ASP855a, MET790, ILE789, LEU788 |
PHE856:O-:TCM:O4 (2.00 Å) |
|
|
|
|
|
PHE856:O-:TCM:O3 (2.83 Å) |
|
|
|
|
|
ASP855:OD1-:TCM:O3
(3.21 Å) |
| Lead 4 | 01578 | −0.32 | VAL726, ALA743,
ILE744, LYS745, | NIL |
|
|
|
| LEU788, LEU777,
MET790, MET766, |
|
|
|
|
| ASP855, CYS755,
THR854, PHE856, |
|
|
|
|
| GLN791, LEU792,
MET793, LEU844 |
|
The second ranked TCM was artesunate, which
interacted with the ATP binding pocket of mutated EGFR only. This
binding pocket comprises ALA743, LEU792, VAL726, LEU844, LEU718,
MET766, PHE856, CYS775, LEU777, LEU858, THR854, ASP855 and ARG841,
and has a ΔG of −5.82 kcal/mol (Fig.
4B).
The third ranked TCM was daidzin, which was
demonstrated to have a ΔG of −1.32 kcal/mol. The binding pocket
daidzin with mutated EGFR comprises the following amino acids:
ALA743, VAL726, LEU792, LEU718, MET793, LEU777, LEU844, THR854,
CYS755, MET766, PHE856, LEU858, ASP855, MET790, ILE789 and LEU788.
Of these, daidzin forms hydrogen bonds with 4 amino acids: THR854,
CYS755, PHE856 and ASP855. The O atoms at the 3rd, 4th, 6th and 7th
positions of daidzin form hydrogen bonds. Fig. 4C provides a 3D illustration of the
interactions.
The fourth ranked TCM compound was emetine, which
formed no non-covalent bonds with the mutated EGFR ATP binding
domain. The hydrophobic binding pocket comprises the following
amino acids: VAL726, ALA743, ILE744, LYS745, LEU788, LEU777,
MET790, MET766, ASP855, CYS755, THR854, PHE856, GLN791, LEU792,
MET793 and LEU844. The binding energy of the complex is −0.32
kcal/mol. Fig. 4D illustrates the
hydrophobic interactions between emetine with the ATP binding
domain.
Drug treatment and cell proliferation
assay
To investigate the effect of the top ranked TCM
compound, nardosinon, under in vitro conditions, the PC9GR4
cell line was used. These cells are derived from PC9 cells and are
resistant to gefitinib treatment due to the T790M mutation. Cells
were treated with nardosinon at 5 different concentrations (0.001,
0.01, 0.1, 1 and 10 µM). A total of five independent experiments
were performed for the 72-h assay with the different concentrations
of nardosinon. The procedure was repeated for the H2347 cell line,
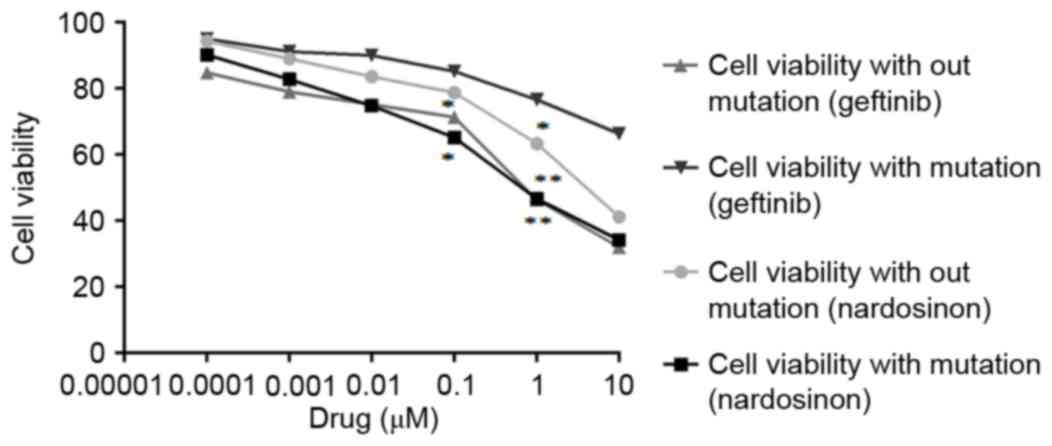
representing NSCLC with wild type EGFR. Fig. 5 illustrates the cell viability (%)
vs. drug concentration of the two cell lines with and without the
T790M mutation. The compound of interest, nardosinon (at a
concentration >0.1 µg) had a significantly greater effect
compared with geftinib on the viability of cells with the T790M
mutation (P<0.01), indicating that it could overcome the drug
resistance caused by the T790M mutation in NSCLC patients.
Following 72 h treatment, an IC50 of 225 mg/ml was calculated via
MTT assay.
The T790M mutation is in the protein kinase domain
of the EGFR and activates the ATP affinity of the EGFR kinase
(20). Blocking this activity may
have therapeutic potential. The difference in physical and chemical
properties, including size, charge, and hydrophobicity value caused
by the amino acid change from threonine to methionine at codon 790,
increased the activity in the kinase domain. In the present study,
computer-aided drug design of a pharmacological agent for the
treatment of patients with NSCLC and the T790M mutation has
revealed a potentially effective TCM compound, nardosinon.
Nardosinon is a compound found in the root extract of
Nardostachys jatamansi (21).
This plant is abundant in China and has a long history of medicinal
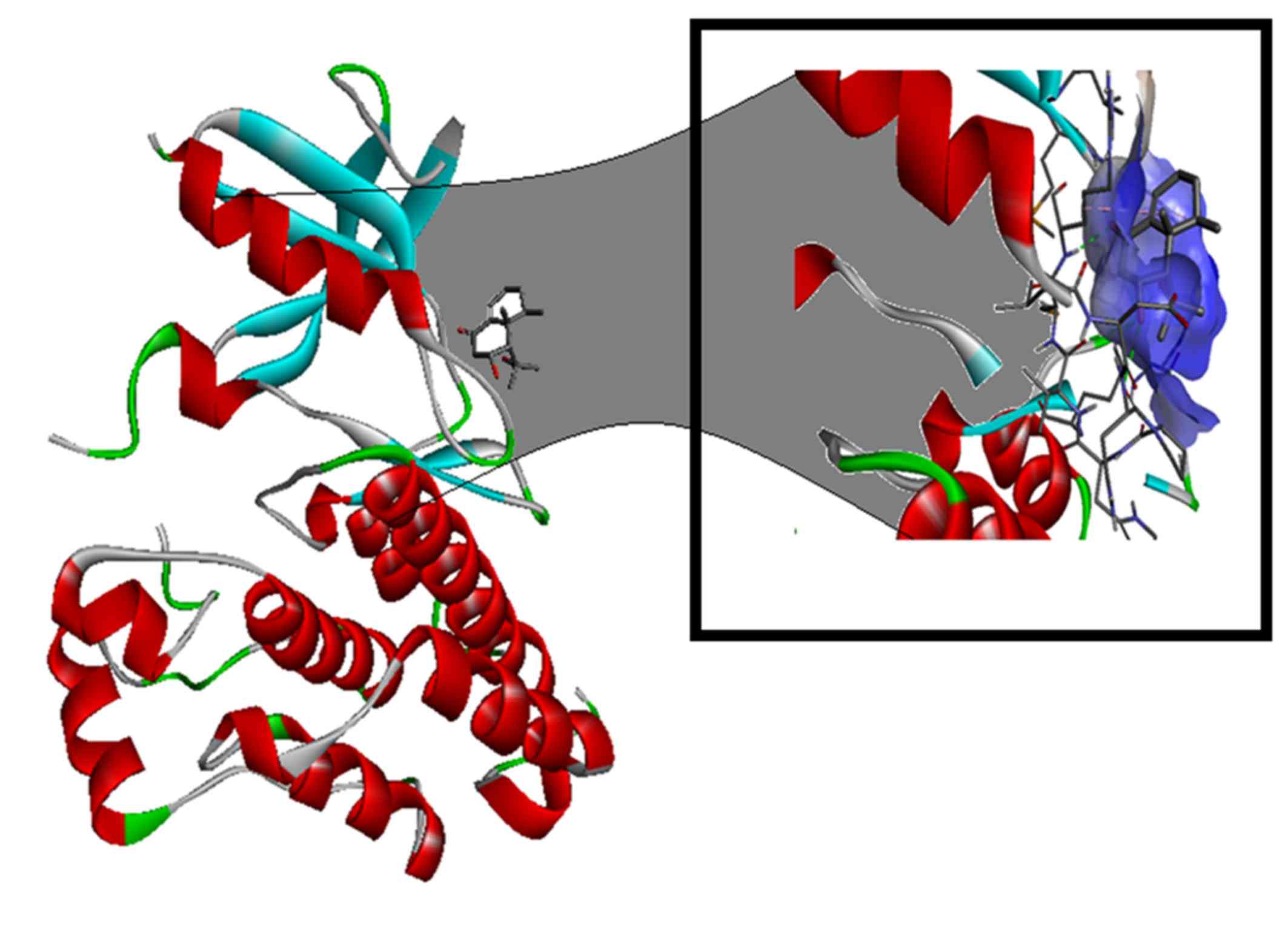
use (22–25). Nardosinon is able to bind to the ATP
binding site of the EGFR (Fig. 6)
and the results from the in vitro studies of the nardosinon
compound in two different cell lines indicates that it is may be
developed as a pharmacological agent capable of treating NSCLC in
patients with T790M EGFR variation.
References
|
1
|
Kobayashi S, Boggon TJ, Dayaram T, Jänne
PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG and Halmos
B: EGFR mutation and resistance of non-small-cell lung cancer to
gefitinib. N Engl J Med. 352:786–792. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang J, Jiang Y, Liang H, Li P, Xiao H, Ji
J, Xiang W, Shi JF, Fan YG, Li L, et al: Attributable causes of
cancer in China. Ann Oncol. 23:2983–2999. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Scagliotti GV, De Marinis F, Rinaldi M,
Crinò L, Gridelli C, Ricci S, Matano E, Boni C, Marangolo M, Failla
G, et al: Phase III randomized trial comparing three platinum-based
doublets in advanced non-small-cell lung cancer. J Clin Oncol.
20:4285–4291. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mendelsohn J and Baselga J: Status of
epidermal growth factor receptor antagonists in the biology and
treatment of cancer. J Clin Oncol. 21:2787–2799. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Miller VA, Kris MG, Shah N, Patel J,
Azzoli C, Gomez J, Krug LM, Pao W, Rizvi N, Pizzo B, et al:
Bronchioloalveolar pathologic subtype and smoking history predict
sensitivity to gefitinib in advanced non-small-cell lung cancer. J
Clin Oncol. 22:1103–1109. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu YL, Zhou C, Hu CP, Feng J, Lu S, Huang
Y, Li W, Hou M, Shi JH, Lee KY, et al: Afatinib versus cisplatin
plus gemcitabine for first-line treatment of Asian patients with
advanced non-small-cell lung cancer harbouring EGFR mutations
(LUX-Lung 6): An open-label, randomised phase 3 trial. Lancet
Oncol. 15:213–222. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gervas P, Ivanova A, Vasiliev N, Ananina
O, Zharkova O, Rogovieva O, Verzhbitskaya N, Didichuk I,
Cheremisina O, Popova N, et al: Frequency of EGFR mutations in
non-small cell lung cancer patients: Screening data from west
siberia. Asian Pac J Cancer Prev. 16:689–792. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chao TT, Wang CY, Lai CC, Chen YL, Tsai
YT, Chen PT, Lin HI, Huang YC, Shiau CW, Yu CJ and Chen KF: TD-19,
an erlotinib derivative, induces epidermal growth factor receptor
wild-type nonsmall-cell lung cancer apoptosis through
CIP2A-mediated pathway. J Pharmacol Exp Ther. 351:352–358. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Linardou H, Dahabreh IJ, Bafaloukos D,
Kosmidis P and Murray S: Somatic EGFR mutations and efficacy of
tyrosine kinase inhibitors in NSCLC. Nat Rev Clin Oncol. 6:352–366.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Helena AY, Arcila ME, Rekhtman N, Sima CS,
Zakowski MF, Pao W, Kris MG, Miller VA, Ladanyi M and Riely GJ:
Analysis of tumor specimens at the time of acquired resistance to
EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers.
Clin Cancer Res. 19:2240–2247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gandhi J, Zhang J, Xie Y, Soh J,
Shigematsu H, Zhang W, Yamamoto H, Peyton M, Girard L, Lockwood WW,
et al: Alterations in genes of the EGFR signaling pathway and their
relationship to EGFR tyrosine kinase inhibitor sensitivity in lung
cancer cell lines. PLoS One. 4:e45762009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fan XX, Wong MP, Cao ZW, Li N, Wu JL, Zhou
H, Jiang Z, Liu L and Leung E: Abstract 3193: Apoptotic effect of a
single compound derived from natural product in Gefitinib-resistant
non-small cell lung cancer cells. Cancer Res. 74:2014. View Article : Google Scholar
|
|
13
|
Chikan NA, Bhavaniprasad V, Anbarasu K,
Shabir N and Patel TN: From natural products to drugs for
epimutation computer-aided drug design. Appl Biochem Biotechnol.
170:164–175. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wood ER, Truesdale AT, McDonald OB, Yuan
D, Hassell A, Dickerson SH, Ellis B, Pennisi C, Horne E, Lackey K,
et al: A unique structure for epidermal growth factor receptor
bound to GW572016 (Lapatinib) Relationships among protein
conformation, inhibitor off-rate, and receptor activity in tumor
cells. Cancer Res. 64:6652–6659. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guex N and Peitsch MC: SWISS-MODEL and the
Swiss-PdbViewer: An environment for comparative protein modeling.
Electrophoresis. 18:2714–2723. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Trott O and Olson AJ: AutoDock Vina:
Improving the speed and accuracy of docking with a new scoring
function, efficient optimization, and multithreading. J Comput
Chem. 31:455–461. 2010.PubMed/NCBI
|
|
17
|
Seeliger D and de Groot BL: Ligand docking
and binding site analysis with PyMOL and Autodock/Vina. J Comput
Aided Mol Des. 24:417–422. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Morris GM, Huey R, Lindstrom W, Sanner MF,
Belew RK, Goodsell DS and Olson AJ: AutoDock4 and AutoDockTools4:
Automated docking with selective receptor flexibility. J Comput
Chem. 30:2785–2791. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lipinski CA, Lombardo F, Dominy BW and
Feeney PJ: Experimental and computational approaches to estimate
solubility and permeability in drug discovery and development
settings. Adv Drug Deliv Rev. 46:3–26. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yun CH, Mengwasser KE, Toms AV, Woo MS,
Greulich H, Wong KK, Meyerson M and Eck MJ: The T790M mutation in
EGFR kinase causes drug resistance by increasing the affinity for
ATP. Proc Natl Acad Sci USA. 105:2070–2075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bae GS, Kim MS, Park KC, Koo BS, Jo IJ,
Choi SB, Lee DS, Kim YC, Kim TH, Seo SW, et al: Effect of
biologically active fraction of Nardostachys jatamansi on
cerulein-induced acute pancreatitis. World J Gastroenterol.
18:3223–3234. 2012.PubMed/NCBI
|
|
22
|
Li W, Shi J, Li Q, Duan HH and Tang MK:
Nardosinone reduces neuronal injury induced by oxygen-glucose
deprivation in primary cortical cultures. Yao Xue Xue Bao.
48:1422–1429. 2013.(In Chinese). PubMed/NCBI
|
|
23
|
Ju SM, Lee J, Kang JG, Jeong SO, Park JH,
Pae HO, Lee GS, Kim WS, Lyu YS and Jeon BH: Nardostachys chinensis
induces granulocytic differentiation with the suppression of cell
growth through p27(Kip1) protein-related G0/G1 phase arrest in
human promyelocytic leukemic cells. Pharm Biol. 53:1002–1009. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
More SV, Koppula S, Kim IS, Kumar H, Kim
BW and Choi DK: The role of bioactive compounds on the promotion of
neurite outgrowth. Molecules. 17:6728–6753. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ahmad A, Sattar MZ, Rathore HA, Fatima T,
Khan SA, Afzal S, Lazhari M, Ahmed FU and Eseyin O: Pharmacological
importance of Nardostachys jatamansi DC: A potential therapeutic
agent in different pathological ailments. J Chem Pharmaceutical
Res. 5:431–438. 2013.
|