Introduction
Although hypertension left ventricular hypertrophy
is the compensation for excessive pressure load by means of
structural changes to a certain extent, more and more studies
showed that continuous myocardial hypertrophy easily leads to the
heart failure (1,2). In recent years, plenty of studies have
analyzed mechanism of formation for left ventricular hypertrophy
respectively from three aspects, including extracellular
stimulation signal, intracellular signal transduction and gene
transcription. Moreover, a many significant signaling pathways have
been identified, such as small G proteins, mitogen-activated
protein kinase (MAPK) and calcineurin (CaN). However, the specific
occurrence and development mechanism of ventricular hypertrophy is
still not fully clarified (3–6).
Therefore, to determine the active factors that could regulate
myocardial hypertrophy and reveal the cause of myocardial
hypertrophy at the molecular level is important.
TRB3 gene was found in Drosophila, and
reported to be a regulatory protein, for the process of sugar,
lipid metabolism and adipocyte differentiation (7,8). In
addition, relevant study results showed that the TRB3 gene could
also act on the mitogen activated protein kinase signaling pathway
to inhibit cell proliferation and induce apoptosis (9,10). The
apoptosis of endothelial cells was closely associated with the
occurrence and development of cardiac and cerebral vascular disease
(11). Therefore, it was suggested
that if the TRB3 gene was related to the formation of myocardial
hypertrophy thereby, affecting polymorphism genetic variation of
TRB3 gene expression, it may be associated with the development
mechanism of myocardial hypertrophy.
MicroRNAs are a class of non-coding RNA, which have
the function of regulating and controlling (12,13). The
polymorphic variation in target sequence usually affects the
recognition and binding of the seed sequence and target sequence,
thus affecting the expression of the target gene. Therefore, we
hypothesized that polymorphism in microRNAs and TRB3 gene site may
be associated with the formation mechanism of hypertension left
ventricular hypertrophy. In order to prove the inference, we first
identified by the means of PCR, the polymorphic site of TRB3 gene
SNP, and its association with hypertension left ventricular
hypertrophy was investigated. After determining the polymorphic
site bioinformatics was used to explore interacting microRNAs.
Patients and methods
Patient data
One hundred twenty-six patients, admitted to to the
First Affiliated Hospital of Nanchang University with confirmed
hypertension from January 2014 to December 2015, were selected for
the present study. They were divided into two groups. The left
ventricular hypertrophy group of 43 cases aged 55–73 years
(average, 66.3 years) and non-left ventricular hypertrophy group of
83 cases aged 49–75 years (average, 58.4 years). Additionally, 31
healthy people were selected as control, aged 40–60 years (average,
55.8 years). The clinical data are shown in Table I. This study was approved by the
Ethics Committee of the First Affiliated Hospital of Nanchang
University. Signed written informed consents were obtained from all
participants before the study.
 | Table I.Clinical data of different groups of
patients with hypertension. |
Table I.
Clinical data of different groups of
patients with hypertension.
|
| Hypertension
group |
|
|---|
|
|
|
|
|---|
| Clinical
features | Left ventricular
hypertrophy group | Non-left ventricular
hypertrophy group | Control group |
|---|
| Body weight (kg) | 75.3±1.2 | 73.4±2.3 | 70.1±0.9 |
| Systolic pressure
(mmHg) |
161.5±6.5a |
153.1±4.3a |
123.5±3.6a,b |
| Diastolic pressure
(mmHg) |
110.5±3.5a |
100.8±5.8a | 60.2±4.1a,b |
| Triglyceride
(mmol/l) | 1.93±1.2 | 1.88±1.5 | 1.42±0.5 |
| Fasting plasma
glucose (mmol/l) | 6.12±0.6 | 6.87±0.4 | 5.93±0.5 |
| Smoking rate (%) | 17 | 22 | 10 |
| Alcohol intake rate
(%) | 30 | 28 | 15 |
PCR amplification
Primers were synthesized by Shenzhen Huada Gene
Technology Co., Ltd. (Shenzhen, China), and the amplification
sequence was as described in literature (14), covering the second and third exon.
Reaction conditions were in accordance with literature (15).
Gene sequencing
Fifteen subjects were randomly selected from the
control and the two groups for a total of 45 cases, whose PCR
amplification products were directly sequenced. Also, their DNA
variation sequences were analyzed, according to literature
(16).
Genetic balance and frequency
statistical test
The analytical method was as previously described
(17).
Bioinformatics analysis
The bioinformatics analytical method was as
described in literature (18).
Pictar database (http://www.pictar.org) was used to scan the TRB3
polymorphic site, in order to find the microRNAs that have specific
binding.
Luciferase activity
determinations
The specific activity determination method was
selected according to literature (19).
Statistical analysis
SPSS version 13.0 statistical software (SPSS, Inc.,
Chicago, IL, USA) was used for data analysis. Chi-square test was
adopted to compare classification statistics data. Test level was
α0.05. P<0.05 was considered to indicate a statistically
significanct difference.
Results
Detection of polymorphic site SNP of
TRB3 gene
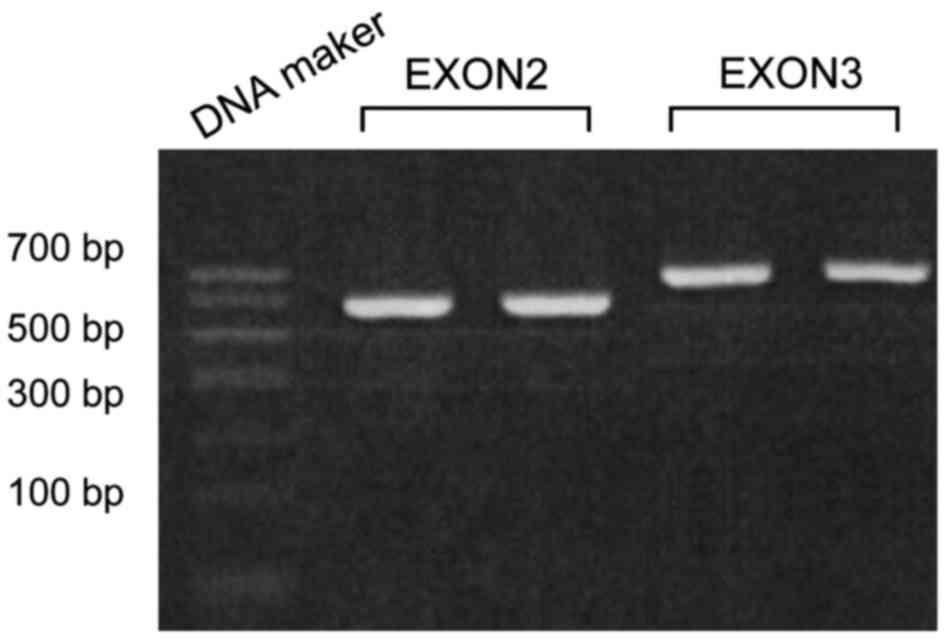
Direct PCR was applied to explore SNP of TRB3 gene.
The electrophoretogram of exon2 and exon3 after PCR amplification
is shown in Fig. 1, and sequencing
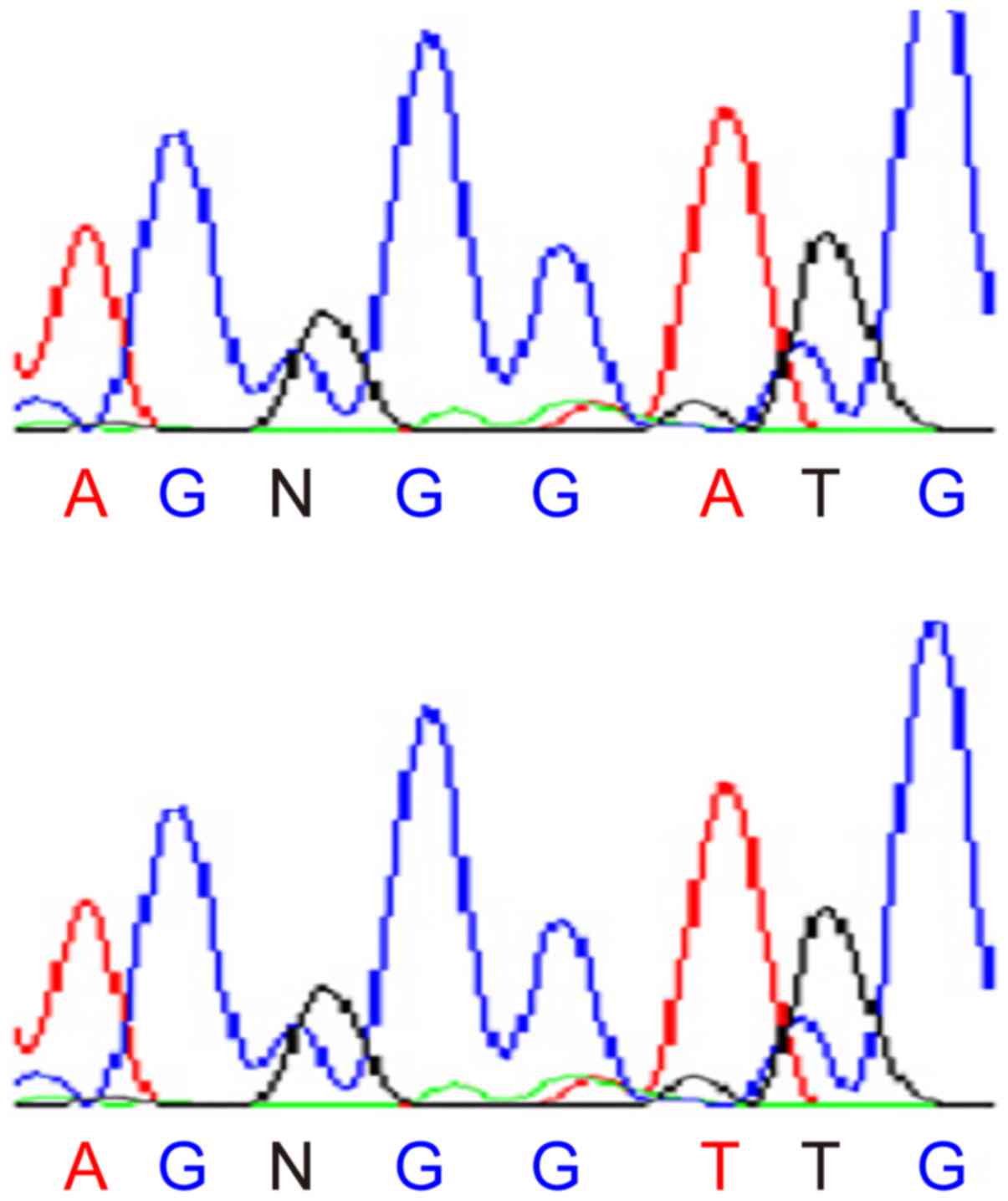
results are shown in Fig. 2.
After the amplified fragments were purified forward
sequencing was performed, DNA sequencing results of exon2 were:
5′-TCACTAAAATCAAATCCCTTTTTTT(A/T)AAT ATCCAATCNAGTATATCCCAAA-3′. The
SNP site (A/T). By means of the NCBI database, it was concluded
that this gene was consistent with the SNP site rs6186912 of TRB3
gene. Similarly, DNA sequencing results of exon3 were:
5′-AATCCCTTTTTTTNAATATCCAATC(C/T)AGTATATC CCAAAATGACATGAATA-3′.
The SNP site (C/T). By means of NCBI database, it
was concluded that this gene was consistent with the SNP site
rs6186923 of the TRB3 gene.
Genotyping
Ten patients with hypertension of left ventricular
hypertrophy, 10 cases of non left ventricular hypertrophy and 10
cases of healthy controls were randomly selected. The sequences
were used as primer for PCR, and then the product was sequenced and
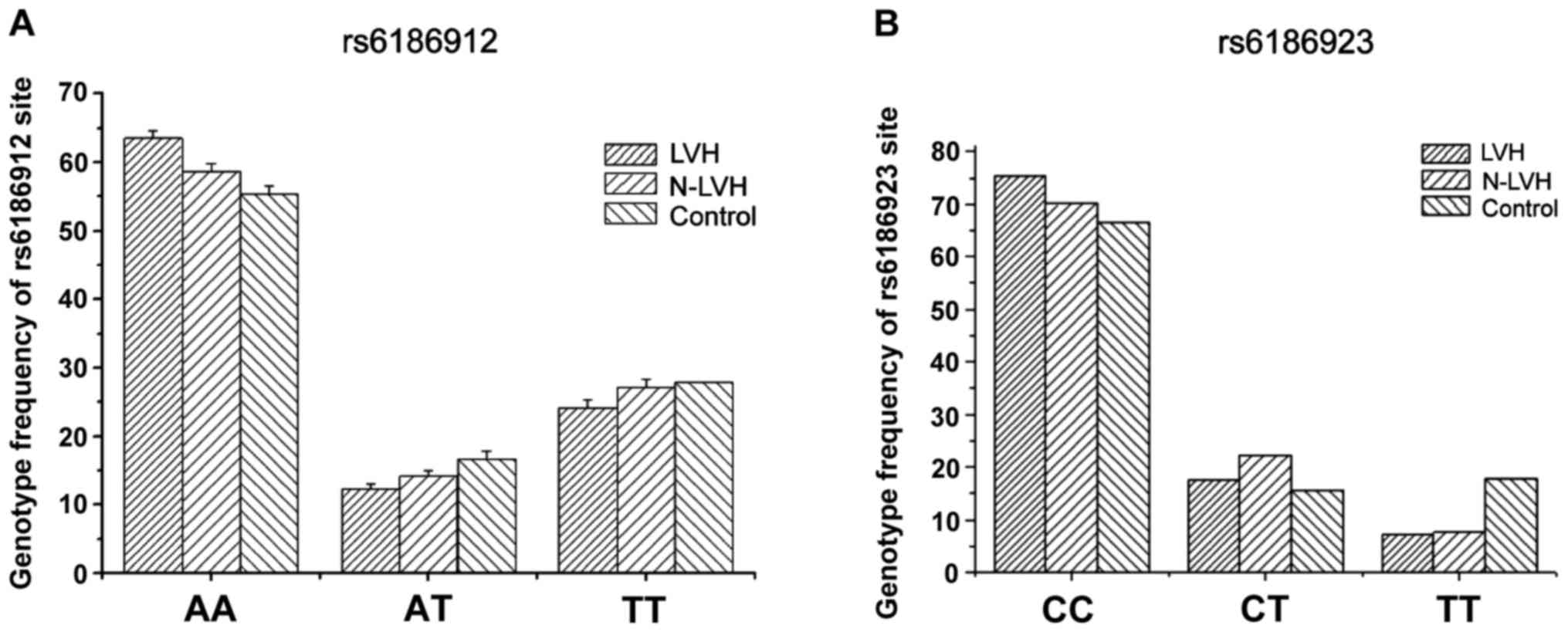
analyzed. The results are shown in Table II. The comparison of AA, AT and TT
genotype frequency of rs6186912 site between each of the two groups
of hypertension and control group had no statistical significance
(P>0.05) (Fig. 3A). While
comparison of CC, CT and TT of rs6186923 site had significant
difference (Fig. 3B). T allele of
left ventricular hypertrophy group was evidently lower than that of
non-left ventricular hypertrophy group and normal group (15.4 vs.
22.3%, P<0.05), but comparison of T allele between non-left
ventricular hypertrophy group and normal group had no distinct
difference.
 | Table II.SNP genotype and allele frequency
distributions of gene sequencing results in different patient
groups and control group. |
Table II.
SNP genotype and allele frequency
distributions of gene sequencing results in different patient
groups and control group.
|
| Patient groups
(%) |
|
|---|
|
|
|
|
|---|
|
| Left ventricular
hypertrophy group | Non left ventricular
hypertrophy group | Control group
(%) |
|---|
| rs6186912 |
| AA | 63.5 | 58.7 | 55.4 |
| AT | 12.3 | 14.2 | 16.7 |
| TT | 24.2 | 27.1 | 27.9 |
| Allele |
| A allele | 19.5 | 21.3 | 22.1 |
| T allele | 80.5 | 78.7 | 77.9 |
| rs6186923 |
| CC | 75.3 | 70.1 | 66.6 |
| CT | 17.5 | 22.3 | 15.6 |
| TT | 7.2 | 7.6 | 17.8 |
| Allele |
| C allele | 84.6 | 77.7 | 74.4 |
| T allele | 15.4 | 22.3 | 25.6 |
Bioinformatics analysis
After determining the association between rs6186923
SNPs gene and hypertension left ventricular hypertrophy, this
experiment was designed to find the corresponding microRNAs action
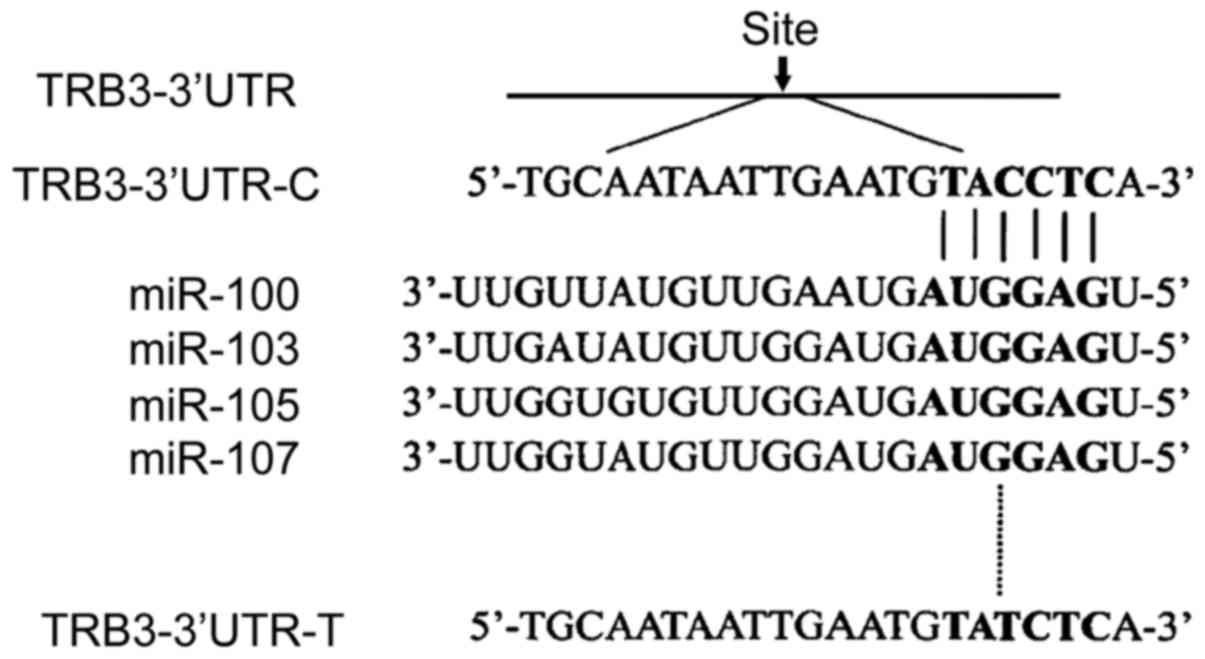
site by bioinformatics method. Through the analysis of the online
software Pictar, it was observed that TRB3 polymorphic site
rs6186923 could be combined with miR-100, miR-103, miR-105 and
miR-107, as shown in Fig. 4. The C
allele, T allele and target sequence mRNA were conformed to
Watson-Crick matching principle. According to the analysis of
Pictar, it was noted that thermodynamic free energy of binding
between rs6186923 and miR-100 target sequence was the highest.
Therefore, to carry out the action which adopts miR-100 target
sequence and rs6186923 was the next step.
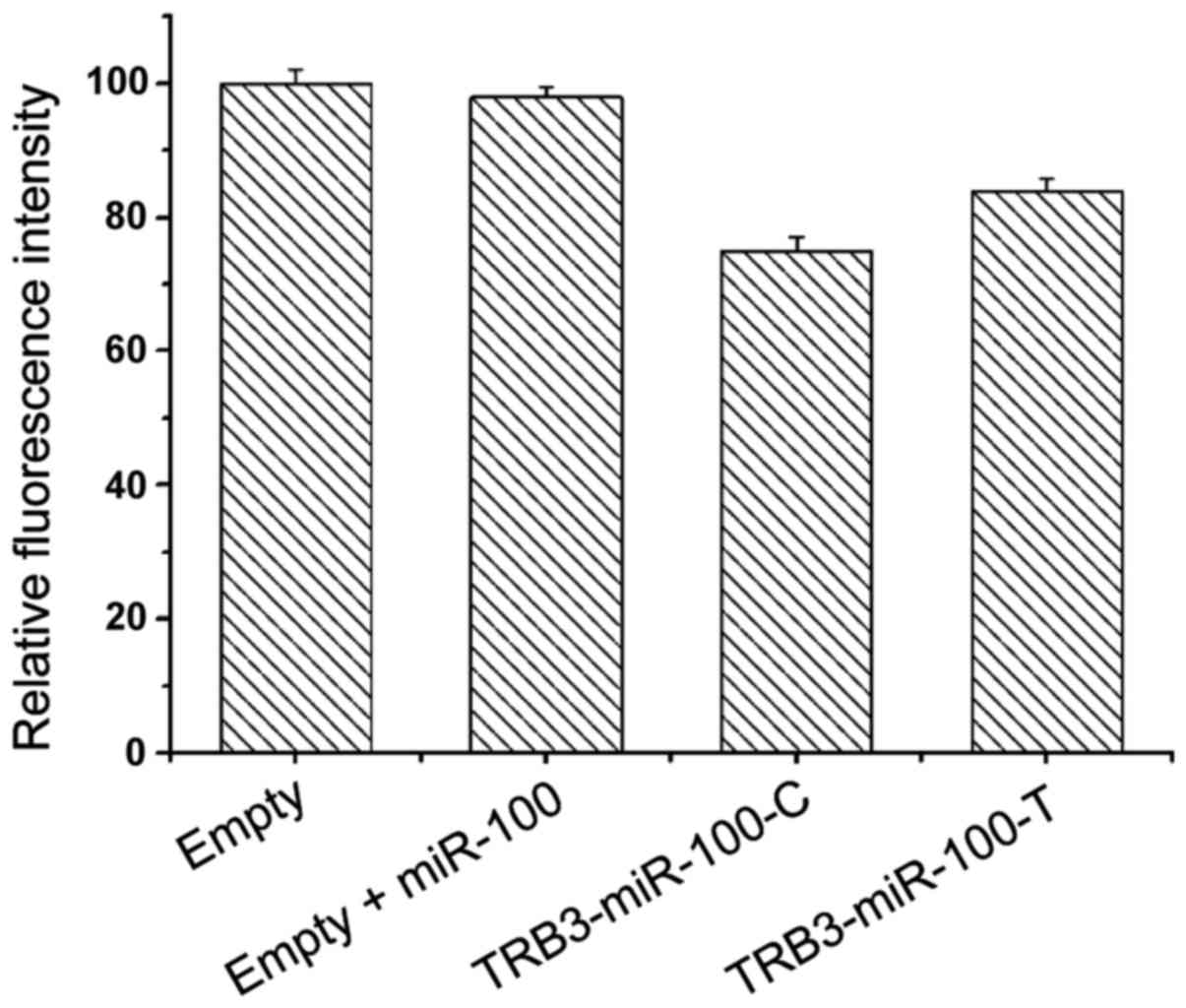
Luciferase test
In order to verify the interaction between the
miR-100 target sequence and TRB3 gene rs6186923, the SNP sequence
was cloned into the 3′UTR region of the reporter gene luciferase.
The principle was that if miR-100 could act on rs6186923 gene, the
translation of reporter gene luciferase would be inhibited, thereby
reducing the fluorescence signal, so the intensity of fluorescence
signal could be used to reflect the inhibitory effect of miR-100
target sequence on rs6186923. The experiment results are shown in
Table III and Fig. 5.
 | Table III.Luciferase test results. |
Table III.
Luciferase test results.
|
| Blank | Blank+miR-100 | TRB3-miR100-C | TRB3-miR100-T |
|---|
| Fluorescence signal
(%) | 100 | 98±1.5 | 75±2.1 | 84±1.7 |
Discussion
In recent years, the enhancement of people's living
standard and the changes in diet, have resulted in elevation of the
incidences of cardiovascular disease (20). Hypertension left ventricular
hypertrophy is a risk factor, which affects the incidence of
cardiovascular disease (21),
however, the mechanism of its formation is still not clear.
Therefore, the purpose of this study was to investigate the
formation mechanism of hypertension left ventricular hypertrophy at
the molecular level, which would lay the foundation for further
study. Prior to the beginning of this study, by the means of
consulting literature it was noted that TRB3 gene is a significant
regulatory factor, which may be associated with the occurrence and
development of cardio-cerebral vascular disease. In recent years,
microRNAs have been observed to play a key role in the regulation
of myocardial hypertrophy. Based on this, we speculated that the
association between TRB3 gene and ventricular hypertrophy, would
allow to explore the formation mechanism of ventricular
hypertrophy.
Therefore, in the present study, SNPs sites of TRB3
gene were investigated by direct PCR method. The PCR products were
then purified and sequenced, and compared with NCBI database. It
was observed that sequencing results of exon2 and exon3 were
corresponding, respectively, with rs6186912 and rs6186923, showing
that SNPs sites were accurate. Then, for the genotypes, the study
investigated the association between rs6186912, rs6186923 and
patients with hypertension left ventricular hypertrophy. The
experiment indicated that the comparison of AA, AT and TT genotype
frequency of rs6186912 site between the groups of hypertension and
control had no statistical significant difference (P>0.05). This
confirmed that SNP site was not directly related to myocardial
hypertrophy. However, comparison of CC, CT and TT of rs6186923 site
had significant differences. T allele of left ventricular
hypertrophy group was evidently lower than that of non-left
ventricular hypertrophy group and normal group (P<0.05), but
comparison of T allele between non-left ventricular hypertrophy
group and normal group showed no distinct difference, which
indicated that this SNP was closely related to hypertension and
myocardial hypertrophy.
After determining the association between TRB3
polymorphism gene and hypertension left ventricular hypertrophy,
corresponding microRNAs were explored by bioinformatics. Through
the analysis of the online software Pictar, it was concluded that
there were 4 microRNAs: miR-100, miR-103, miR-105 and miR-107 that
could act on the rs6186923SNP site of TRB3. Further it was noted
that thermodynamic free energy of binding between rs6186923 and
miR-100 target sequence was the highest. Therefore, miR-100 was
used for subsequent analysis as microRNAs to interact on rs6186923.
Then luciferase test was applied to investigate the effect of
miR-100 on rs6186923, showing that the expression of TRB3-miR100-C
gene and TRB3-miR100-T gene luciferase were significantly reduced,
which confirmed the effect of miR-100 gene on rs6186923.
In conclusion, microRNA-miR100 affected hypertension
left ventricular hypertrophy by mediating polymorphic site of TRB3
gene rs6186923. However, verification of miR-100 on rs6186923 by
western blot needs to be further evaluated in future studies.
References
|
1
|
Nanayakkara G, Viswaprakash N, Zhong J,
Kariharan T, Quindry J and Amin R: PPARγ activation improves the
molecular and functional components of I(to) remodeling by
angiotensin II. Curr Pharm Des. 19:4839–4847. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Subramanian V, Golledge J, Heywood EB,
Bruemmer D and Daugherty A: Regulation of peroxisome
proliferator-activated receptor-γ by angiotensin II via
transforming growth factor-β1-activated p38 mitogen-activated
protein kinase in aortic smooth muscle cells. Arterioscler Thromb
Vasc Biol. 32:397–405. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Angeloni C and Hrelia S: Quercetin reduces
inflammatory responses in LPS-stimulated cardiomyoblasts. Oxid Med
Cell Longev. 2012:8371042012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Takahashi K, Sasaki T, Mammoto A, Takaishi
K, Kameyama T, Tsukita S and Takai Y: Direct interaction of the Rho
GDP dissociation inhibitor with ezrin/radixin/moesin initiates the
activation of the Rho small G protein. J Biol Chem.
272:23371–23375. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bonni A, Brunet A, West AE, Datta SR,
Takasu MA and Greenberg ME: Cell survival promoted by the Ras-MAPK
signaling pathway by transcription-dependent and -independent
mechanisms. Science. 286:1358–1362. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Matsumura T, Kinoshita H, Ishii N, Fukuda
K, Motoshima H, Senokuchi T, Taketa K, Kawasaki S, Nishimaki-Mogami
T, Kawada T, et al: Telmisartan exerts antiatherosclerotic effects
by activating peroxisome proliferator-activated receptor-γ in
macrophages. Arterioscler Thromb Vasc Biol. 31:1268–1275. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Du K, Herzig S, Kulkarni RN and Montminy
M: TRB3: a tribbles homolog that inhibits Akt/PKB activation
by insulin in liver. Science. 300:1574–1577. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ohoka N, Yoshii S, Hattori T, Onozaki K
and Hayashi H: TRB3, a novel ER stress-inducible gene, is induced
via ATF4-CHOP pathway and is involved in cell death. EMBO J.
24:1243–1255. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qi L, Heredia JE, Altarejos JY, Screaton
R, Goebel N, Niessen S, Macleod IX, Liew CW, Kulkarni RN, Bain J,
et al: TRB3 links the E3 ubiquitin ligase COP1 to lipid metabolism.
Science. 312:1763–1766. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Panicker SR, Sreenivas P, Babu MS,
Karunagaran D and Kartha CC: Quercetin attenuates monocyte
chemoattractant protein-1 gene expression in glucose primed aortic
endothelial cells through NF-kappaB and AP-1. Pharmacol Res.
62:328–336. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Meléndez GC, McLarty JL, Levick SP, Du Y,
Janicki JS and Brower GL: Interleukin 6 mediates myocardial
fibrosis, concentric hypertrophy, and diastolic dysfunction in
rats. Hypertension. 56:225–231. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kozomara A and Griffiths-Jones S: miRBase:
annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res. 42:D68–D73. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Farazi TA, Hoell JI, Morozov P and Tuschl
T: MicroRNAs in human cancer. Adv Exp Med Biol. 774:1–20. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hoshino A, Matoba S, Iwai-Kanai E,
Nakamura H, Kimata M, Nakaoka M, Katamura M, Okawa Y, Ariyoshi M,
Mita Y, et al: p53-TIGAR axis attenuates mitophagy to exacerbate
cardiac damage after ischemia. J Mol Cell Cardiol. 52:175–184.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li YC, Liu YY, Hu BH, Chang X, Fan JY, Sun
K, Wei XH, Pan CS, Huang P, Chen YY, et al: Attenuating effect of
post-treatment with QiShen YiQi Pills on myocardial fibrosis in rat
cardiac hypertrophy. Clin Hemorheol Microcirc. 51:177–191.
2012.PubMed/NCBI
|
|
16
|
Hou J and Kang YJ: Regression of
pathological cardiac hypertrophy: signaling pathways and
therapeutic targets. Pharmacol Ther. 135:337–354. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lv D, Liu J, Zhao C, Sun Q, Zhou Q, Xu J
and Xiao J: Targeting microRNAs in pathological hypertrophy and
cardiac failure. Mini Rev Med Chem. 15:475–478. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Y and Zhang Z: Computational biology in
microRNA. Wiley Interdiscip Rev RNA. 6:435–452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liang Y, Qiu X, Xu RZ and Zhao XY:
Berbamine inhibits proliferation and induces apoptosis of KU812
cells by increasing Smad3 activity. J Zhejiang Univ Sci B.
12:568–574. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Eilat-Adar S, Sinai T, Yosefy C and Henkin
Y: Nutritional recommendations for cardiovascular disease
prevention. Nutrients. 5:3646–3683. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhong C, Luzhan Z, Genshan M, Jiahong W,
Xiaoli Z and Qi Q: Monocyte chemoattractant protein-1-2518 G/A
polymorphism, plasma levels, and premature stable coronary artery
disease. Mol Biol Rep. 37:7–12. 2010. View Article : Google Scholar : PubMed/NCBI
|