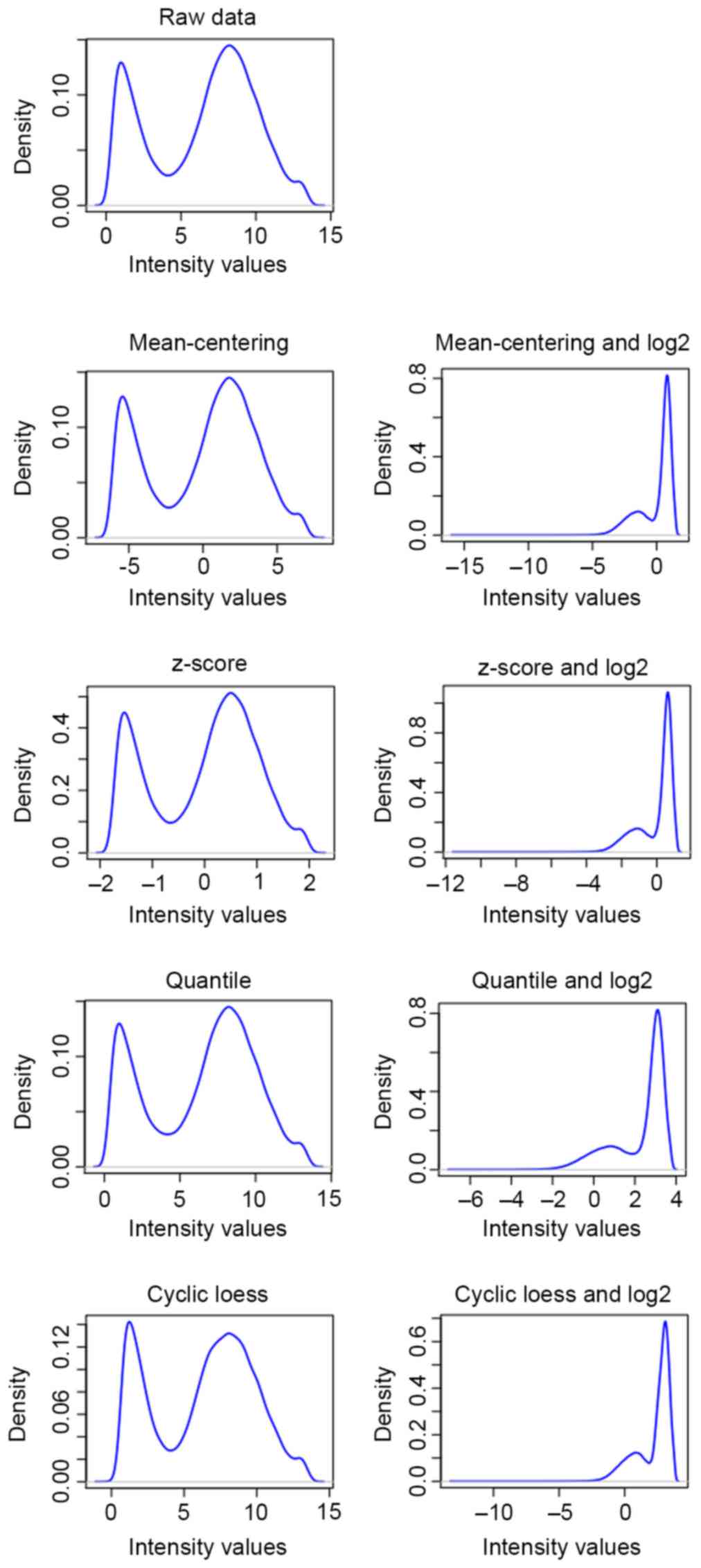
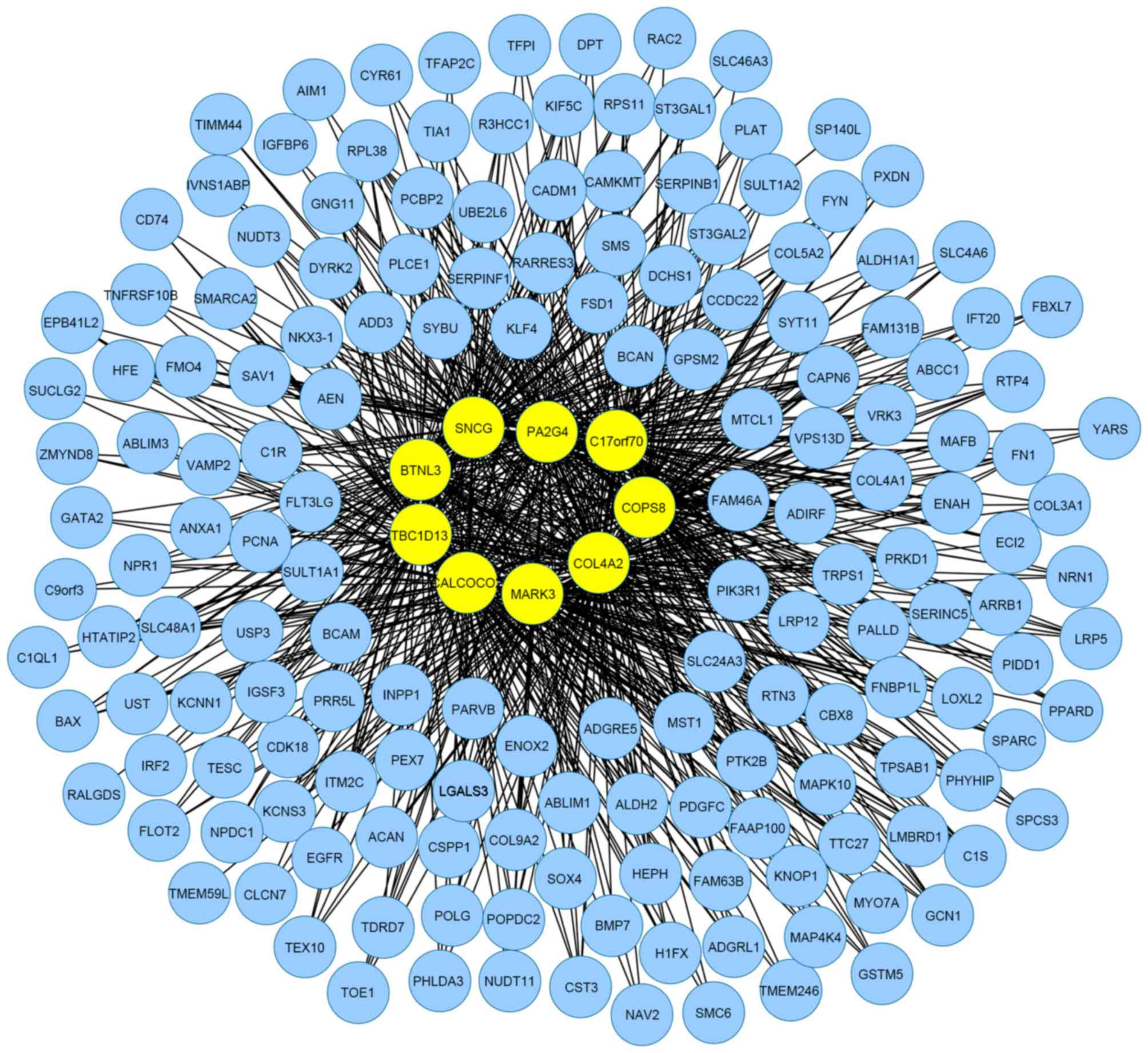
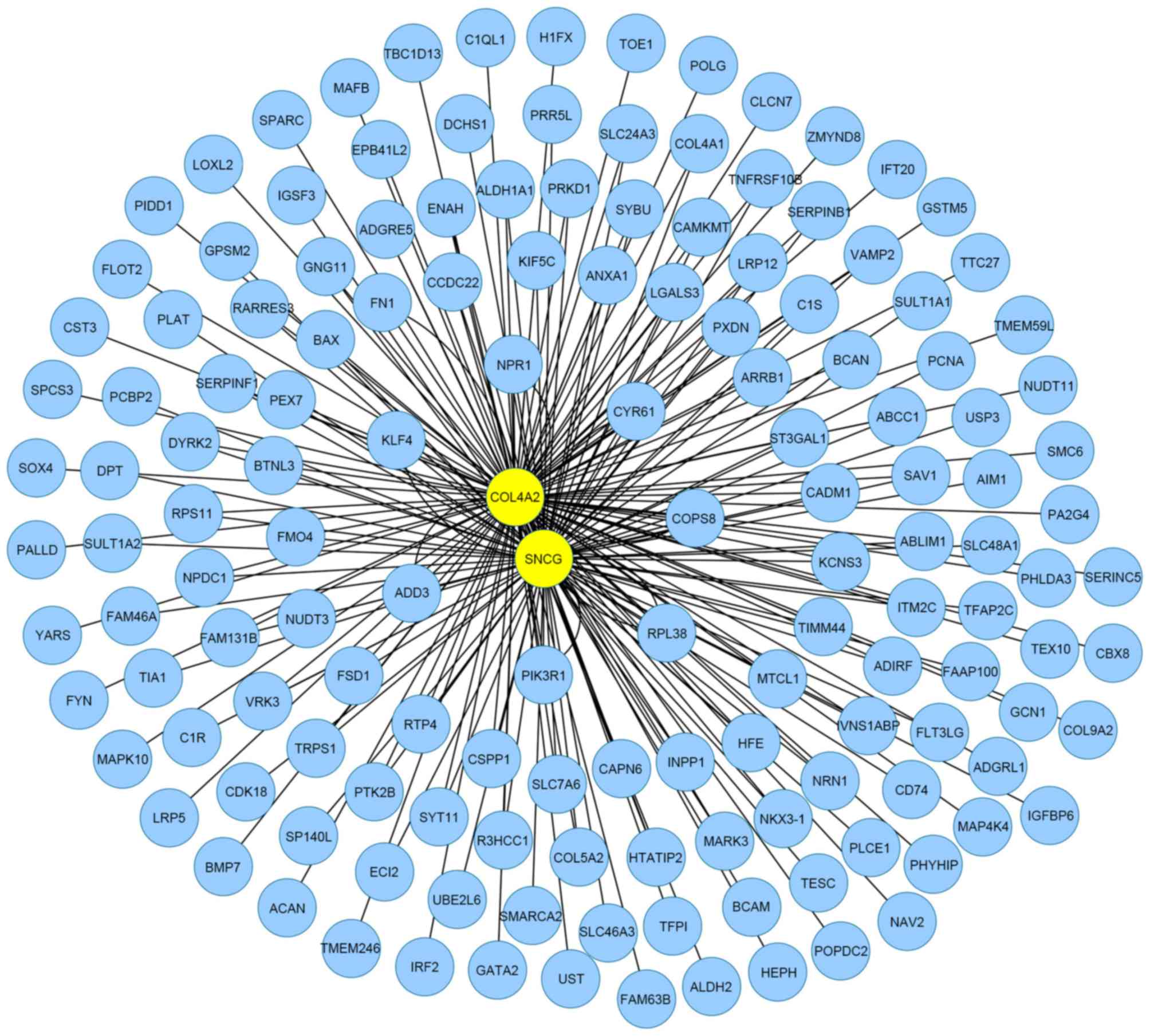
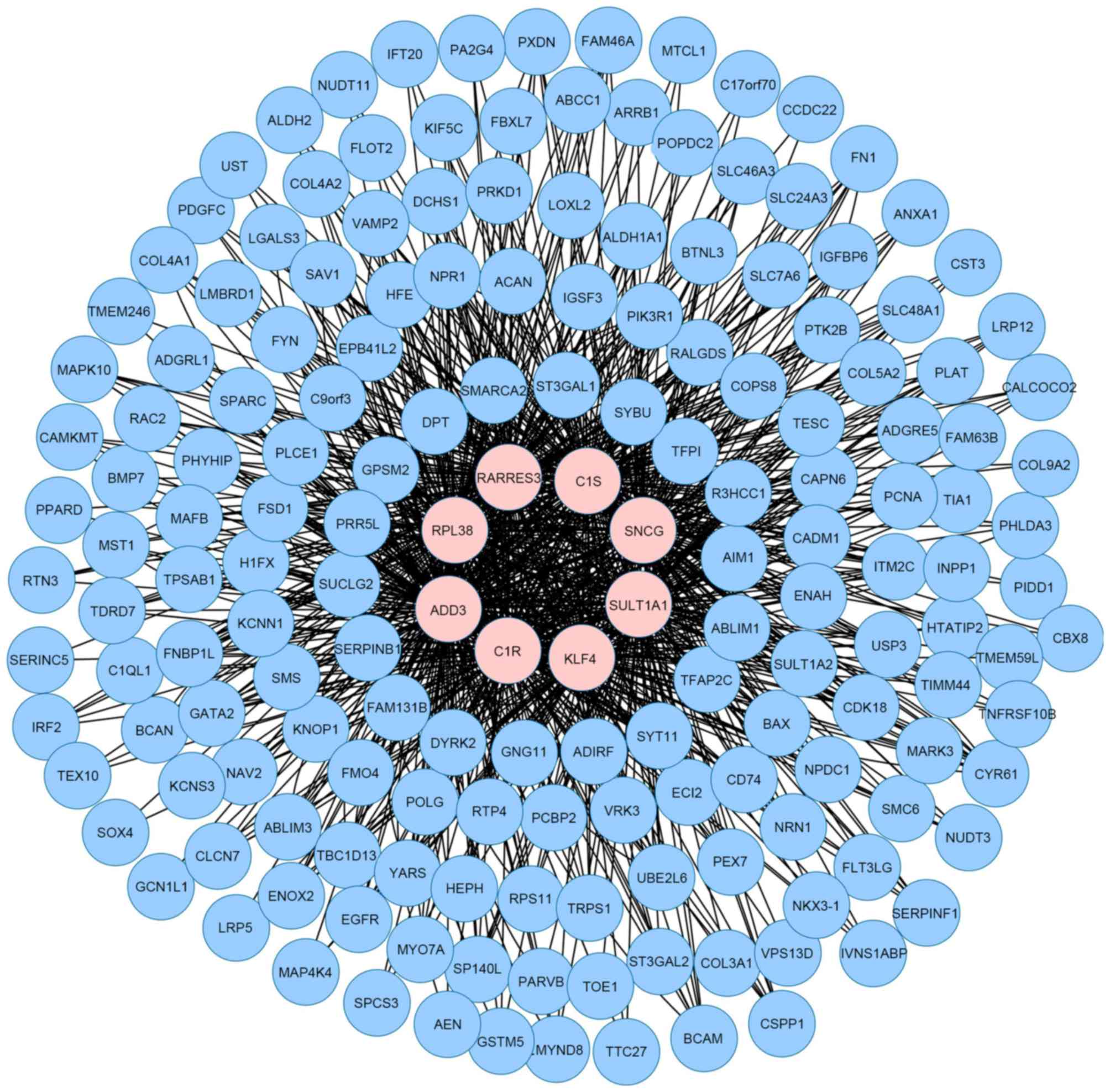
|
1
|
Frey BJ and Dueck D: Clustering by passing
messages between data points. Science. 315:972–976. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bodenhofer U, Kothmeier A and Hochreiter
S: APCluster: An R package for affinity propagation clustering.
Bioinformatics. 27:2463–2464. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kiddle SJ, Windram OP, McHattie S, Mead A,
Beynon J, Buchanan-Wollaston V, Denby KJ and Mukherjee S: Temporal
clustering by affinity propagation reveals transcriptional modules
in Arabidopsis thaliana. Bioinformatics. 26:355–362. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Woźniak M, Tiuryn J and Dutkowski J:
MODEVO: Exploring modularity and evolution of protein interaction
networks. Bioinformatics. 26:1790–1791. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang CW, Chen KT and Lu CL: iPARTS: An
improved tool of pairwise alignment of RNA tertiary structures.
Nucleic Acids Res. 38:(Web Server issue). W340–W347. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sakellariou A, Sanoudou D and Spyrou G:
Combining multiple hypothesis testing and affinity propagation
clustering leads to accurate, robust and sample size independent
classification on gene expression data. BMC bioinformatics.
13:2702012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Krzanowski WJ and Lai Y: A criterion for
determining the number of groups in a data set using sum-of-squares
clustering. Biometrics. 44:23–34. 1988. View Article : Google Scholar
|
|
8
|
Liu ZP, Wang Y, Zhang XS and Chen L:
Network-based analysis of complex diseases. IET Syst Biol. 6:22–33.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen L, Wang RS and Zhang XS:
Reconstruction of Gene Regulatory NetworksBiomolecular Networks.
John Wiley & Sons, Inc.; pp. 47–87. 2009, View Article : Google Scholar
|
|
10
|
Wylie D, Shelton J, Choudhary A and Adai
AT: A novel mean-centering method for normalizing microRNA
expression from high-throughput RT-qPCR data. BMC Res Notes.
4:5552011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cheadle C, Vawter MP, Freed WJ and Becker
KG: Analysis of microarray data using Z score transformation. J Mol
Diagn. 5:73–81. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ballman KV, Grill DE, Oberg AL and
Therneau TM: Faster cyclic loess: Normalizing RNA arrays via linear
models. Bioinformatics. 20:2778–2786. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pollard K, Dudoit S and van der Laan M:
Multiple testing procedures: R multtest package and applications to
genomics. UC Berkeley Division of Biostatistics Working Paper
Series. Working Paper 164. http://www.bepress.com/ucbbiostat/paper1642004.
|
|
15
|
Westfall PH and Young SS: Resampling-Based
Multiple Testing: Examples and Methods for P-Value Adjustment. John
Wiley & Sons; New York: 1993
|
|
16
|
Sakellariou A, Sanoudou D and Spyrou G:
Investigating the minimum required number of genes for the
classification of neuromuscular disease microarray data. IEEE Trans
Inf Technol Biomed. 15:349–355. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jeffery IB, Higgins DG and Culhane AC:
Comparison and evaluation of methods for generating differentially
expressed gene lists from microarray data. BMC Bioinformatics.
7:3592006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
R Development Core Team, . R: A Language
and Environment for Statistical Computing. R Foundation for
Statistical Computing; Vienna: 2012
|
|
19
|
Chang CC and Lin CJ: LIBSVM: A library for
support vector machines. ACM Transactions on Intelligent Systems
and Technology (TIST). 2:272011.
|
|
20
|
Aha DW, Kibler D and Albert MK:
Instance-based learning algorithms. Machine Learning. 6:37–66.
1991. View Article : Google Scholar
|
|
21
|
Huang J and Ling CX: Using AUC and
accuracy in evaluating learning algorithms. IEEE Transactions on
Knowledge and Data Engineering. 17:299–310. 2005. View Article : Google Scholar
|
|
22
|
Baldi P, Brunak S, Chauvin Y, Andersen CA
and Nielsen H: Assessing the accuracy of prediction algorithms for
classification: An overview. Bioinformatics. 16:412–424. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kantardjieff KA and Rupp B: Matthews
coefficient probabilities: Improved estimates for unit cell
contents of proteins, DNA and protein-nucleic acid complex
crystals. Protein Sci. 12:1865–1871. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Butte AJ and Kohane IS: Mutual information
relevance networks: Functional genomic clustering using pairwise
entropy measurements. Pac Symp Biocomput. 418–429. 2000.PubMed/NCBI
|
|
25
|
Faith JJ, Hayete B, Thaden JT, Mogno I,
Wierzbowski J, Cottarel G, Kasif S, Collins JJ and Gardner TS:
Large-scale mapping and validation of Escherichia coli
transcriptional regulation from a compendium of expression
profiles. PLoS Biol. 5:e82007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Meyer PE, Lafitte F and Bontempi G: Minet:
AR/Bioconductor package for inferring large transcriptional
networks using mutual information. BMC Bioinformatics. 9:4612008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Csardi G and Nepusz T: Igraph-package.
2008, http://cneurocvs.rmki.kfki.hu/igraph/doc/R/aaa-igraph-package.html
|
|
28
|
Haythornthwaite C: Social network
analysis: An approach and technique for the study of information
exchange. Library & Information Science Research. 18:323–342.
1996. View Article : Google Scholar
|
|
29
|
Wasserman S: Social network analysis:
Methods and applications. Cambridge Univ Press. 25:111994.
|
|
30
|
Barthelemy M: Betweenness centrality in
large complex networks. Eur Physical J B. 38:163–168. 2004.
View Article : Google Scholar
|
|
31
|
Schank T and Wagner D: Approximating
clustering-coefficient and transitivity. J Graph Algorithms
Applications. 9:265–275. 2005. View Article : Google Scholar
|
|
32
|
Islam MS, Protic O, Giannubilo SR, Toti P,
Tranquilli AL, Petraglia F, Castellucci M and Ciarmela P: Uterine
leiomyoma: Available medical treatments and new possible
therapeutic options. J Clin Endocrinol Metab. 98:921–934. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bulun SE: Uterine fibroids. N Engl J Med.
369:1344–1355. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zivanovic O, Jacks LM, Iasonos A, Leitao
MM Jr, Soslow RA, Veras E, Chi DS, Abu-Rustum NR, Barakat RR,
Brennan MF and Hensley ML: A nomogram to predict postresection
5-year overall survival for patients with uterine leiomyosarcoma.
Cancer. 118:660–669. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lavedan C, Leroy E, Dehejia A, Buchholtz
S, Dutra A, Nussbaum RL and Polymeropoulos MH: Identification,
localization and characterization of the human gamma-synuclein
gene. Hum Genet. 103:106–112. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhuang Q, Liu C, Qu L and Shou C:
Synuclein-γ promotes migration of MCF7 breast cancer cells by
activating extracellular-signal regulated kinase pathway and
breaking cell-cell junctions. Mol Med Rep. 12:3795–3800.
2015.PubMed/NCBI
|
|
37
|
Mori K, Akers S, Bshara W, et al: Gamma
synuclein expression in ovarian cancer. Gynecol Oncol. 131:2722013.
View Article : Google Scholar
|
|
38
|
Guo C, Sun L, Liu Q, Xie YB and Wang X:
SNCG expression and clinical significance in colorectal cancer
liver metastasis. Zhonghua Wei Chang Wai Ke Za Zhi. 15:625–628.
2012.(In Chinese). PubMed/NCBI
|
|
39
|
Shi L, Zheng S, Zhao Y, Wang JW and Zhan
XH: Expressions of SNCG in gastric adenocarcinoma and its clinical
significances. Pro Mod Bio. 11:1532–1535. 2011.
|
|
40
|
Min L, Ma R, Yuan H, Liu CY, Dong B, Zhang
C, Zeng Y, Wang L, Guo JP, Qu LK, et al: Combined expression of
metastasis related markers Naa10p, SNCG and PRL-3 and its
prognostic value in breast cancer patients. Asian Pac J Cancer
Prev. 16:2819–2826. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Singh VK and Jia Z: Targeting
synuclein-gamma to counteract drug resistance in cancer. Expert
Opin Ther Targets. 12:59–68. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mhawech-Fauceglia P, Wang D, Syriac S,
Godoy H, Dupont N, Liu S and Odunsi K: Synuclein-γ (SNCG) protein
expression is associated with poor outcome in endometrial
adenocarcinoma. Gynecol Oncol. 124:148–152. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Morgan J, Hoekstra AV, Chapman-Davis E,
Hardt JL, Kim JJ and Buttin BM: Synuclein-gamma (SNCG) may be a
novel prognostic biomarker in uterine papillary serous carcinoma.
Gynecol Oncol. 114:293–298. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kuo DS, Labelle-Dumais C and Gould DB:
COL4A1 and COL4A2 mutations and disease: Insights into pathogenic
mechanisms and potential therapeutic targets. Hum Mol Genet.
21:R97–R110. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Weston G, Trajstman AC, Gargett CE,
Manuelpillai U, Vollenhoven BJ and Rogers PA: Fibroids display an
anti-angiogenic gene expression profile when compared with adjacent
myometrium. Mol Hum Reprod. 9:541–549. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Poncelet C, Fauvet R, Feldmann G, Walker
F, Madelenat P and Darai E: Prognostic value of von Willebrand
factor, CD34, CD31, and vascular endothelial growth factor
expression in women with uterine leiomyosarcomas. J Surg Oncol.
86:84–90. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gilden M, Malik M, Britten J, Delgado T,
Levy G and Catherino WH: Leiomyoma fibrosis inhibited by liarozole,
a retinoic acid metabolic blocking agent. Fertil Steril.
98:1557–1562. 2012. View Article : Google Scholar : PubMed/NCBI
|