Introduction
Polycystic ovary syndrome (PCOS) is a female
endocrine disease in which high levels of androgen are produced.
Symptoms include abnormalities of menstruation, hirsutism, acne,
pelvic pain and difficulty achieving pregnancy (1–3).
Patients may even present diabetes mellitus, obesity, obstructive
sleep apnea, heart disease, emotional disorders, endometrial
carcinoma and other complications (4,5). The
pathogenesis mechanisms of PCOS are not yet clear. In 2013, a
summary on the pathogenesis of PCOS found the incidence of PCOS not
only being influenced by the living environment of patients, but
also by genetics (6). Later, a study
group found a possible influence of IL-6 in pig ovary cells in the
development of PCOS. Their experiment results suggest that IL-6 may
influence the expression of key mRNAs in ovarian cells (7). Studies on the genetic influence on PCOS
are still lacking; this research aimed at discovering associations
to SNPs in a wide genome analysis.
Genome-wide association studies (GWAS) are commonly
applied to analyze the correlation between DNA polymorphisms and
specific disease traits. Such analyses allow one to investigate the
distribution characteristics of alleles in large population groups.
GWAS usually focuses on the correlation characteristics for single
nucleotide polymorphisms (SNPs). Patients in the experiment group
were classified according to the clinical manifestations, while the
genotype was not given priority. Each participant provided a DNA
sample and millions of genetic variations were read by SNP arrays.
SNP variations found more frequently in affected individuals than
in controls were chosen for further analyses (8–11). In
recent years, researchers have investigated the correlation between
genomes and diseases (12,13). The use of GWAS integrates molecular
biology, genetics and statistics and may reveal pathogenic
mechanisms of diseases at a molecular level. This investigation
adopted the GWAS method to gather information on SNPs that may be
associated with PCOS and may help elucidate pathogenic mechanisms
of the disease.
Materials and methods
To confirm the PCOS diagnosis, previous experiments
also detected the prolactin, luteinizing hormone and insulin
resistance levels in patients. According to the Rotterdam standard
proposed by the European Society for Human Reproduction and
Embryology (14), persons with over
two items exceeding the normal level were defined as having PCOS.
During the first experimental stage 200 female PCOS patients who
came to the hospital for treatment were enrolled in the study. The
age of the patients ranged from 22 to 30 years, averaging 28.3±3.6
years. The age of the 228 healthy individuals enrolled in the
control group ranged from 19 to 33 years, averaging 25.7±5.3 years.
At the 2nd experimental stage, 200 further female PCOS patients who
came to other hospitals in the same province were selected. The
patient ages ranged from 19 to 33 years, averaging 25.7±5.3 years.
The ages of 1,400 healthy individuals in the control group ranged
from 19 to 35 years, averaging 26.6±4.9 years.
Inclusion criteria for the PCOS group: the diagnosis
of PCOS conformed to the Rotterdam standards proposed in 2003,
details are as follows: ⅰ) infrequent menstruation or amenorrhea;
ⅱ) clinical or biochemical manifestations of hyperandrogenism; and
ⅲ) vaginal ultrasonography revealed at least one ovary with >12
follicles of 2–8 mm. Patients who met two of the above conditions
and had other diseases, like adrenocortical hyperplasia, Cushings
syndrome, and androgen secreting tumors excluded were considered to
be affected with PCOS.
Inclusion criteria for the control group: ⅰ) normal
menstrual cycle; ⅱ) normal basic endocrine detection results; ⅲ)
vaginal ultrasonography revealed no organic lesions at uterus or
ovary (and the ovary showed no polycystic manifestations); ⅳ) no
administration of hormonal medicines within three months and other
endocrine abnormalities could be excluded; and ⅴ) no family history
of diabetes.
Blood collection
On the 3rd to 5th day of the menstrual cycle, elbow
vein blood was extracted for determinations of hormone levels,
biochemical indexes and an oral glucose tolerance test (OGTT).
Height, weight, waistline and hipline were measured on the day of
blood extraction. The waistline was the maximum abdomen perimeter
at the level of the navel. Hipline was the maximum hip perimeter,
within the space from the waist to the thigh. The body mass index
(BMI) was calculated as the weight (kg)/height2
(m2). The weight hip ratio (WHR) was calculated as the
measurements of the waistline/hipline. The homeostatic model
assessment and insulin resistance (HOMA-IR) was calculated as
[fasting insulin (mU/l) × fasting blood glucose (mmol/l)]/22.5.
Finally, every research subject signed an informed consent
form.
Blood samples of 5 ml venous blood in EDTA were
collected from each participant. The samples were centrifuged for 4
h according to standard methods to, respectively, separate plasma,
white and red cells into 1.5 ml centrifuge tubes. Samples not
immediately processed were stored at −80°C with cryopreservation
for future use.
Genome extraction
DNA extraction kits (Qiagen, Hilden, Germany) were
used to extract genomic DNA. This method produces a high yield, is
easy and time-saving and needs only low consumption of cells (only
200 µl of the WBC layer were used). The DNA concentration yields
ranged between 20 and 50 ng/µl and their purity (measured with
ultraviolet 260 OD/280 OD) was between 1.6 and 2.0, meeting
research needs.
PCR reaction
DNA was extracted from blood samples of research
subjects, and amplicons were generated by PCR using published
primers and protocols (15). The
sequence of the primers used is shown in Table I.
 | Table I.PCR primers. |
Table I.
PCR primers.
| Primer name | Sequence |
|---|
| 1-F |
AGTCGATGATGCTAGCTGA |
| 1-R |
CGTAGCTAGCTAGCTACG |
| 2-F |
CTAGCTAGATAGCTAGCTACG |
| 2-R |
CGATGCATATTAGCTACGATGC |
SNP selection
The selection of SNP sites was completed with a
Human Mapping 6.0 chip according to published methods (16). The chip included over 900,000 SNP
sites of genetic variation. The primary screening was performed
with a matrix-assisted laser desorption/ionization time of flight
mass spectrometry (MALDI-TOF-MS). The genotype of SNPs was read
according to the published report (16), after hybridization with the gene
chip.
Statistical analysis
The SAS software version 9.0. (SAS Institute Inc.,
Cary, NC, USA) was used for statistical analyses, according to the
analysis methods proposed in the literature (17).
Results
Extraction and PCR amplification of
genomes
A GWAS was performed. A stable PCR amplification
method as established by adjusting the denaturation temperature to
94°C, the annealing temperature to 45°C and the cycle number to 35
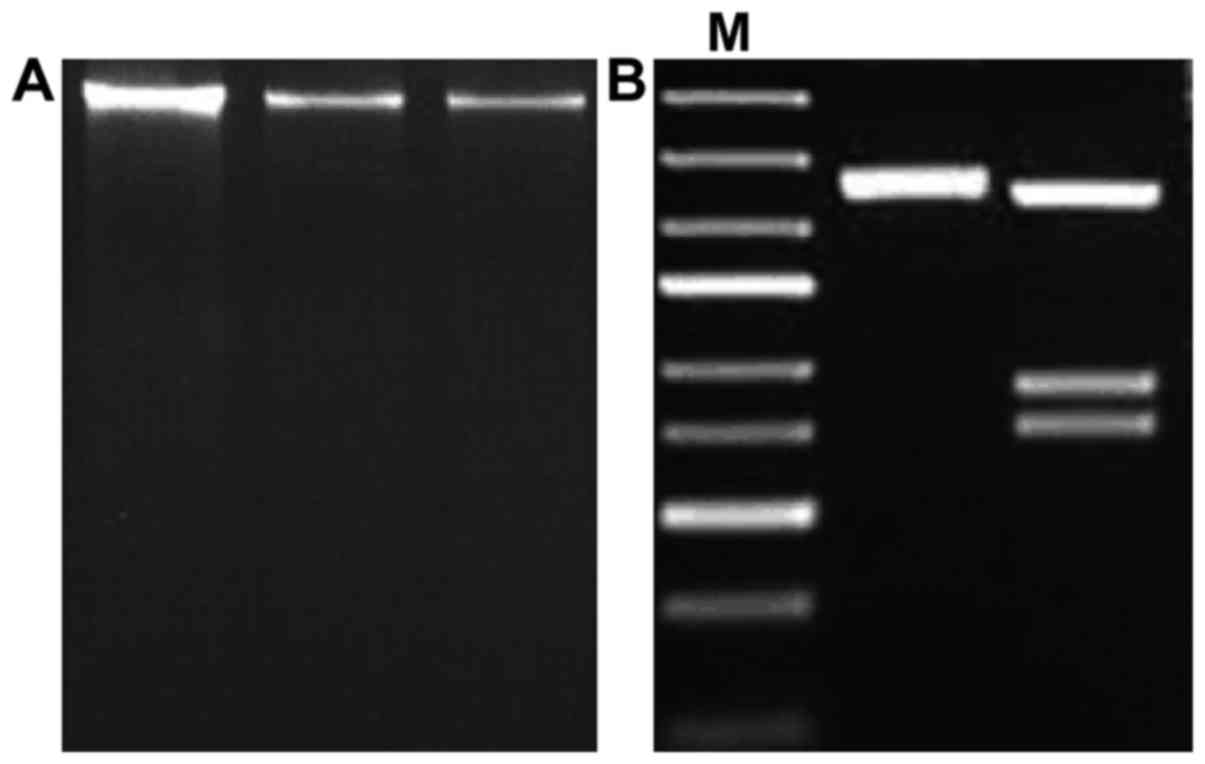
times. The optimal PCR results can be seen in Fig. 1, as well as gel electrophoresis of
whole DNA samples extracted.
Analyses of SNP sites related to PCOS
during the 1st experimental stage
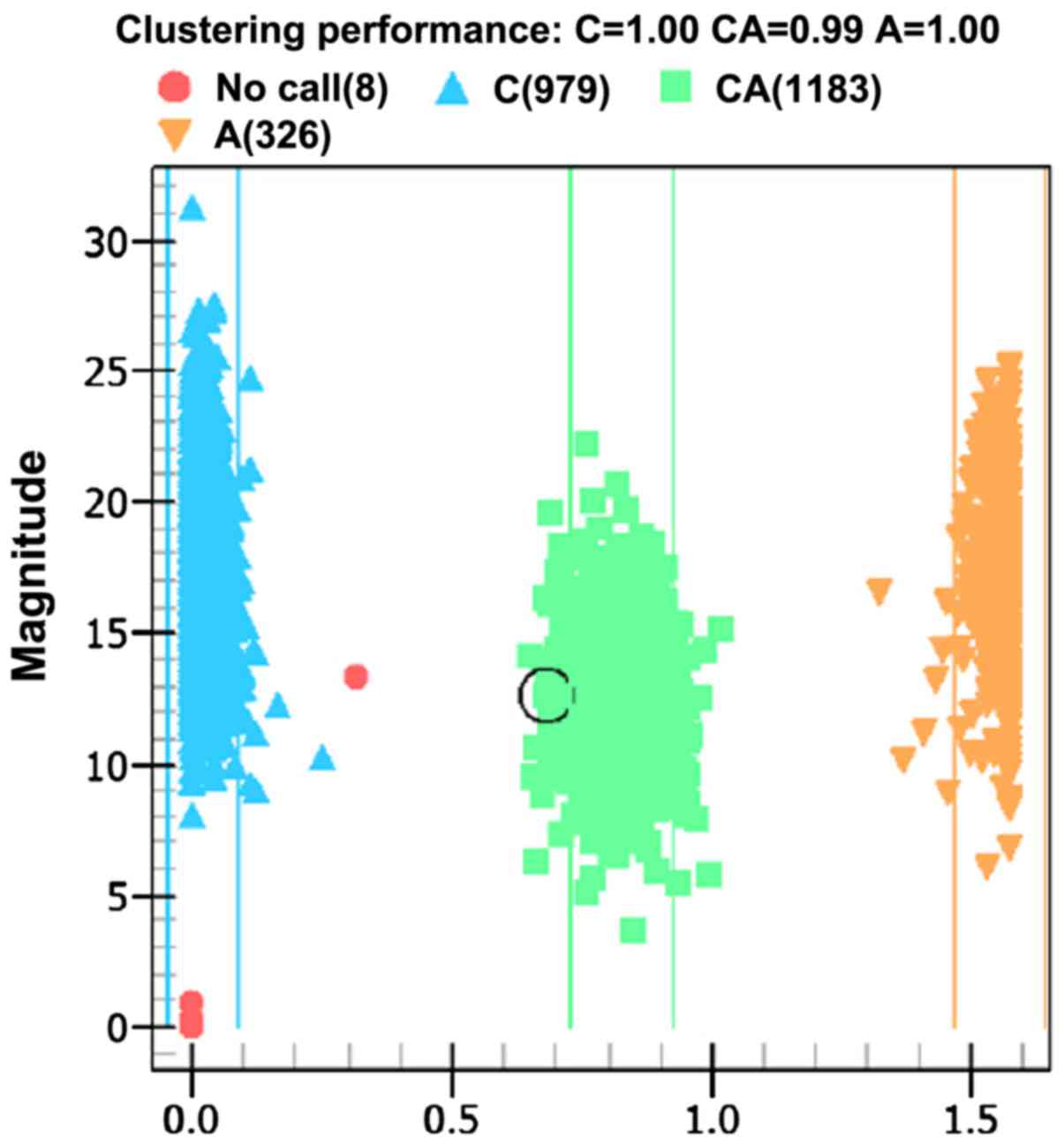
Ionization time-of-flight mass spectrometry was used
to analyze the products of PCR amplification and its allele genes
are shown in the distribution plot of Fig. 2.
Firstly, after reviewing relevant literature and
NCBI data, 10 SNP sites that may correlate with PCOS were selected
and then verified by mass spectrometry as shown in Table II.
 | Table II.Correlation between 10 SNP sites and
PCOS by comparison of allele frequency between patients and control
groups. |
Table II.
Correlation between 10 SNP sites and
PCOS by comparison of allele frequency between patients and control
groups.
|
|
|
| Allele frequency |
|
|---|
|
|
|
|
|
|
|---|
| SNPs | Gene | Allele | PCOS group | Control group | P-value |
|---|
| rs346795081 | THADA | A/C | 0.426 | 0.358 | <0.05 |
| rs346796267 | THADA | C/G | 0.343 | 0.339 | >0.05 |
| rs346797283 | DENND1A | A/C | 0.324 | 0.321 | >0.05 |
| rs346803513 | DENND1A | A/T | 0.447 | 0.412 | <0.05 |
| rs346982406 | DENND1A | A/T | 0.128 | 0.125 | >0.05 |
| rs346996092 | TOX3 | C/T | 0.268 | 0.257 | >0.05 |
| rs346999236 | TOX3 | C/T | 0.321 | 0.295 | <0.05 |
| rs347021263 | YAP1 | A/T | 0.179 | 0.186 | >0.05 |
| rs347026725 | YAP1 | A/T | 0.351 | 0.350 | >0.05 |
| rs347029500 | YAP1 | C/T | 0.342 | 0.325 | <0.05 |
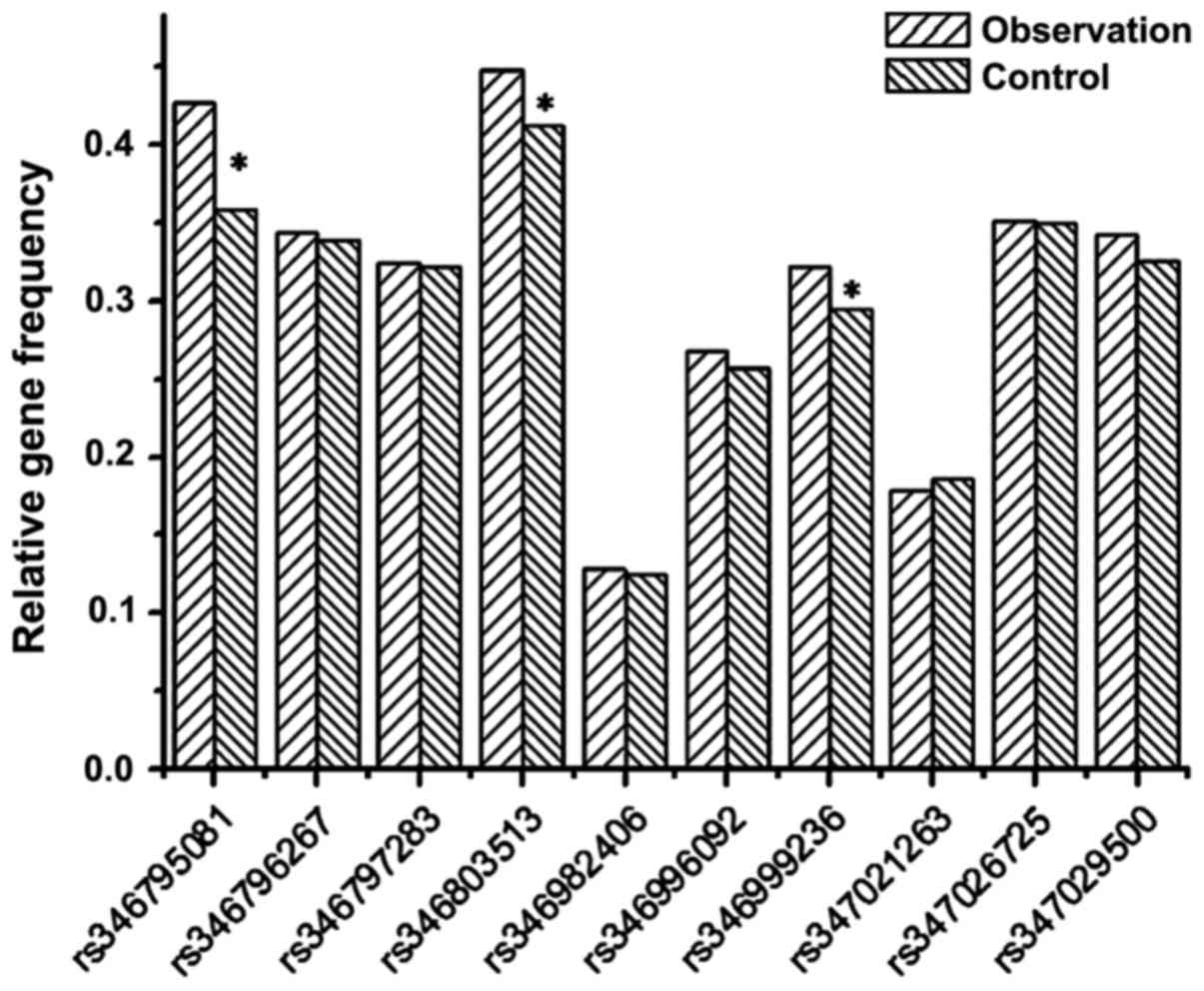
The table shows that among 10 SNP sites, only
frequencies of alleles at rs346795081 on THADA gene, rs346803513 on
DENND1A gene, rs346999236 on the TOX3 gene and rs347029500 on the
YAP1 gene were significantly different between the patient and
control groups (P<0.05). Fig. 3
shows a graph of the relative frequencies for all the 10 alleles in
the patient and control groups.
Sample amplification verification
during the 2nd stage
Four SNP sites were selected during the 1st stage.
These sites were deemed relevant to the pathogenesis of PCOS. To
verify the accuracy of the allele selection, PCR was used to
amplify the same regions from control and DNA samples of PCOS
patients. A correlation analysis was carried out to find a link
between the occurrence of the four SNP sites and PCOS, and the
results are shown in Table
III.
 | Table III.Correlation between the four SNP sites
and the occurrence of PCOS. |
Table III.
Correlation between the four SNP sites
and the occurrence of PCOS.
|
|
| Allele frequency |
|
|---|
|
|
|
|
|
|---|
| SNPs | Research stage | PCOS group | Control group | P-value |
|---|
| rs346795081 | 1st stage | 0.426 | 0.358 | <0.05 |
|
| 2nd stage | 0.433 | 0.321 | <0.05 |
|
| Total | 0.429 | 0.339 | <0.05 |
| rs346803513 | 1st stage | 0.447 | 0.412 | <0.05 |
|
| 2nd stage | 0.451 | 0.425 | <0.05 |
|
| Total | 0.449 | 0.418 | <0.05 |
| rs346999236 | 1st stage | 0.321 | 0.295 | <0.05 |
|
| 2nd stage | 0.317 | 0.286 | <0.05 |
|
| Total | 0.319 | 0.290 | <0.05 |
| rs347029500 | 1st stage | 0.342 | 0.325 | <0.05 |
|
| 2nd stage | 0.333 | 0.328 | >0.05 |
|
| Total | 0.337 | 0.326 | >0.05 |
The sample size was increased to 1,200 cases during
the 2nd stage of the study. Results show the correlation between
rs347029500 and PCOS had a P>0.05, which rules out statistical
significance. Considering the results of the 1st and 2nd
experimental stages, only the differences of allele frequencies at
three SNP sites, that is, rs346795081, rs346803513 and rs346999236
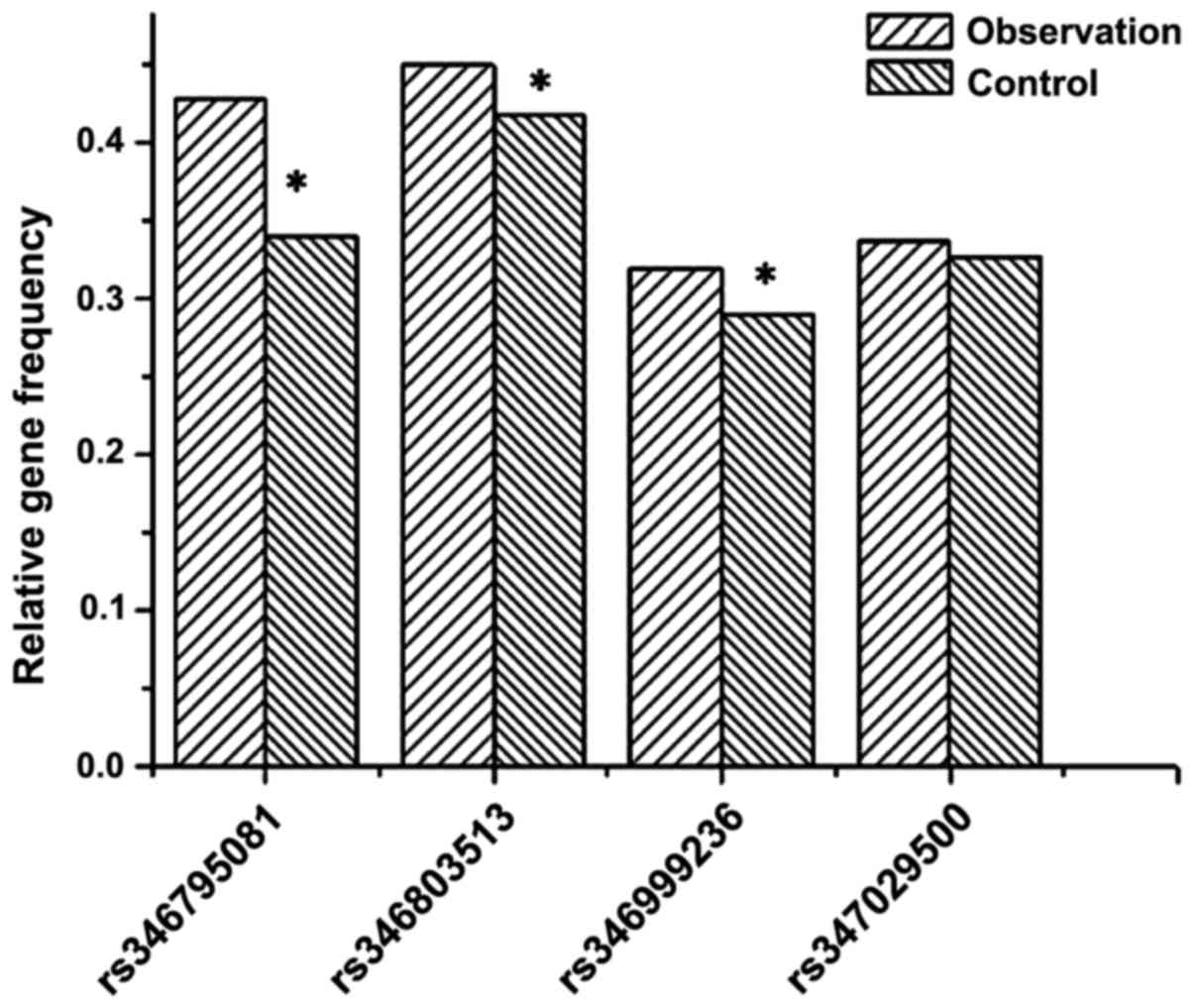
were still significantly different (P<0.05). Fig. 4 shows the differences between the
allele frequencies in the patient and control groups are only
significant at rs346795081, rs346803513 and rs346999236
(P<0.05).
Discussion
With the current scientific and technological
developments, the third generation sequencing technology has made
gene sequencing no longer a difficult problem. High throughput and
highly effective sequencing can often provide abundant information
(18). Ensuring that the processing
and analysis of the data obtained give accurate results has become
a biggest challenge. GWAS are based on high throughput sequencing
and are combined with statistics to select and sort the large
amount of data got from sequencing (19). GWAS can thus predict or help infer
effective information and instruct on the research direction
(20). This study detected SNPs that
may be correlated with the manifestation of PCOS GWAS, the selected
SNPs will help lay a foundation for future experiments elucidating
the role of different genes in the pathogenesis of PCOS (21).
The experiment was divided into four parts. The
first part involved the selection of patients and the acquisition
of DNA from blood samples by solid phase extraction. Protein
components with strong polarity were removed first and then the
purity of the DNA after elution of solid phase extraction column
was determined by electrophoresis. Next, the optimal parameters for
PCR were established.
For genotyping, the third part of the study,
ionization time-of-flight mass spectrometry was adopted. Different
SNP sites are found in fragments of different length. The
nucleotide length defines the molecular weight. Spectrometry
identifies SNP sites by difference in molecular weight.
There is literature indicating that the frequency of
SNP alleles in the patient group vs. the frequency in the control
group can imply a correlation with the patients disease (17). We compared the gene frequency at
different SNP sites between the PCOS and the control group and
selected SNP sites that are significantly different between the two
groups.
The final part of the study was concerned with the
specific data analysis. This part was divided into two stages. The
1st stage comprised literature review and comparison with NCBI data
bank. A total of 10 SNP sites that were likely to be correlated
with PCOS were selected in order to ensure the efficiency of the
analysis, few first conducted comparisons and analyses of DNA
sequencing results within a small group of patients to exclude some
irrelevant SNPs and facilitate the analysis. According to the
results at this 1st stage, we analyzed the genomes of 200 PCOS
patients and the control group and found that, among 10 SNP sites,
only the alleles at rs346795081, rs346803513, rs346999236 and
rs347029500 were significantly different. Differences at other SNP
sites lacked statistical significance and were not taken into
account for the next stage of the study.
After exclusion of six SNP sites during the 1st
stage, the experimental sample size was increased to 1,200 cases to
verify the correlation between four SNP sites and PCOS. The results
of the second analysis revealed no significant differences at the
SNP rs347029500 site. However, the other three SNP sites,
rs346795081, rs346803513 and rs346999236, were closely correlated
with the occurrence of PCOS.
To conclude, three SNP sites were found to correlate
closely with the occurrence of PCOS. This information can provide
reference for further analysis and future studies on how specific
genes regulate the onset and progress of PCOS.
Acknowledgements
This study was supported by the Changzhou Municipal
Science and Technology Bureau (no. CJ20140021).
References
|
1
|
Trikudanathan S: Polycystic ovarian
syndrome. Med Clin North Am. 99:221–235. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bostancı MS, Akdemir N, Cinemre B,
Cevrioglu AS, Özden S and Ünal O: Serum irisin levels in patients
with polycysticovary syndrome. Eur Rev Med Pharmacol Sci.
19:4462–4468. 2015.PubMed/NCBI
|
|
3
|
Al-Zubeidi H and Klein KO: Randomized
clinical trial evaluating metformin versus oral contraceptive pills
in the treatment of adolescents with polycystic ovarian syndrome. J
Pediatr Endocrinol Metab. 28:853–858. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yakubov Y and Mandel L: Bilateral parotid
swelling in polycystic ovarian syndrome. J Oral Maxillofac Surg.
74:991–994. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang B1, Sun ZJ, Chen B, Zhang J, Zhao H,
Li CW, Cao YK and Cao F: Statin ameliorates endothelial dysfunction
and insulin resistance in Tibet women with polycystic ovary
syndrome. Eur Rev Med Pharmacol Sci. 20:1185–1191. 2016.PubMed/NCBI
|
|
6
|
Silfeler Benk D, Kurt Keskin R, Kaya OA,
Yengil E, Hamamci B, Okyay AG and Beyazit A: Demodex folliculorum
in polycystic ovary syndrome patients. Eur Rev Med Pharmacol Sci.
19:1141–1145. 2015.PubMed/NCBI
|
|
7
|
Ting W, Yanyan Q, Jian H, Keqin H and Duan
M: The relationship between insulin resistance and CpG island
methylation of LMNA gene in polycystic ovary syndrome. Cell Biochem
Biophys. 67:1041–1047. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gondro C: Genome wide association
studiesPrimer to Analysis of Genomic Data Using R. Springer
International Publishing; Cham, Switzerland: pp. 73–103. 2015,
View Article : Google Scholar
|
|
9
|
Zhou X and Stephens M: Efficient
algorithms for multivariate linear mixed models in genome-wide
association studies. Nat Methods. 11:407–409. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou PP, Li Y, Ma ZD, Li ZY, Chen FY and
Jiang YX: Single nucleotide polymorphisms in the promoter region of
mir-133a-1 and in pre-mir-152 rs1707 may contribute to the risk of
asthma in a Chinese Han population. Eur Rev Med Pharmacol Sci.
20:2642–2649. 2016.PubMed/NCBI
|
|
11
|
Taşan M, Musso G, Hao T, Vidal M, MacRae
CA and Roth FP: Selecting causal genes from genome-wide association
studies via functionally coherent subnetworks. Nat Methods.
12:154–159. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lerchbaum E, Schwetz V, Giuliani A and
Obermayer-Pietsch B: Assessment of glucose metabolism in polycystic
ovary syndrome: HbA1c or fasting glucose compared with the oral
glucose tolerance test as a screening method. Hum Reprod.
28:2537–2544. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Legro RS, Arslanian SA, Ehrmann DA, Hoeger
KM, Murad MH, Pasquali R and Welt CK: Endocrine Society: Diagnosis
and treatment of polycystic ovary syndrome: An Endocrine Society
clinical practice guideline. J Clin Endocrinol Metab. 98:4565–4592.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Belosi C, Selvaggi L, Apa R, Guido M,
Romualdi D, Fulghesu AM and Lanzone A: Is the PCOS diagnosis solved
by ESHRE/ASRM 2003 consensus or could it include ultrasound
examination of the ovarian stroma? Hum Reprod. 21:3108–3115. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ecklund LC and Usadi RS: Endocrine and
reproductive effects of polycystic ovarian syndrome. Obstet Gynecol
Clin North Am. 42:55–65. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Abbott DH, Nicol LE, Levine JE, Xu N,
Goodarzi MO and Dumesic DA: Nonhuman primate models of polycystic
ovary syndrome. Mol Cell Endocrinol. 373:21–28. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li R, Zhang Q, Yang D, Li S, Lu S, Wu X,
Wei Z, Song X, Wang X, Fu S, et al: Prevalence of polycystic ovary
syndrome in women in China: A large community-based study. Hum
Reprod. 28:2562–2569. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bili E, Dampala K, Iakovou I, Tsolakidis
D, Giannakou A and Tarlatzis BC: The combination of ovarian volume
and outline has better diagnostic accuracy than prostate-specific
antigen (PSA) concentrations in women with polycystic ovarian
syndrome (PCOs). Eur J Obstet Gynecol Reprod Biol. 179:32–35. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mutharasan P, Galdones E, Bernabé Peñalver
B, Garcia OA, Jafari N, Shea LD, Woodruff TK, Legro RS, Dunaif A
and Urbanek M: Evidence for chromosome 2p16.3 polycystic ovary
syndrome susceptibility locus in affected women of European
ancestry. J Clin Endocrinol Metab. 98:E185–E190. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Glueck CJ, Woo JG, Khoury PR, Morrison JA,
Daniels SR and Wang P: Adolescent oligomenorrhea (age 14–19) tracks
into the third decade of life (age 20–28) and predicts increased
cardiovascular risk factors and metabolic syndrome. Metabolism.
64:539–553. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bassiouny YA, Rabie WA, Hassan AA and
Darwish RK: Association of the luteinizing
hormone/choriogonadotropin receptor gene polymorphism with
polycystic ovary syndrome. Gynecol Endocrinol. 30:428–430. 2014.
View Article : Google Scholar : PubMed/NCBI
|