Introduction
Prostate cancer is a common malignant tumor in the
urinary system of middle aged and elderly males and has a high
incidence and mortality rate (1).
The mechanisms of prostate cancer include the interaction between
growth factors and epithelia-stroma (2), altered expression of tumor suppressor
genes and oncogenes (3), DNA
methylation (4) and androgen
receptor mutations (5). Tumor
protein D52 like 2 (TPD52L2), also known as TPD54 and D54, is a
member of the tumor protein D52 (TPD52) family (6). The most important physiological
function of the TPD52 family is its participation in the regulation
of Ca2+-dependent vesicular transport processes
(7). In addition, the overexpression
of proteins in the TPD52 family in breast cancer (8) and hepatocellular carcinoma (9) suggests that the TPD52 family may serve
an important role in the onset and development of tumors. A
previous study on brain glioma cells demonstrated that knockout of
TPD52L2 inhibits the growth and colony formation of cells by
arresting cells in the G0/G1 phase (10). Similarly, it has been demonstrated
that knockout of TPD52L2 in hepatic cells inhibits their growth
(11).
microRNA (miR) is a type of small non-encoding
single-strand RNA 18–24 nucleotides long that is present in
eukaryote cells and viruses (12).
It has been suggested that miRNA serves regulatory roles in the
onset and development of prostate cancer, including the role of a
tumor-suppressor (13). Furthermore,
a previous study demonstrated that miR-15/miR-16 suppressed the
activation of several oncogenes by inhibiting the transcription and
expression of B-cell lymphoma-2, myeloid cell leukemia-1, cyclin D1
and Wnt family member 3A (14). It
was also demonstrated that miR-23a downregulated the expression of
glutaminase in prostate cancer cells, inhibited the catabolic
metabolism of glutamine and inhibited tumor cell growth (15). miR-503 was first identified in human
retinoblastoma tissues (16) and
downregulation of miR-503 expression has been identified in liver,
brain, lung, and colon tumors (17).
miR-503 specifically targets B-cell lymphoma-2 and increases the
sensitivity of A549/CDDP cells to cisplatin-induced apoptosis in
small cell lung cancer cells (18).
The present study investigated the expression of miR-503 and its
effect on the proliferation and migration of prostate cancer
cells.
Materials and methods
Patients
A total of 20 male patients with prostate cancer
(mean age, 56.2 years) who had undergone radical prostatectomy at
The People's Hospital of Rizhao (Shandong, China) between January
2016 and April 2016 were included in the present study. Prostate
tissues were obtained from patients and pathologically confirmed as
prostate cancer according to 2005 International Society of
Urological Pathology (ISUP) consensus conference (19). None of the patients received
radiotherapy or chemotherapy prior to surgery. Prostate cancer
tissues and tumor-adjacent normal tissues (control) were removed
and frozen at −80°C within 10 min following excision. All
procedures were approved by the Ethics Committee of People's
Hospital of Rizhao (Shandong, China) and written informed consent
was obtained from all patients or patient families.
Bioinformatics
TargetScan (http://www.targetscan.org) was used to predict the
miRNA molecule that interacts with TPD52L2 in order to understand
the regulatory mechanism of TPD52L2 in prostate cancer.
Cells and cell transfection
DU145 cells (The Cell Bank of Type Culture
Collection of Chinese Academy of Sciences, Shanghai, China) were
cultured in Dulbecco's Modified Eagle's medium (DMEM) supplemented
with 10% fetal bovine serum (FBS). Cells were divided into a
DU145+miR-503 mimic group, a DU145+miR-503 inhibitor group, a
negative control transfected (NC) group and a blank (control)
group. Log-phase DU145 cells (2×105) were seeded in
24-well plates containing DMEM supplemented with 10% FBS without
antibiotics 1 day prior to transfection. Once 70% confluence was
reached, 7.5 µl miR-NC, miR-503 mimics and miR-503 inhibitor
plasmids (Shanghai GenePharma Co., Ltd, Shanghai, China) and 7.5 µl
Lipofectamine 2000 (Thermo Fisher Scientific, Inc., Waltham, MA,
USA) were added to two individual vials each containing 125 µl
serum-free DMEM. The two vials were mixed 5 min later, left for a
further 5 min and the mixture was subsequently added to the cells.
Following 6 h incubation, the old medium was discarded and fresh
DMEM supplemented with 10% FBS was added. The cells were then
cultured under normal conditions at 37°C for 48 h prior to use.
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
A total of 100 mg tissue was ground into powder
using liquid nitrogen prior to the addition of 1 ml TRIzol (Thermo
Fisher Scientific, Inc.) for lysis. A total of 2×105
cells were trypsinized and mixed with 1 ml TRIzol for lysis. Total
RNA was extracted using the phenol chloroform method (20). The purity of RNA was determined by
A260/A280 using ultraviolet spectrophotometry (Nanodrop ND1000,
Thermo Fisher Scientific, Inc., Wilmington, DE, USA). cDNA was
synthesized by reverse transcription using the Reverse
Transcription system (Takara Biotechnology Co., Ltd., Dalian,
China) from 1 µg RNA and stored at −20°C.
The expression of miR-503 was determined using a
SYBR PrimeScript miRNA RT-PCR kit (Takara Biotechnology Co.). U6
was used as the internal reference. The reaction system (25 µl)
contained 12.5 µl SYBR Premix Ex Taq, 1 µl PCR Forward Primer, 1 µl
Uni-miR qPCR Primer, 2 µl template and 8.5 µl double distilled
water. The primer sequences used were as follows: miR-503, forward,
5′-TGCGGTAGCAGCGGGAACAGTTC-3′ (reverse primer was Uni-miR qPCR
Primer that was provided by the kit and its sequence is a
commercial secret); and U6, forward 5′-TGCGGGTGCTCGCTTCGGCAGC-3′
and reverse 5′-AACGCTTCACGAATTTGCGT-3′. The reaction protocol was
as follows: Initial denaturation at 95°C for 30 sec; 40 cycles of
95°C for 5 sec and 60°C for 20 sec (Bio-Rad IQ5 Realtime PCR;
Bio-Rad Laboratories, Inc., Hercules, CA, USA). The
2−ΔΔCq method was used to calculate the relative
expression of miR-503 against U6 (21). Each sample was tested in
triplicate.
A SYBR Green RT-qPCR kit (Kapa Biosystems, Inc.,
Wilmington, MA, USA) was used according to the manufacturer's
protocol to detect the mRNA expression of TPD52L2 in prostate
tissues and DU145 cells. GADPH was used as an internal reference.
The reaction system (20 µl) was composed of 10 µl SYBR EX Taq-Mix,
0.5 µl upstream primer, 0.5 µl downstream primer, 1 µl cDNA and 8
µl double diluted water. The primer sequences were as follows:
TPD52L2 mRNA, forward, 5′-TTCACAGGCAGGACAGAAGA-3′ and reverse,
5′-TTGAAGGTCGCAGAGTTCCT-3′; GADPH, forward,
5′-AAGGTGAAGGTCGGATCA-3′ and reverse, 5′-GGAAGATGGTGATGGGATTT-3′.
The PCR thermocycling conditions were as follows: Initial
denaturation at 95°C for 10 min; 40 cycles of denaturation at 95°C
for 1 min, annealing at 60°C for 40 sec and elongation at 72°C for
30 sec and final elongation at 72°C for 1 min (iQ5; Bio-Rad
Laboratories, Inc.). The 2−ΔΔCq method was used to
calculate the relative expression of TPD52L2 mRNA against GADPH.
Each sample was tested in triplicate.
Western blot analysis
A total of 50 mg tissue was ground using liquid
nitrogen. DU145 cells were trypsinized and collected prior to being
mixed with 100 µl precooled radioimmunoprecipitation assay lysis
buffer (600 µl; 50 mM Tris-base, 1 mM EDTA, 150 mM sodium chloride,
0.1% sodium dodecyl sulfate, 1% TritonX-100, 1% sodium
deoxycholate; Beyotime Institute of Biotechnology, Shanghai, China)
for 50 min on ice. The mixture was centrifuged at 12,000 × g at 4°C
for 5 min. The supernatant was used to determine protein
concentration using a bicinchoninic acid protein concentration
determination kit [RTP7102, Real-Times (Beijing) Biotechnology Co.,
Ltd., Beijing, China]. Protein samples (50 µg) were mixed with 2×
sodium dodecyl sulfate loading buffer prior to denaturation in a
boiling water bath for 5 min. Subsequently, samples (10 µl) were
subjected to 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis at 100 V. The resolved proteins were transferred to
polyvinylidene difluoride membranes on ice (300 mA, 1.5 h) and
blocked with 5 g/l skimmed milk at room temperature for 1 h.
Membranes were incubated with rabbit anti-human TPD52L2 polyclonal
primary antibody (1:2,000; catalogue no. ab194938) and rabbit
anti-human GAPDH primary antibody (1:2,000; catalogue no. ab37168;
both Abcam Cambridge, UK) at 4°C overnight. Following three
extensive washes with phosphate-buffered saline and Tween 20 for 15
min, membranes were incubated with polyclonal goat anti-rabbit
horseradish peroxidase-conjugated secondary antibody (1:1,000;
catalogue no. ab6721; Abcam) for 1 h at room temperature prior to
three washes with phosphate-buffered saline and Tween 20 for 15
min. Membranes were developed using an enhanced chemiluminescence
detection kit (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for
imaging. Image lab v3.0 software (Bio-Rad Laboratories, Inc.) was
used to analyze imaging signals. The relative content of TPD52L2
protein was expressed as the ratio of TPD52L2/GAPDH.
Transwell assay
Transwell chambers (Corning Inc., Corning, NY, USA)
were used to evaluate the migration ability of DU145 cells.
Transfected cells were trypsinized and re-suspended to a density of
5×105 cells/ml using DMEM containing 0.1% bovine serum
albumin (Sigma-Aldrich; Merck KGaA). The cell suspension (200 µl;
5×105 cells) was added into the migration chamber
(Hyclone, GE Healthcare Life Sciences; Logan, UT, USA). A total of
750 µl DMEM supplemented with 20% fetal bovine serum was added to
the lower chamber. After 4 h, cells in the migration chamber were
wiped using a cotton swab. Cells that had migrated to the other
side of the chamber were fixed with 100% methanol at room
temperature for 10 min. Following staining at room temperature with
0.1% crystal violet for 15 min, the number of cells was counted
under a microscope (DM1000; Leica Microsystems GmbH, Wetzlar,
Germany).
MTT assay
Following transfection, DU145 cells were seeded into
96-well plates at a density of 3,000 cells/well. Each experiment
was conducted in triplicate. At 24, 48, and 72 h following
transfection, 20 µl MTT (5 g/l; JRDUN Biotechnology, Shanghai,
China) was added to each well. Following incubation at 37°C for 4 h
dimethyl sulfoxide was used to dissolve purple formazan. The
absorbance of each well was measured at 492 nm with a microplate
reader and cell proliferation curves were subsequently plotted.
Flat plate colony formation test
DU145 cells from the DU145+miR-503 mimic,
DU145+miR-503 inhibitor, NC and control groups were trypsinized and
washed twice with phosphate-buffered saline. The number of cells
was counted following staining with trypan blue at room temperature
and the cells were prepared into a suspension with a density of
0.5×103 cells/ml. The cell suspension (2 ml) was
inoculated into 6-well plates, followed by incubation under normal
conditions at 37°C. Half the medium was replenished on day 5. The
medium was discarded on day 14 and cells were washed once with
phosphate-buffered saline. Cells were fixed with methanol at room
temperature for 10 min and stained with crystal violet at room
temperature for 5 min. Following extensive washing with
phosphate-buffered saline, the cells were observed under a
microscope (DM1000; Leica Microsystems GmbH) and five fields were
selected for colony counting. The colony formation rate was then
calculated using the following equation: colony formation rate =
(number of clones) / (number of seeded cells) ×100% (22).
Statistical analysis
Statistical analysis was performed using SPSS 16.0
(SPSS, Inc., Chicago, IL, USA). Measurement data were expressed as
mean ± standard deviation and data from two groups were compared
using a Student's t-test. P<0.05 indicated a statistically
significant difference.
Results
TPD52L2 expression is elevated in
prostate cancer tissues
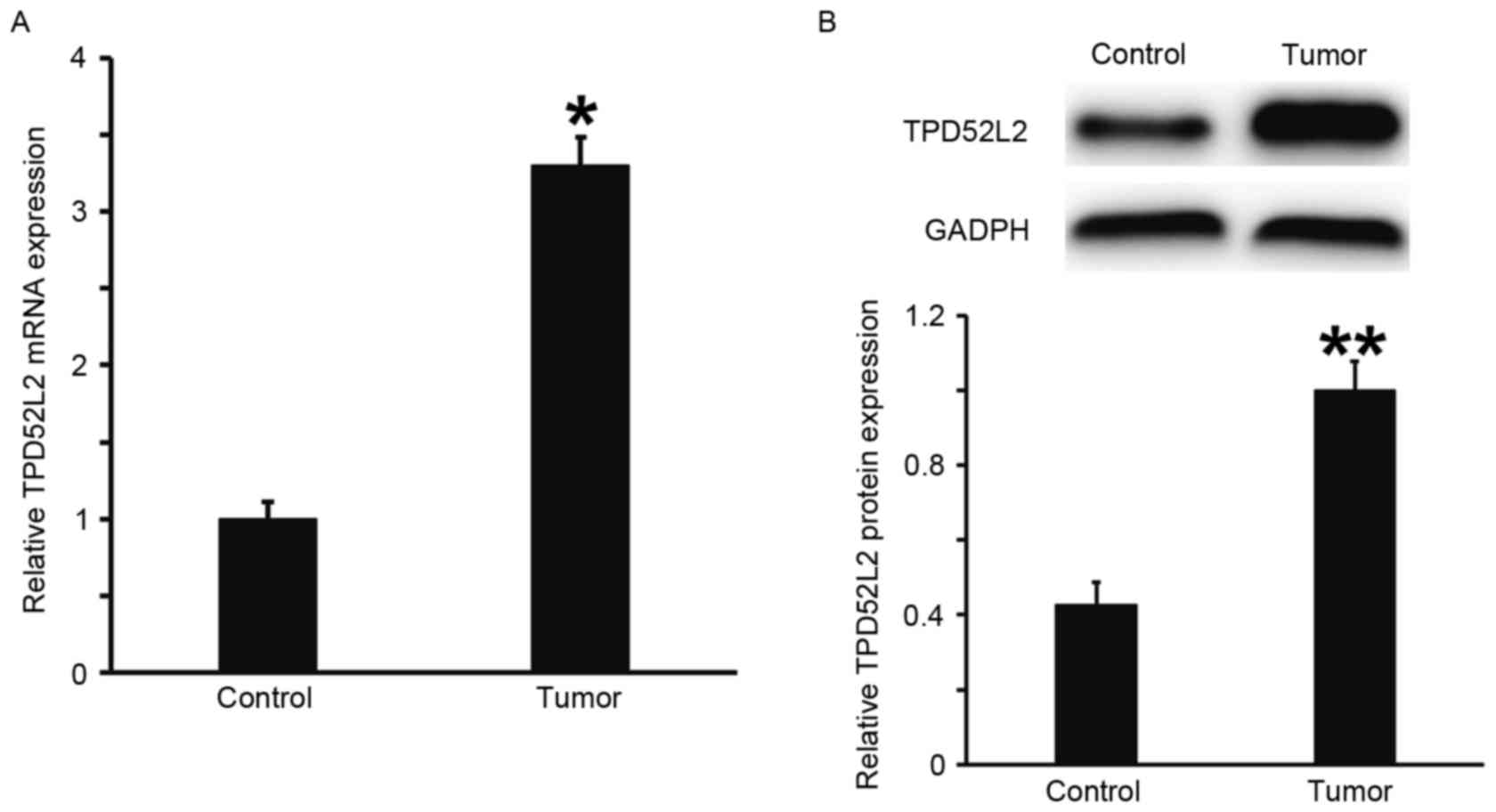
RT-qPCR and western blot analysis were performed to
measure the expression of TPD52L2 mRNA and protein in prostate
tumor tissues. The results demonstrated that the levels of TPD52L2
mRNA and protein in prostate cancer tissues were significantly
increased compared with those from tumor-adjacent normal tissues
(P<0.05; Fig. 1A and B). This
suggests that the expression of TPD52L2 is elevated in prostate
cancer tissues.
Expression of miR-503 in prostate
cancer tissues is reduced, which may be due to the direct
interaction between miR-503 and TPD52L2
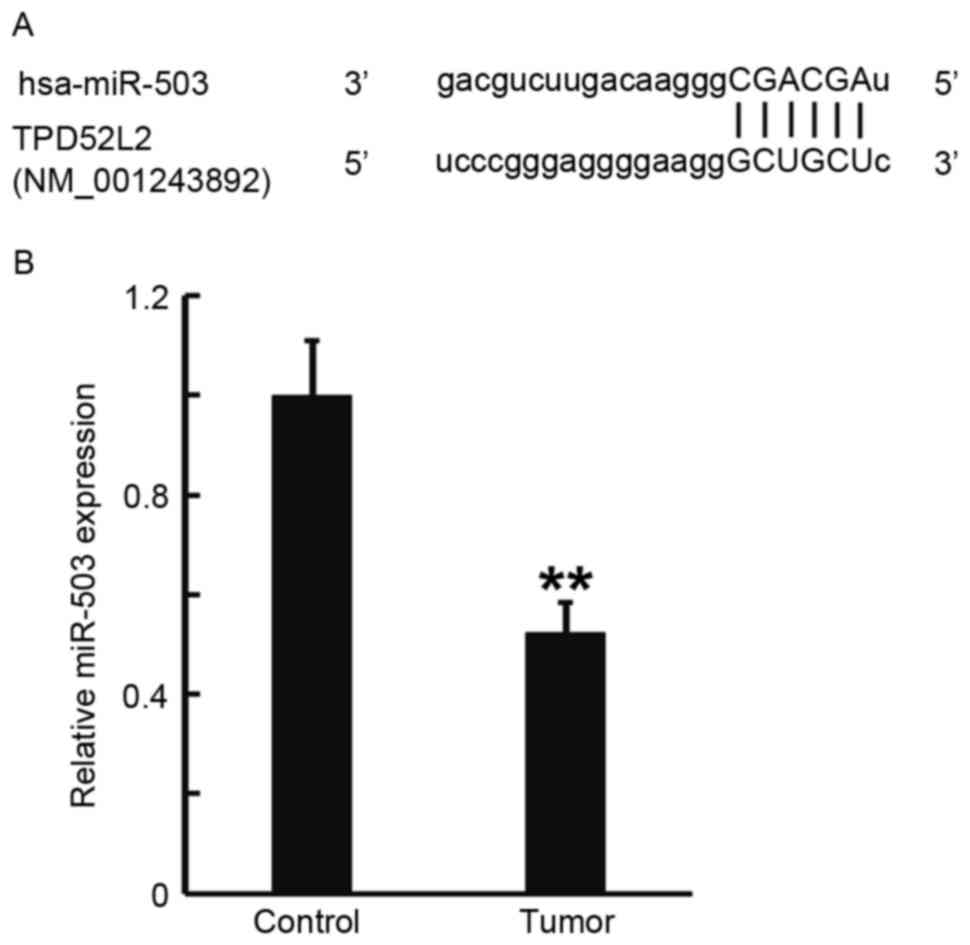
TargetScan was used to predict the direct
interaction between miR-503 and TPD52L2, and RT-qPCR was performed
to measure the expression of miR-503 in prostate cancer tissues.
TargetScan identified the 3′-untranslated region (3′UTR) of TPD52L2
as a direct target of miR-503 (Fig.
2A). RT-qPCR data determined that miR-503 levels in prostate
cancer tissues were significantly decreased compared with those in
tumor-adjacent normal tissues (P<0.01; Fig. 2B). These results indicate that the
expression of miR-503 in prostate cancer tissues was decreased,
which may be due to the direct interaction of miR-503 with
TPD52L2.
Overexpression of miR-503 inhibits the
transcription and translation of TPD52L2
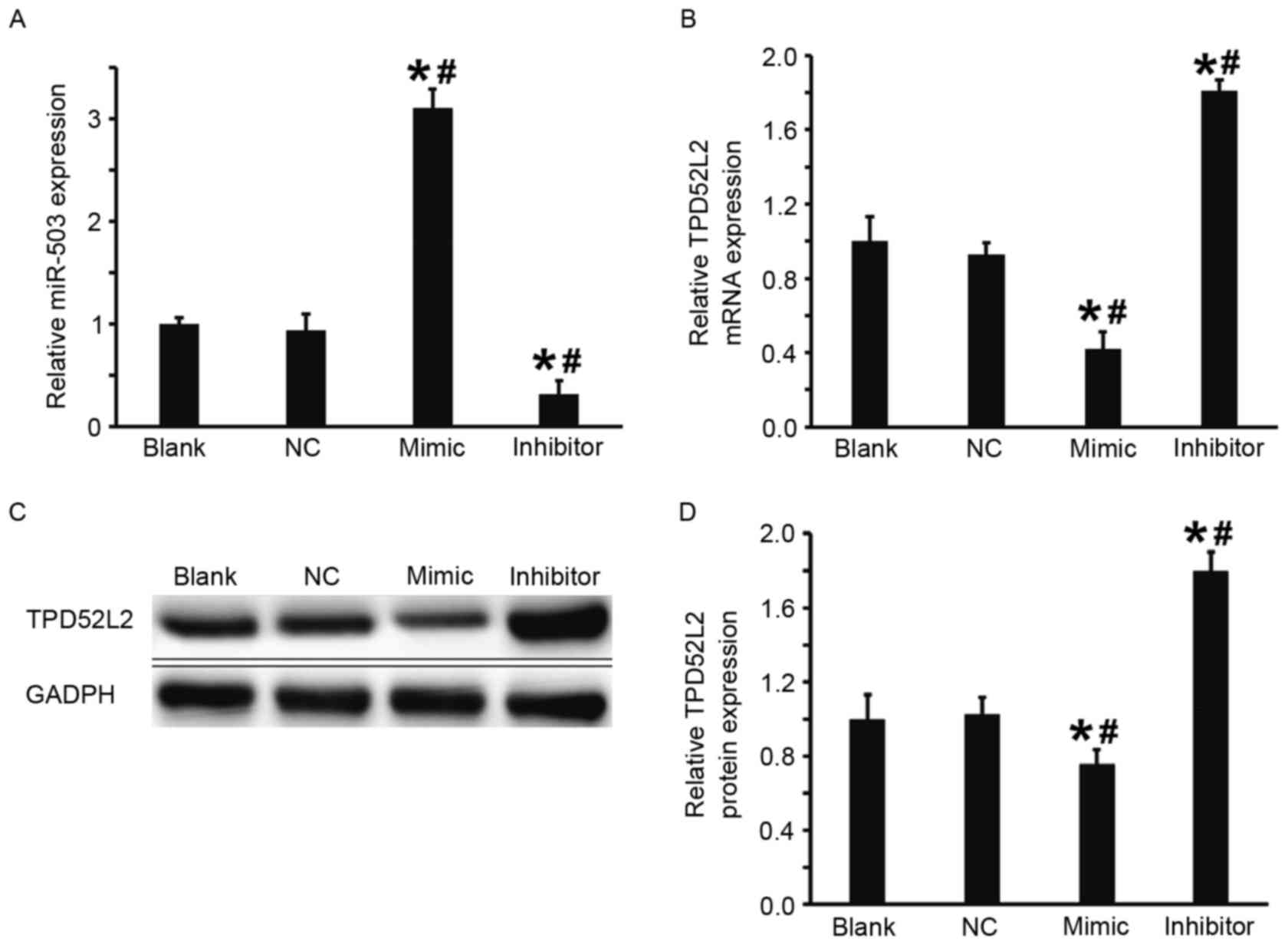
The DU145 cells were transfected with miR-503 mimic
or miR-503 inhibitor to examine the regulation of TPD52L2 gene
expression by miR-503 in DU145 cells. Compared with the blank and
NC groups, the expression of miR-503 in DU145 cells transfected
with miR-503 mimic was significantly increased and in DU145 cells
transfected with miR-503 inhibitor it was significantly decreased
(both P<0.05; Fig. 3A).
Furthermore, the results from RT-qPCR and western blot analysis
demonstrated that in comparison with the blank and NC groups,
levels of TPD52L2 mRNA and protein in the DU145+miR-503 mimic group
were significantly decreased and in the DU145+miR-503 inhibitor
group they were significantly increased (all P<0.05; Fig. 3B-D). These results suggest that
overexpression of miR-503 inhibits the transcription and
translation of TPD52L2.
Overexpression of miR-503 suppresses
the migration ability of DU145 cells
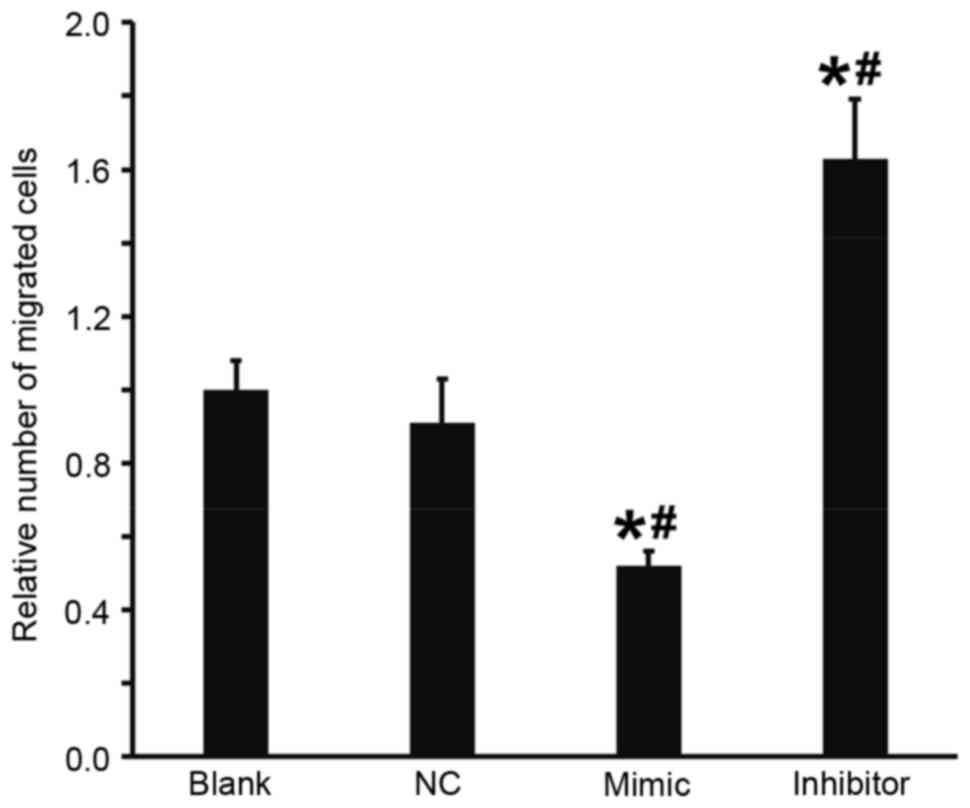
A Transwell assay was performed to investigate the
effect of miR-503 on the migration of DU145 cells. The results
demonstrated that, compared with the blank and NC groups, the
number of migrated cells in the DU145+miR-503 mimic group was
significantly decreased and in the DU145+miR-503 inhibitor group it
was significantly increased (all P<0.05; Fig. 4). These results indicate that the
overexpression of miR-503 suppresses the migration ability of DU145
cells.
miR-503 inhibits the proliferation of
DU145 cells
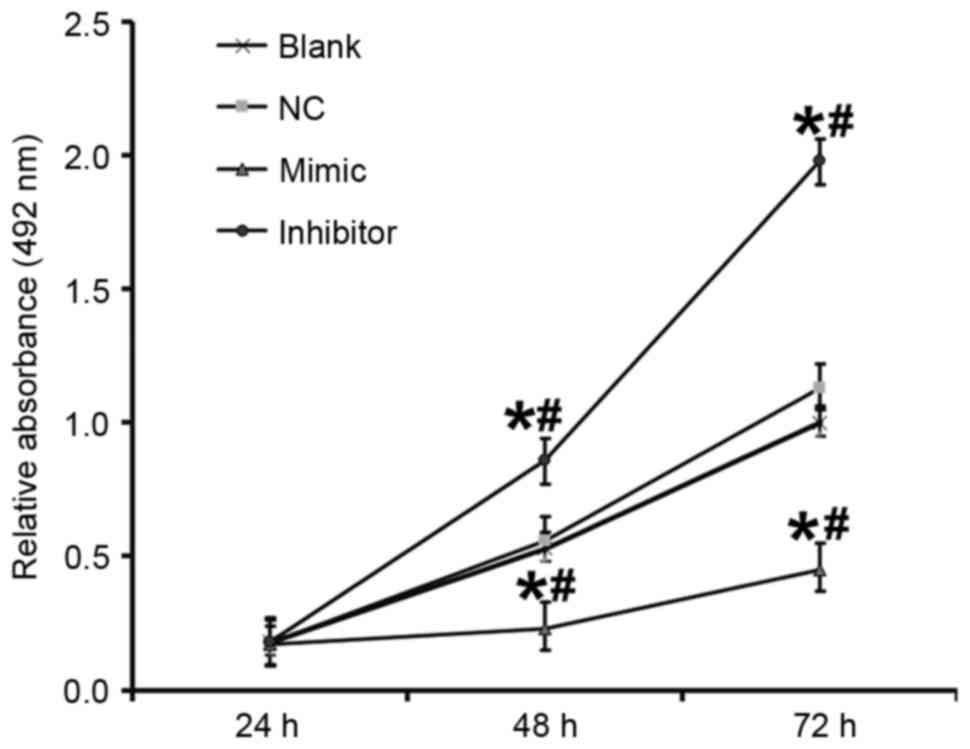
An MTT assay was performed to determine the effect
of miR-503 on the proliferation of DU145 cells. Compared with the
blank and NC groups, the absorbance of cells in the DU145+miR-503
mimic group was significantly decreased and in the DU145+miR-503
inhibitor group it was significantly increased at 48 and 72 h (all
P<0.05; Fig. 5). This suggests
that miR-503 inhibits the proliferation of DU145 cells.
Overexpression of miR-503 reduces the
colony formation ability of DU145 cells
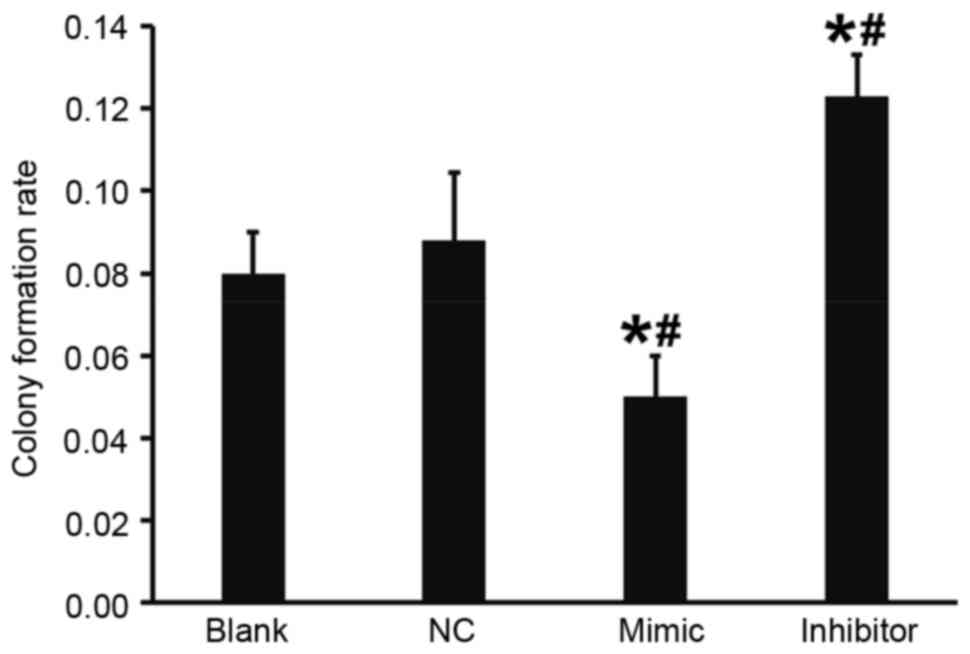
A flat plate colony formation test was used to
determine how miR-503 affects the colony formation ability of DU145
cells. Colony counting demonstrated that compared with the blank
and NC groups, the colony formation rate of DU145 cells in the
DU145+miR-503 mimic group was significantly decreased and in the
DU145+miR-503 inhibitor group it was significantly increased (both
P<0.05; Fig. 6). These results
indicate that the overexpression of miR-503 reduces the colony
formation ability of DU145 cells.
Discussion
miRNA acts as an oncogene or tumor-suppressor gene
by binding to the 3′-UTR seeding sequence of mRNA (23). It has been demonstrated that miR-503
is downregulated in hepatic tissues and cells and inhibits tumor
angiogenesis by acting on fibroblast growth factor 2 and vascular
endothelial growth factor A (24),
suggesting that it is an important tumor-suppressor factor.
However, the mechanism of action of miR-503 in prostate cancer
remains unclear.
The results of the present study demonstrate that
the expression of miR-503 in prostate cancer tissues is
significantly lower than in tumor-adjacent normal tissues and
bioinformatic analysis indicated that TPD52L2 is a potential target
gene of miR-503. Ummanni et al (25) demonstrated that TPD52 promotes αVβ3
integrin-mediated cell migration in prostate cancer by activating
the phosphatidylinositol 3-kinase/Akt signaling pathway and the
mitochondrial apoptotic response as an upstream response element.
The present study demonstrated that the expression of TPD52L2 mRNA
and protein in prostate cancer tissues was higher than in normal
tissues, suggesting that miR-503 may be associated with TPD52L2.
Transfection with miR-503 mimic or miR-503 inhibitor induces the
overexpression or inhibition of miR-503 expression in DU145 cells,
respectively. The overexpression of miR-503 in DU145 cells inhibits
the expression of TPD52L2, and by contrast, inhibition of miR-503
elevates the expression of TPD52L2. The present study on cellular
function has demonstrated that DU145 cells overexpressing miR-503
exhibit decreased migration, proliferation and colony formation,
whereas cells with reduced miR-503 expression have improved
migration, proliferation and colony formation functions. Previous
studies have identified that miR-503 is abnormally expressed in
retinal glioblastoma, adenoma, breast cancer, non-small cell lung
cancer and hepatic cancer. Furthermore, it has been demonstrated
that miR-503 targets phosphoinositide-3-kinase, p85 or inhibitor of
nuclear factor κB kinase subunit β, regulates the Rho guanine
nucleotide exchange factor 19, DDHD domain containing 2 and F-box
and WD repeat domain containing 7 genes and initiates apoptosis,
thus promoting the growth and differentiation of tumors and
inhibiting the invasion and metastasis of tumors (18,26–31).
This indicates that miR-503 serves important roles in different
types of cancer.
In conclusion, the present study demonstrated that
miR-503 expression in prostate cancer cells is negatively
associated with the migration ability of cells and suggested that
miR-503 may serve important roles in the onset and development of
prostate cancer by targeting TPD52L2. Therefore, the current study
provides novel insights into the mechanisms of prostate cancer.
Acknowledgements
The present study was supported by the People's
Hospital of Rizhao. The authors also wish to thank Professor
Shuguang Yang and Professor Zhaojun Ding for their valuable
help.
References
|
1
|
Grönberg H: Prostate cancer epidemiology.
Lancet. 361:859–864. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wong YC and Wang YZ: Growth factors and
epithelial-stromal interactions in prostate cancer development. Int
Rev Cytol. 199:65–116. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Taylor BS, Schultz N, Hieronymus H,
Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva
B, et al: Integrative genomic profiling of human prostate cancer.
Cancer Cell. 18:11–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Devaney JM, Wang S, Funda S, Long J,
Taghipour DJ, Tbaishat R, Furbert-Harris P, Ittmann M and
Kwabi-Addo B: Identification of novel DNA-methylated genes that
correlate with human prostate cancer and high-grade prostatic
intraepithelial neoplasia. Prostate Cancer Prostatic Dis.
16:292–300. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marcelli M, Ittmann M, Mariani S,
Sutherland R, Nigam R, Murthy L, Zhao Y, DiConcini D, Puxeddu E,
Esen A, et al: Androgen receptor mutations in prostate cancer.
Cancer Res. 60:944–949. 2000.PubMed/NCBI
|
|
6
|
Boutros R, Fanayan S, Shehata M and Byrne
JA: The tumor protein D52 family: Many pieces, many puzzles.
Biochem Biophys Res Commun. 325:1115–1121. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thomas DD, Martin CL, Weng N, Byrne JA and
Groblewski GE: Tumor protein D52 expression and Ca2+-dependent
phosphorylation modulates lysosomal membrane protein trafficking to
the plasma membrane. Am J Physiol Cell Physiol. 298:C725–C739.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Balleine RL, Fejzo MS, Sathasivam P,
Basset P, Clarke CL and Byrne JA: The hD52 (TPD52) gene is a
candidate target gene for events resulting in increased 8q21 copy
number in human breast carcinoma. Genes Chromosomes Cancer.
29:48–57. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang Y, Chen CL, Pan QZ, Wu YY, Zhao JJ,
Jiang SS, Chao J, Zhang XF, Zhang HX, Zhou ZQ, et al: Decreased
TPD52 expression is associated with poor prognosis in primary
hepatocellular carcinoma. Oncotarget. 7:6323–6334. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Z, Sun J, Zhao Y, Guo W, Lv K and
Zhang Q: Lentivirus-mediated knockdown of tumor protein D52-like 2
inhibits glioma cell proliferation. Cell Mol Biol (Noisy-le-grand).
60:39–44. 2014.PubMed/NCBI
|
|
11
|
Pan ZY, Yang Y, Pan H, Zhang J, Liu H,
Yang Y, Huang G, Yin L, Huang J and Zhou WP: Lentivirus-mediated
TPD52L2 depletion inhibits the proliferation of liver cancer cells
in vitro. Int J Clin Exp Med. 8:2334–2341. 2015.PubMed/NCBI
|
|
12
|
Almeida MI, Reis RM and Calin GA: MicroRNA
history: Discovery, recent applications, and next frontiers. Mutat
Res. 717:1–8. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Porkka KP, Pfeiffer MJ, Waltering KK,
Vessella RL, Tammela TL and Visakorpi T: MicroRNA expression
profiling in prostate cancer. Cancer Res. 67:6130–6135. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bonci D, Coppola V, Musumeci M, Addario A,
Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C,
et al: The miR-15a-miR-16-1 cluster controls prostate cancer by
targeting multiple oncogenic activities. Nat Med. 14:1271–1277.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gao P, Tchernyshyov I, Chang TC, Lee YS,
Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT and
Dang CV: c-Myc suppression of miR-23a/b enhances mitochondrial
glutaminase expression and glutamine metabolism. Nature.
458:762–765. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao JJ, Yang J, Lin J, Yao N, Zhu Y,
Zheng J, Xu J, Cheng JQ, Lin JY and Ma X: Identification of miRNAs
associated with tumorigenesis of retinoblastoma by miRNA microarray
analysis. Childs Nerv Syst. 25:13–20. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Watahiki A and Wang Y, Morris J, Dennis K,
O'Dwyer HM, Gleave M, Gout PW and Wang Y: MicroRNAs associated with
metastatic prostate cancer. PLoS One. 6:e249502011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qiu T, Zhou L, Wang T, Xu J, Wang J, Chen
W, Zhou X, Huang Z, Zhu W, Shu Y and Liu P: miR-503 regulates the
resistance of non-small cell lung cancer cells to cisplatin by
targeting Bcl-2. Int J Mol Med. 32:593–598. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Epstein JI, Allsbrook WC Jr, Amin MB and
Egevad LL: ISUP Grading Committee: The 2005 International Society
of Urological Pathology (ISUP) consensus conference on Gleason
grading of prostatic carcinoma. Am J Surg Pathol. 29:1228–1242.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chomczynski P and Sacchi N.: Single-step
method of RNA isolation by acid guanidinium
thiocyanate-phenol-chloroform extraction. Anal Biochem.
162:156–159. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li W, Ma H and Sun J: microRNA-34a/c
function as tumor suppressors in Hep-2 laryngeal carcinoma cells
and may reduce GALNT7 expression. Mol Med Rep. 9:1293–1298. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou B, Ma R, Si W, Li S, Xu Y, Tu X and
Wang Q: MicroRNA-503 targets FGF2 and VEGFA and inhibits tumor
angiogenesis and growth. Cancer Lett. 333:159–169. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ummanni R, Teller S, Junker H, Zimmermann
U, Venz S, Scharf C, Giebel J and Walther R: Altered expression of
tumor protein D52 regulates apoptosis and migration of prostate
cancer cells. FEBS J. 275:5703–5713. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou J, Tao Y, Peng C, Gu P and Wang W:
miR-503 regulates metastatic function through Rho guanine
nucleotide exchanger factor 19 in hepatocellular carcinoma. J Surg
Res. 188:129–136. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang Y, Liu L, Zhang Y, Guan H, Wu J, Zhu
X, Yuan J and Li M: MiR-503 targets PI3K p85 and IKK-β and
suppresses progression of non-small cell lung cancer. Int J Cancer.
135:1531–1542. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang T, Ge G, Ding Y, Zhou X, Huang Z, Zhu
W, Shu Y and Liu P: MiR-503 regulates cisplatin resistance of human
gastric cancer cell lines by targeting IGF1R and BCL2. Chin Med J
(Engl). 127:2357–2362. 2014.PubMed/NCBI
|
|
29
|
Li L, Sarver AL, Khatri R, Hajeri PB,
Kamenev I, French AJ, Thibodeau SN, Steer CJ and Subramanian S:
Sequential expression of miR-182 and miR-503 cooperatively targets
FBXW7, contributing to the malignant transformation of colon
adenoma to adenocarcinoma. J Pathol. 234:488–501. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu Y, Guo L, Liu J, Liu R, Liu M and Chen
J: The reversing and molecular mechanisms of miR-503 on the
drug-resistance to cisplatin in A549/DDP cells. Zhongguo Fei Ai Za
Zhi. 17:1–7. 2014.(In Chinese). PubMed/NCBI
|
|
31
|
Polioudakis D, Abell NS and Iyer VR:
miR-503 represses human cell proliferation and directly targets the
oncogene DDHD2 by non-canonical target pairing. BMC Genomics.
16:402015. View Article : Google Scholar : PubMed/NCBI
|