Introduction
Glyphosate is a phosphonomethyl amino acid
derivative that is the active ingredient in a number of herbicides
(1). Glyphosate-based herbicides are
the most highly utilized agrochemicals in the world, particularly
on genetically modified plants (2).
Extensive evidence demonstrates that the large scale application of
glyphosate causes high amounts of residue in water and soil
(3). This residue was not previously
considered to pose any risks to human health (4). However, recently, glyphosate has been
demonstrated to induce embryo-toxic and neurotoxic effects in in
vitro and in vivo studies (5,6). The
teratogenic effect of glyphosate on early morphogenesis in embryos
raises concerns regarding the clinical phenomenon including, birth
defects and behavior disorders, seen in children exposed to
glyphosate in the countryside (2,7).
Therefore, the safety of glyphosate remains controversial.
In vitro studies have revealed that
glyphosate can pass through the blood brain barrier and placental
barrier (8). Evidence has also
indicated that glyphosate causes widespread apoptotic
neurodegeneration, as well as effects on neuronal development and
axon growth (9). Results from the
Childhood Autism Risks from Genetics and Environment study provide
further evidence for an association between neurodevelopmental
disorders (NDDs) and gestational organophosphate exposure,
particularly glyphosate (10).
However, the detailed mechanism of glyphosate neurotoxicity in the
developing brain is not well understood. Similarly, while the exact
etiology of NDDs remains unknown, novel studies have provided
insight into the possible role of environmental and epigenetic
factors in the etiology of NDDs (11,12).
MicroRNAs (miRNAs) are small, non-coding RNAs that
are recognized as endogenous regulators of post-transcriptional
gene expression (13). They are
involved in numerous biological processes, including the cell
cycle, cell proliferation, the cellular response to stress
(14) and the regulation of gene
expression (15). Increasing
evidence indicates that miRNAs are dysregulated in numerous
diseases, including NDDs such as attention deficit hyperactivity
disorder (ADHD) and autism spectrum disorder (ASD). Multiple
circulating miRNAs have been demonstrated to be differentially
expressed in child patients with a range of NDDs compared with
healthy children (16). Development
of the prefrontal cortex (PFC), which is the brain region most
affected in ADHD (17), may be
associated with regulation of gene expression by miRNAs, which are
numerous in the brain (18). A
previous study by the current group indicated that miRNA let-7d was
elevated in the serum of ADHD subjects (19), as well as in the PFC of spontaneously
hypertensive rats, which were used as an ADHD model (20).
Considering the pivotal role of miRNAs in the
regulation of gene expression and neurodevelopment dysfunction, a
miRNA microarray method was used in the present study to
investigate miRNA expression changes in the PFC of mouse offspring
following glyphosate exposure during pregnancy and lactation.
Furthermore, certain significantly altered miRNAs associated with
brain development were selected to perform bioinformatics analysis.
This included target gene prediction, Gene Ontology (GO) term
enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway analysis. The aim of the present study was to reveal the
potential function of these miRNAs in the PFC of mice offspring, as
well as the mechanism of glyphosate neurotoxicity in the developing
brain.
Materials and methods
Sample preparation
All procedures were approved by the Institutional
Animal Care and Use Committee of Hangzhou Medical College
(Hangzhou, China) and conformed to the guidelines for ethical
treatment of animals. Experiments required collecting RNA samples
from PFCs isolated from postnatal day (PND) 28 male mice, with the
day of birth considered as PND 0. The pesticide used in the present
study was a commercial formulation marketed in China as
Roundup® (Monsanto Company, St. Louis, MO, USA),
containing 48 g glyphosate isopropylamine salt per 100
cm3 of product (equivalent to 35.6% w/v of Glyphosate
acid).
All experiments were performed in accordance with
the China Council of Animal Care and approved by the Hangzhou
Medical College Animal Care Committee. A total of 18 pregnant ICR
mice (age, 9–11 weeks; weight 40–50 g; Shanghai Laboratory Animal
Center, Chinese Academy of Sciences, Shanghai, China) were randomly
divided into two groups, with each group consisting of 8 pregnant
mice. All mice were given free access to food and water and were
maintained in a 12 h light/dark cycle in a temperature-controlled
breeding room (21°C) with 45–60% humidity and <66±2 dB room
noise level. Each group was used for the miRNA microarray assay and
the polymerase chain reaction (PCR) array. In the control group,
pregnant mice were provided with purified water. In the
glyphosate-treated group, pregnant mice were provided with drinking
water containing 0.38% glyphosate (1% Roundup®) during
pregnancy and lactation, equivalent to 50 mg of glyphosate/kg/day.
This dose corresponded with 1/20th of the glyphosate
no-observed-adverse-effect level, as described previously (4). The mothers received treatment from
embryonic day (E) 14 to PND 7 and were then provided with normal
drinking water. The offspring received it indirectly via pregnancy
and lactation and weaning occurred on PND 21, they were then
provided with normal drinking water. A total of 8 offspring (4
females and 4 males) from each group were sacrificed on PND 28, the
brains were quickly removed, and the PFC was isolated on an ice
pad.
Total RNA extraction
Total RNA was isolated using TRIzol (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) and purified with
an RNeasy Mini kit (Qiagen GmbH, Hilden, Germany), according to the
manufacturer'sprotocol. The concentration of RNA was determined by
measuring the absorbance at 260 nm (A260) by a NanoDrop
spectrophotometer (ND-1000, NanoDrop Technologies; Thermo Fisher
Scientific, Inc.), the value of A260/A280 provided an estimate of
the purity of RNA. When the RNA samples complied with an A260/A280
ratio of 1.8–2.0, the RNA analysis could proceed and RNA integrity
was determined by 1.2% agarose gel electrophoresis.
miRNA microarray hybridization
Profiling of miRNA expression was performed using
miRCURY LNA™ microRNA Array v19.0, 7th generation, hsa, mmu &
rno (Exiqon, Inc., Woburn, MA, USA). The microarray contained 3,100
capture probes, which cover all human, mouse and rat miRNAs
annotated in the miRBase (release 19) (mirbase.org/).
The total isolated RNA was taken from pooled samples of each group.
They were then labeled using the miRCURY™ Power Labeling
kit (Exiqon, Inc.) and hybridized to miRCURY LNA™ miRNAs
Array v19.0, according to the manufacturer's protocol.
Hybridization image scanning was performed using the Axon GenePix
4000B microarray scanner (Molecular Devices, LLC, Sunnyvale, CA,
USA).
miRNA microarray analysis
Scanned images were imported into GenePix Pro 6.0
software (Molecular Devices, LLC) for grid alignment and data
extraction. Replicated miRNAs were averaged, and miRNAs with
intensities ≥30 in all samples were selected for calculating a
normalization factor. Expressed data were normalized using the
median normalization. Then, significant, differentially expressed
miRNAs between the two groups were identified using fold change and
P-values. Differentially expressed miRNAs between two samples were
filtered through fold change. The value of the miRNAs was the
foregound intensity of each probe. The normalized ratio of the
miRNA's foreground and background and other data were deposited in
the NCBI Gene Expression Omnibus and are accessible online
(accession no. GSE100079; www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE100079).
Filtering was performed to identify differentially expressed miRNAs
with fold changes ≥2.0 and P-values ≤0.05.
Reverse transcription-quantitative PCR
(RT-qPCR)
Based on previous research (21,22), 20
miRNAs (mmu-miR-322-5p, mmu-miR-376b-5p, mmu-miR-592-5p,
mmu-miR-142a-5p, mmu-miR-540-5p, mmu-miR-181a-5p, mmu-miR-183-3p,
mmu-miR-470-5p, mmu-miR-19b-3p, mmu-let-7a-5p, mmu-miR-376a-5p,
mmu-miR-381-3p, mmu-miR-3475-3p, mmu-miR-34b-5p, mmu-miR-320-3p,
mmu-miR-484, mmu-miR-93-5p, mmu-miR-375-5p, mmu-miR-34c-5p,
mmu-miR-3098-3p) were selected that are considered to be relevant
to brain development. To validate the accuracy of the miRNA
microarray data, the RNAs were polyadenylated through a poly (A)
polymerase reaction using the MystiCq® microRNA cDNA
Synthesis mix (Sigma-Aldrich; Merck, KgaA, Darmstadt, Germany) and
then reverse transcribed into cDNA by ReadyScript™
reverse transcriptase and oligo-dT adapter primers (Sigma-Aldrich;
Merck KGaA). Individual miRNAs were quantified using SYBR Green
qPCR ReadyMix™. Reverse primers were MystiCq®
Universal PCR Primer (Sigma-Aldrich; Merck KGaA) and the forward
primers were the specific MystiCq miRNA qPCR assay primers,
mmu-miR-34b-5p, 5′-AGGCAGTGTAATTAGCTGATTGT-3′; mmu-miR-322-5p,
5′-CAGCAGCAATTCATGTTTTGGA-3′; mmu-miR-376b-5p,
5′-GTGGATATTCCTTCTATGGTTA-3′ purchased from Sigma-Aldrich (Merck
KGaA) and Wcgene Biotechnology Corporation (Shanghai, China)
synthetic primers as follows: mmu-let-7a-5p,
5′-TGAGGTAGTAGGTTGTATAGTT-3′; mmu-miR-19b-3p,
5′-TGTGCAAATCCATGCAAAACTGA-3′; mmu-miR-34c-5p,
5′-AGGCAGTGTAGTTAGCTGATTGC-3′; mmu-miR-93-5p,
5′-CAAAGTGCTGTTCGTGCAGGTAG-3′; mmu-miR-142a-5p,
5′-CATAAAGTAGAAAGCACTACT-3′; mmu-miR-181a-5p,
5′-AACATTCAACGCTGTCGGTGAGT-3′; mmu-miR-183-3p,
5′-GTGAATTACCGAAGGGCCATAA-3′; mmu-miR-320-3p,
5′-AAAAGCTGGGTTGAGAGGGCGA-3′; mmu-miR-375-5p,
5′-GCGACGAGCCCCTCGCACAAAC-3′; mmu-miR-376a-5p,
5′-GGTAGATTCTCCTTCTATGAGT-3′; mmu-miR-381-3p,
5′-TATACAAGGGCAAGCTCTCTGT-3′; mmu-miR-470-5p,
5′-TTCTTGGACTGGCACTGGTGAGT-3′; mmu-miR-484,
5′-TCAGGCTCAGTCCCCTCCCGAT-3′; mmu-miR-540-5p,
5′-CAAGGGTCACCCTCTGACTCTGT-3′; mmu-miR-592-5p,
5′-ATTGTGTCAATATGCGATGATGT-3′; mmu-miR-3098-3p,
5′-TTCTGCTGCCTGCCTTTAGGA-3′; mmu-miR-3475-3p,
5′-TCTGGAGGCACATGGTTTGAA-3′; U1, 5′-CTTACCTGGCAGGGGAGATA-3′. The
protocol of miRNA RT-qPCR array analysis was as previously
described (23) and as specified on
the Wcgene website (wcgene.com). The mouse U1 small
nuclear rna gene were used to normalize expression.
The2−∆∆Cq method (24)
was used to determine differences in expression level between the
glyphosate and the control group. P<0.05 was considered to
indicate a statistically significant difference. The miRNA RT-qPCR
array experiments were conducted at Wcgene Biotechnology
Corporation (Shanghai, China).
Target gene prediction and
bioinformatics analysis
Based on existing research and the RT-qPCR results
the potential function of 11 miRNAs was explored (mmu-miR-142a-5p,
mmu-miR-181a-5p, mmu-miR-19b-3p, mmu-miR-322-5p, mmu-miR-470-5p,
mmu-miR-540-5p, mmu-miR-320-3p, mmu-miR-324-5p, mmu-miR-34b-5p,
mmu-miR-484, mmu-miR-93-5p), TargetScan (targetscan.org/vert_71/), miRanda (microrna.org/) and PicTar (pictar.org/) software was used to predict target
mRNAs. Then, miRNA function was explored further using the GO
database (geneontology.org/) as an analysis
tool for target genes of the predicted miRNAs. The pathways of the
miRNA targets were then explored using the KEGG functional
annotation analysis (genome.jp/kegg/). The results indicated that a large
number of the target genes were involved in the Wnt and Notch
signaling pathways.
Wnt and Notch signaling pathway PCR
array
On the basis of the KEGG functional annotation
analysis, the mouse Wnt and Notch signaling pathway RT2
profiler™ PCR array plates (Wcgene Biotechnology Corporation) were
used, which contained 84 key genes involved in the Wnt pathway and
26 key genes involved in the Notch pathway. The reaction was
performed according to the manufacturer's protocol. Real-time PCR
was performed using SYBR-Green Master mix (Qiagen GmbH) and
processed in the GeneAmp 5700 Sequence Detection system (Applied
Biosystems; Thermo Fisher Scientific). The data was exported to and
analyzed by Wcgene Biotechnology Corporation.
Statistical analysis
SPSS version 16.0 (SPSS, Inc., Chicago, IL, USA) was
used to perform statistical analyses. Data are presented as the
mean ± standard error of the mean. Aspin-Welch's t-test was applied
to identify genes and miRNAs that demonstrated a significant
differential expression upon exposure to glyphosate. P<0.05 was
considered to indicate a statistically significant difference.
Results
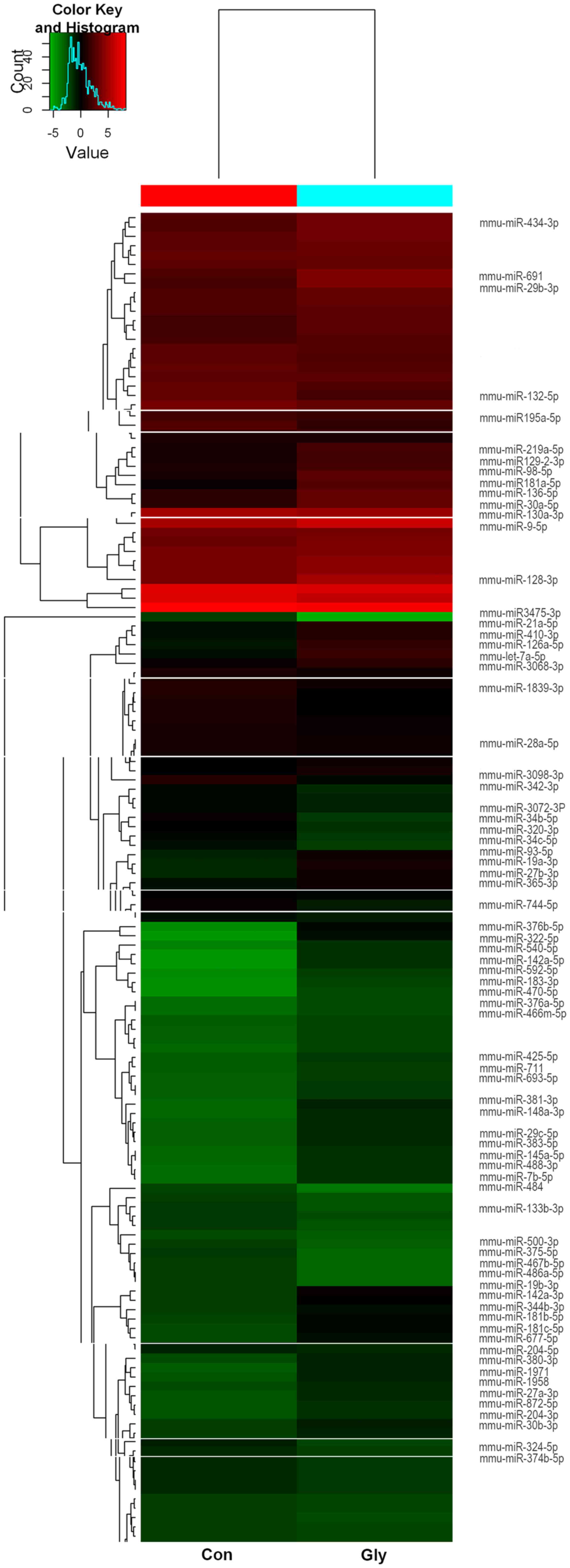
miRNA expression analysis
The results of the miRCURY LNA™ miRNA
microarray assay identified 74 miRNAs that were differentially
expressed in the two groups with P<0.01 and signal values
>500. In the glyphosate group, 55 miRNAs were upregulated and 19
miRNAs were downregulated compared with the control group (Table I and Fig.
1).
 | Table I.Differentially expressed miRNAs in
the prefrontal cortex between the glyphosate and control groups
analyzed by microarray at a signal value >500 and P<0.01. |
Table I.
Differentially expressed miRNAs in
the prefrontal cortex between the glyphosate and control groups
analyzed by microarray at a signal value >500 and P<0.01.
| A, Upregulated
miRNAs in the glyphosate group |
|---|
|
|---|
| miRNA | Control group | Glyphosate
group | Fold-change |
|---|
| mmu-miR-711 | 68 | 82 | 2.0276 |
| mmu-miR-27b-3p | 126.5 | 274.5 | 3.55381 |
| mmu-miR-381-3p | 65.5 | 111 | 4.75526 |
| mmu-miR-425-5p | 69 | 89.5 | 2.20282 |
| mmu-miR-872-5p | 79.5 | 99 | 2.13158 |
| mmu-miR-592-5p | 62.5 | 96 | 10.3658 |
| mmu-miR-434-3p | 891.5 | 1,777 | 2.12625 |
|
mmu-miR-181a-5p | 223 | 985.5 | 5.69772 |
|
mmu-miR-130a-3p | 354 | 772.5 | 2.49714 |
| mmu-miR-30a-5p | 457 | 1,319.5 | 3.24703 |
| mmu-miR-27a-3p | 80.5 | 100 | 2.45888 |
|
mmu-miR-374b-5p | 65 | 87 | 2.58045 |
|
mmu-miR-181c-5p | 85.5 | 170 | 4.27632 |
| mmu-miR-136-5p | 466 | 1,436.5 | 3.49801 |
| mmu-miR-29b-3p | 736 | 2512 | 3.70614 |
| mmu-miR-204-3p | 76 | 98.5 | 2.07237 |
| mmu-miR-7b-5p | 70.5 | 94.5 | 3.6098 |
| mmu-miR-128-3p | 2263 | 5,590 | 2.56915 |
| mmu-miR-1958 | 78.5 | 100 | 2.14266 |
|
mmu-miR-466m-5p | 64 | 73.5 | 2.2359 |
|
mmu-miR-126a-5p | 158.5 | 466 | 4.10526 |
| mmu-miR-98-5p | 319 | 1,104 | 4.10139 |
| mmu-miR-540-5p | 64 | 94.5 | 6.7352 |
|
mmu-miR-376b-5p | 58.5 | 167 | 16.8316 |
| mmu-miR-9-5p | 6,291.5 | 12,794 | 2.09456 |
|
mmu-miR-129-2-3p | 292.5 | 694.5 | 2.66427 |
|
mmu-miR-344b-3p | 83 | 150 | 2.93836 |
| mmu-miR-410-3p | 166 | 364.5 | 2.93628 |
| mmu-miR-470-5p | 64.5 | 72 | 5.21053 |
| mmu-miR-204-5p | 81 | 113 | 2.58348 |
|
mmu-miR-219a-5p | 290 | 741.5 | 2.97483 |
| mmu-miR-29c-5p | 68.5 | 103 | 3.48947 |
| mmu-miR-19b-3p | 86 | 220 | 5.00505 |
| mmu-miR-183-3p | 62 | 78 | 5.57143 |
| mmu-miR-677-5p | 77 | 150 | 3.72895 |
| mmu-miR-19a-3p | 128.5 | 233.5 | 2.6063 |
| mmu-miR-1971 | 78 | 110 | 2.82237 |
|
mmu-miR-142a-3p | 95 | 199.5 | 4.26947 |
|
mmu-miR-148a-3p | 66 | 107.5 | 3.90662 |
|
mmu-miR-142a-5p | 59.5 | 99.5 | 9.88995 |
| mmu-miR-383-5p | 76.5 | 101.5 | 3.37218 |
| mmu-miR-693-5p | 64 | 84 | 2.25219 |
| mmu-let-7a-5p | 171.5 | 590.5 | 4.88616 |
| mmu-miR-21a-5p | 173 | 396 | 3.18842 |
| mmu-miR-488-3p | 67 | 95 | 3.68058 |
|
mmu-miR-181b-5p | 92 | 177.5 | 3.88215 |
|
mmu-miR-145a-5p | 73 | 98.5 | 3.3676 |
|
mmu-miR-376a-5p | 57.5 | 78 | 4.81579 |
| mmu-miR-691 | 962 | 2,353.5 | 2.62274 |
|
mmu-miR-3068-3p | 232 | 413 | 2.14251 |
| mmu-miR-322-5p | 67.5 | 159.5 | 23.8105 |
| mmu-miR-30b-3p | 101 | 124 | 2.22819 |
| mmu-miR-365-3p | 132.5 | 264 | 3.29501 |
| mmu-miR-28a-5p | 75.5 | 106 | 3.66541 |
| mmu-miR-380-3p | 89 | 116.5 | 3.35885 |
|
| B, Downregulated
miRNAs in the glyphosate group |
|
| miRNA | Control
group | Glyphosate
group |
Fold-change |
|
| mmu-miR-744-5p | 239 | 131.5 | 0.47581 |
| mmu-miR-34c-5p | 176.5 | 89.5 | 0.3677 |
|
mmu-miR-486a-5p | 85.5 | 57 | 0.41863 |
| mmu-miR-132-5p | 1,478.5 | 698.5 | 0.47413 |
| mmu-miR-500-3p | 90.5 | 62 | 0.49263 |
| mmu-miR-484 | 80 | 53 | 0.33676 |
|
mmu-miR-467b-5p | 86 | 57.5 | 0.40789 |
|
mmu-miR-3098-3p | 364.5 | 166 | 0.40067 |
| mmu-miR-342-3p | 187 | 100 | 0.47039 |
| mmu-miR-34b-5p | 240.5 | 86 | 0.26139 |
|
mmu-miR-3072-3p | 194 | 113 | 0.49684 |
| mmu-miR-320-3p | 216.5 | 93.5 | 0.33023 |
|
mmu-miR-195a-5p | 1,096.5 | 487.5 | 0.43865 |
| mmu-miR-375-5p | 86 | 57 | 0.36154 |
|
mmu-miR-133b-3p | 111 | 71 | 0.49221 |
| mmu-miR-93-5p | 170 | 84 | 0.34644 |
|
mmu-miR-1839-3p | 379 | 193 | 0.48888 |
|
mmu-miR-3475-3p | 96 | 48 | 0.08435 |
| mmu-miR-324-5p | 155 | 79.5 | 0.40297 |
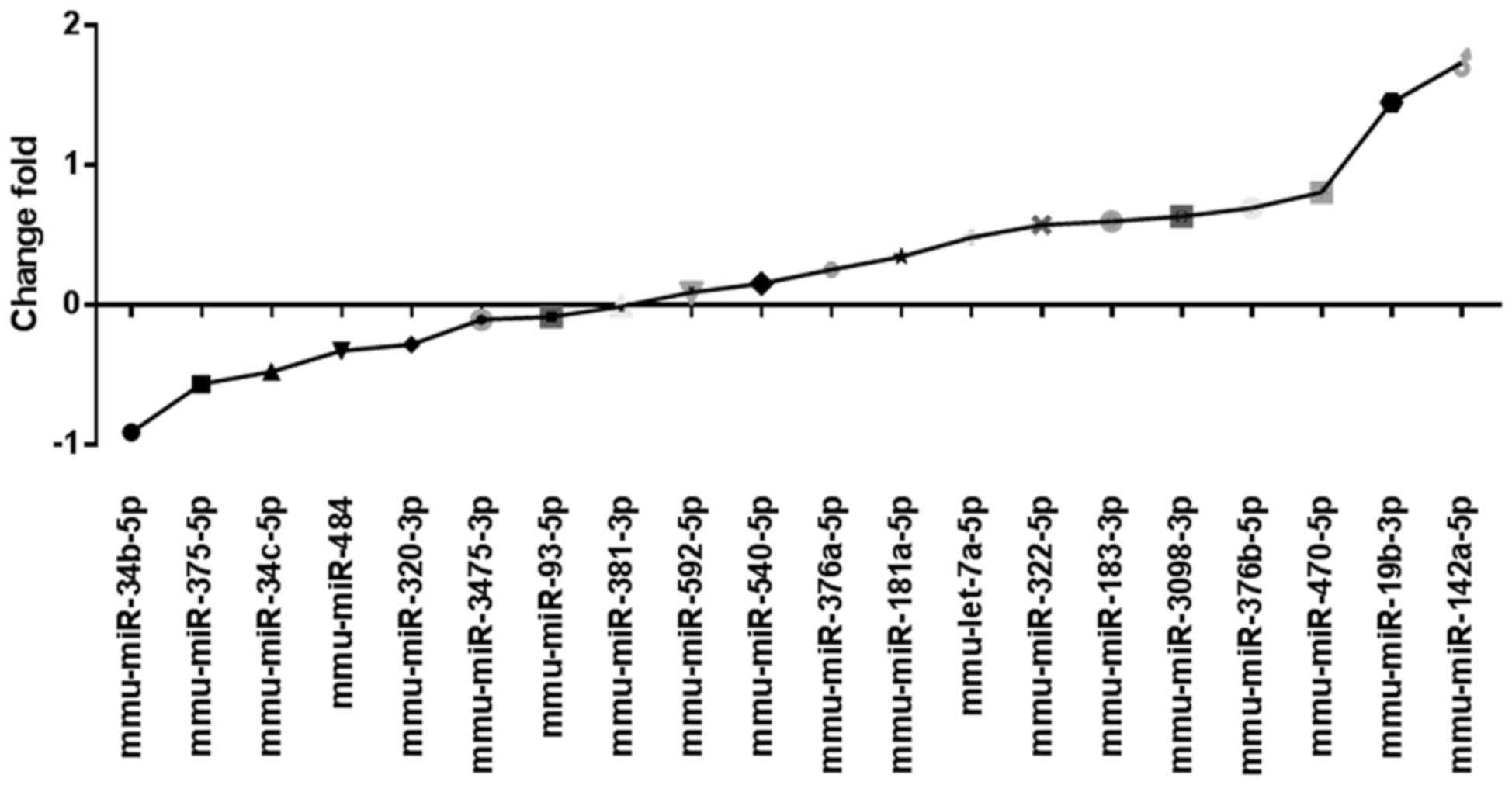
Single validation of miRNA by
qPCR
To confirm the accuracy of the miRNA microarray, 20
differentially expressed, representative miRNAs, which were
considered relevant to brain development were selected based on
previously published studies and used for qPCR, (25,26). The
results demonstated that the expression levels of 12 miRNAs,
including miR-322-5p and miR-19b-3p, were upregulated in the
glyphosate group. The expression levels of 7 miRNAs, including
miR-34b-5p and miR-320-3p, were downregulated (Fig. 2). These results were in accordance
with the miRNA microarray data.
miRNA predicted targets and relevant
bioinformatic analysis
To better recognize the role of miRNA expression in
biosynthesis, three software packages (TargetScan, PicTar and
miRanda) were used for target prediction, GO enrichment analysis
and KEGG annotation analysis. A total of 11 putative target genes
from miRNAs in the two groups were identified (Table II). These genes are associated with
neuronal development, suggesting that miRNAs are involved in
regulating the pathological processes of glyphosate-induced
neurotoxicity through the targeting of these genes.
 | Table II.Predicted miRNA target genes
associated with neuronal or brain development. |
Table II.
Predicted miRNA target genes
associated with neuronal or brain development.
| miRNA | Count | Target genes |
|---|
|
mmu-miR-142a-5p | 4 | Bnip2, Mnat1, Otx2,
Setd2 |
|
mmu-miR-181a-5p | 11 | Ddit4, Dock7, Fos,
Grm5, Inpp5e, Phox2b, Plcl2, Sema4c, Six2, Slc9a6, Tgif2 |
| mmu-miR-19b-3p | 23 | Cntfr, Dlc1,
Fkbp1b, Fosl1, Kcnc4, Kif3a, Neurod1, Palb2, Pla2g10, Ppara, Ptprg,
Raf1, Pon2, Nr3c2, Rap1a, Rfx4, Scn1b, Slc9a6, Sphk2, Tgif1,
Tnfrsf12a, Usp33, Wdr1 |
| mmu-miR-322-5p | 17 | Adrb2, Atp7a,
Atxn2, Cnih2, Dll1, Epha7, Grm7, Katnb1, Kif1b, Lrp6, Omg, Ptprm,
Raf1, Rnf10, Sncg, Stxbp3a, Tgif2 |
| mmu-miR-470-5p | 3 | Bcl11a, Isl1,
Tgif1 |
| mmu-miR-540-5p | 2 | Efna4, Ncoa1 |
| mmu-miR-320-3p | 9 | Adam10, Kcnip4,
Mapk8ip3, Myo10, Pik3ca, Plk3, Prkg1, Ulk1, Vim |
| mmu-miR-324-5p | 3 | Pbx1, Slitrk4,
Unc5c |
| mmu-miR-34b-5p | 10 | Abr, Chl1, Cntnap1,
Crhr1, Foxg1, Jag1, Lef1, Nrn1, Numbl, Notch1 |
| mmu-miR-484 | 6 | Crtc2, Dpysl2,
Kcnj11, Grik4, Sirt2, Strn |
| mmu-miR-93-5p | 17 | Arhgef7, Cdkn1a,
E2f1, Epha5, Foxb1, Kif5a, Lhx8, Map3k5, Myh14, Neurog2, Ski,
Sorl1, Tnfrsf21, Vegfa, Vldlr, Wee1, Wfs1 |
These putative miRNA targets were used as inputs to
perform the GO functional enrichment. The 26 GO terms identified to
be associated with neuronal development are shown in Table III, including neurogenesis
(GO:0050769), neuron differentiation (GO:0030182) and brain
development (GO:0007420). These results demonstrate that the target
genes of miRNAs in mouse PFC may be involved in a wide variety of
pathophysiological processes following glyphosate exposure.
 | Table III.GO functional enrichment for target
genes involved in neuronal or brain development. |
Table III.
GO functional enrichment for target
genes involved in neuronal or brain development.
| GO term | Count | Target genes |
|---|
| GO:0048812, neuron
projection morphogenesis | 18 | IGF1R, NTN1, NUMBL,
ULK1, SLITRK4, NRN1, NEUROG2, CHL1, EPHA5, KIF5A, UNC5C, MAPK8IP3,
FOXB1, FOXG1, VEGFA, VIM, WEE1, CNTNAP1 |
| GO:0048699,
generation of neurons | 31 | NEUROG2, CHL1,
NTN1, PRKG1, FOXG1, VEGFA, NUMBL, IGF1R, ULK1, SLITRK4, NRN1,
EPHA5, KIF5A, UNC5C, MAPK8IP3, FOXB1, SKI, FGFR1, VIM, STRN, LEF1,
LHX8, CNTNAP1, JAG1, DPYSL2, PBX1, MYCN, TNFRSF21, WEE1, SIRT2,
VLDLR |
| GO:0031175, neuron
projection development | 22 | IGF1R, NTN1, NUMBL,
ULK1, SLITRK4, NRN1, NEUROG2, CHL1, EPHA5, KIF5A, UNC5C, MAPK8IP3,
FOXB1, FGFR1, VIM, FOXG1, PRKG1, STRN, WEE1, CNTNAP1, VEGFA,
VLDLR |
| GO:0030182, neuron
differentiation | 27 | IGF1R, NTN1, NUMBL,
ULK1, SLITRK4, NRN1, NEUROG2, CHL1, EPHA5, KIF5A, UNC5C, MAPK8IP3,
WEE1, VLDLR, FOXB1, FGFR1, VIM, FOXG1, PRKG1, STRN, LEF1, LHX8,
CNTNAP1, VEGFA, JAG1, DPYSL2, PBX1 |
| GO:0097485, neuron
projection guidance | 9 | NEUROG2, CHL1,
EPHA5, KIF5A, NTN1, UNC5C, MAPK8IP3, FOXG1, VEGFA |
| GO:0007420, brain
development | 16 | LEF1, NUMBL, ULK1,
EPHA5, FOXB1, FGFR1, SKI, FOXG1, LHX8, NEUROG2, E2F1, PRKG1,
MAPK8IP3, ABR, IGF1R, UNC5C |
| GO:0007417, central
nervous system development | 19 | ABR, FGFR1, IGF1R,
UNC5C, VIM, LEF1, NUMBL, ULK1, EPHA5, FOXB1, SKI, FOXG1, LHX8,
NEUROG2, E2F1, PRKG1, MAPK8IP3, MYCN, TNFRSF21 |
| GO:0030900,
forebrain development | 12 | LEF1, NUMBL, EPHA5,
FOXB1, FGFR1, SKI, FOXG1, LHX8, NEUROG2, E2F1, PRKG1, MAPK8IP3 |
| GO:0036445,
neuronal stem cell division | 3 | FGFR1, NUMBL,
EF1 |
| GO:0050767,
regulation of neurogenesis | 17 | FOXG1, VEGFA, SKI,
FGFR1, VIM, NTN1, DPYSL2, JAG1, PBX1, NEUROG2, MAPK8IP3, ULK1,
MYCN, TNFRSF21, NUMBL, SIRT2, VLDLR |
| GO:0007405,
neuroblast proliferation | 5 | FOXG1, VEGFA,
FGFR1, NUMBL, LEF1 |
| GO:0051960,
regulation of nervous system development | 18 | FOXG1, VEGFA, SKI,
FGFR1, VIM, NTN1, SIRT2, TNFRSF21, DPYSL2, JAG1, PBX1, NEUROG2,
MAPK8IP3, ULK1, MYCN, NUMBL, EPHA5, VLDLR |
| GO:0045666,
positive regulation of neuron differentiation | 8 | BNIP2, FGFR1, IL6,
NCOA1, NEUROD1, PHOX2B, TGIF1, TGIF2 |
| GO:0021872,
forebrain generation of neurons | 5 | FGFR1, NUMBL,
FOXG1, LEF1, LHX8 |
| GO:0045664,
regulation of neuron differentiation | 13 | FGFR1, VIM, NTN1,
FOXG1, JAG1, PBX1, NEUROG2, MAPK8IP3, ULK1, VEGFA, NUMBL, VLDLR,
DPYSL2 |
| GO:0038179,
neurotrophin signaling pathway | 4 | RAP1A, RAF1, DDIT4,
SLC9A6 |
| GO:0021954, central
nervous system neuron development | 4 | FGFR1, FOXG1, LHX8,
NEUROG2 |
| GO:0097485, neuron
projection guidance | 9 | B3GNT1, EFNA4,
USP33, OTX2, SCN1B, PLA2G10, EPHA7, ISL1, PTPRM |
| GO:0021953, central
nervous system neuron differentiation | 6 | ULK1, FGFR1, FOXG1,
LEF1, LHX8, NEUROG2 |
| GO:0001764, neuron
migration | 5 | FOXG1, NEUROG2,
CHL1, NTN1, PRKG1 |
| GO:0050808, synapse
organization | 8 | FGFR2, AFG3L2,
LRRC4, C1QL3, THBS2, LRRTM2, SNCG, SLC9A6 |
| GO:0002052,
positive regulation of neuroblast proliferation | 2 | FOXG1, VEGFA |
| GO:0021885,
forebrain cell migration | 3 | FGFR1, FOXG1,
FOXB1 |
| GO:0045665,
negative regulation of neuron differentiation | 3 | FOXG1, JAG1,
PBX1 |
| GO:0050769,
positive regulation of neurogenesis | 5 | FOXG1, VEGFA, NTN1,
NUMBL, VIM |
| GO:0021846, cell
proliferation in forebrain | 2 | FGFR1, NUMBL |
KEGG functional annotation
analysis
To evaluate the biological pathways involved in
glyphosate-induced neurotoxicity, KEGG pathway annotation of the
miRNA targets (Table IV) was
performed. KEGG pathway analysis revealed certain biological
processes that may be involved in glyphosate-induced neurotoxicity,
and provided useful insights for further investigation of the role
of targeted miRNAs in the neurotoxic effects of glyphosate on the
developing brain.
 | Table IV.KEGG pathway annotations for miRNA
targets. |
Table IV.
KEGG pathway annotations for miRNA
targets.
| Pathway ID | Definition | P-value | Count | Genes |
|---|
| 4014 | Ras signaling
pathway |
1.03134×10−6 | 20 | EFNA4, FASL, FGF15,
FGFR1, FGFR2, GNG5, IGF1R, INSR, MAP2K1, PAK4, PAK6, PLA2G10,
PLA2G3, RAF1, RALA, RAP1A, RASSF1, SHOC2, SOS2, TBK1 |
| 4010 | MAPK signaling
pathway | 0.00018 | 17 | CASP3, FASL, FGF15,
FGFR1, FGFR2, FLNC, FOS, GNA12, IL1A, MAP2K1, MAP3K1, MAP4K3,
MKNK1, RAF1, RAP1A, SOS2, TGFBR1 |
| 4110 | Cell cycle | 0.00055 | 8 | CDKN1A, E2F1, E2F5,
MCM3, RBL1, RBL2, WEE1, YWHAQ |
| 4068 | FoxO signaling
pathway | 0.00087 | 8 | CCNG2, CDKN1A,
FOXG1, IGF1R, PIK3CA, PLK3, RBL2, SLC2A4 |
| 4722 | Neurotrophin
signaling pathway | 0.00337 | 9 | ARHGDIA, FASL,
MAP2K1, MAP3K1, NFKBIA, PRKCD, RAF1, RAP1A, SOS2 |
| 4151 | PI3K-Akt signaling
pathway | 0.01463 | 11 | CDKN1A, CRTC2,
FGFR1, GNB5, IGF1R, LAMB3, OSM, PIK3CA, RBL2, VEGFA, YWHAQ |
| 4360 | Axon guidance | 0.01524 | 8 | EFNA4, EPHA7,
LIMK2, LRRC4, PAK4, PAK6, SEMA4C, SEMA6D |
| 5214 | Glioma | 0.01612 | 4 | CDKN1A, E2F1,
IGF1R, PIK3CA |
| 4730 | Long-term
depression | 0.01771 | 5 | GNA12, IGF1R,
MAP2K1, PPP2R1A, RAF1 |
| 4066 | HIF-1 signaling
pathway | 0.02363 | 5 | CDKN1A, IGF1R,
LDHA, PIK3CA, VEGFA |
| 4520 | Adherens
junction | 0.02465 | 4 | FGFR1, IGF1R, LEF1,
SSX2IP |
| 4024 | cAMP signaling
pathway | 0.02635 | 10 | ADRB2 FOS HCN2 LIPE
MAP2K1 NFKBIA PDE3A PPARA RAF1 RAP1A |
The pathway that was most enriched was Ras, followed
by mitogen-activated protein kinase and the cell cycle. Thus, KEGG
analysis revealed certain biological processes that may be involved
in glyphosate-induced neurotoxicity. In addition, it was identified
that the genes Cdkn1a, Numbl, Notch1 and
Lef1 were involved in the Wnt and Notch signaling pathways.
The Wnt signaling pathway serves a key role in cell cycle
progression and differentiation during dopaminergic neurogenesis in
the midbrain, which is thought to be associated with normal motor
behavior (27). Notch signaling also
appears to regulate dopaminergic neuronal development (28). Therefore, the two pathways may
potentially contribute to neurodegenerative diseases and NDDs.
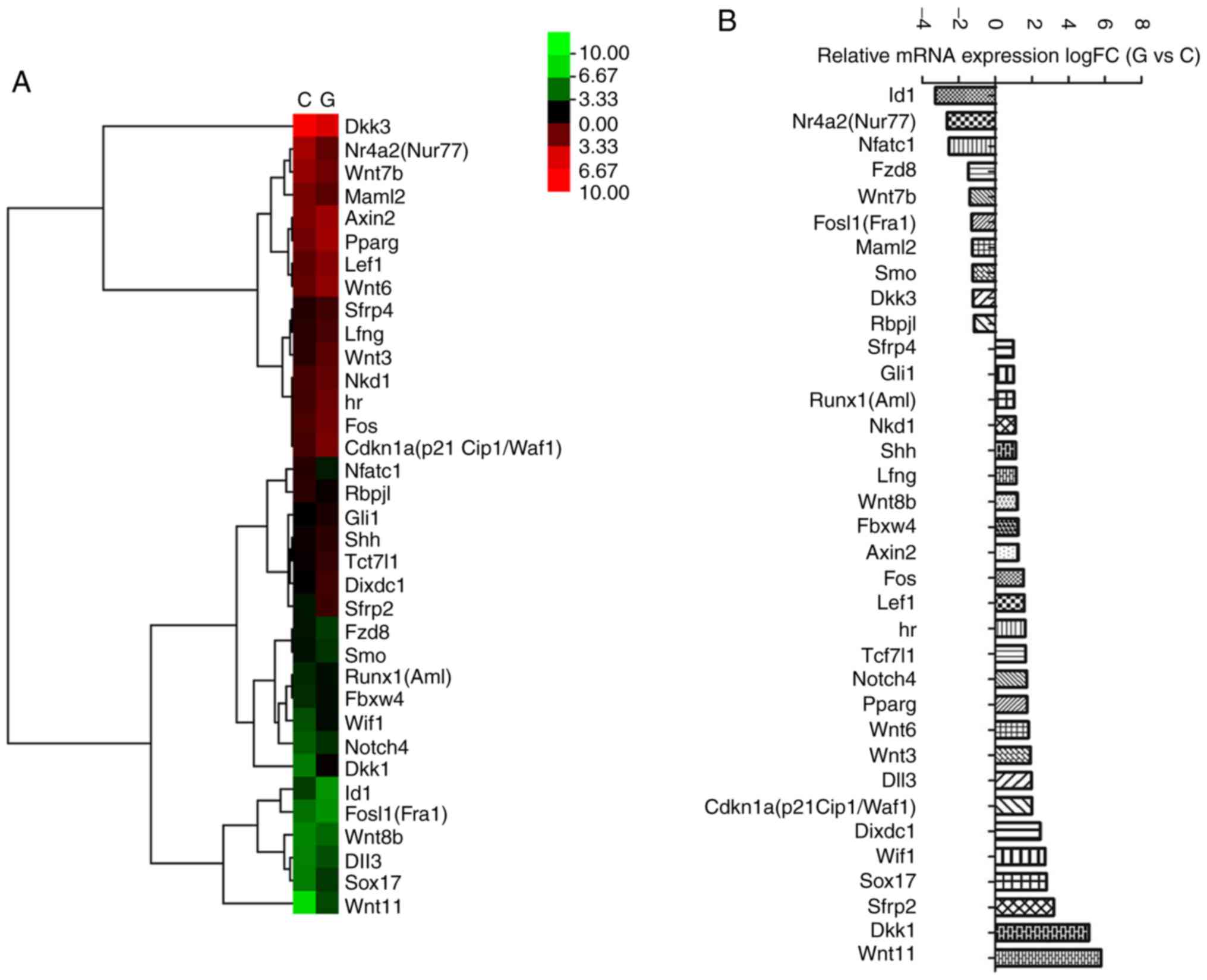
To validate the effects of the Wnt and Notch
signaling pathway, the mouse Wnt and Notch signaling pathway
RT2 profiler™ PCR array was used in the present study.
Fig. 3 presents the differential
expression profile of the Wnt and Notch signaling pathways in the
two groups. Ten transcripts, including Nr4a2 and
Wnt7b, were downregulated by glyphosate exposure, while 25
transcripts, including Dkk1, Dixdc1, Runx1, Shh, Lef1 and
Axin2, were upregulated by glyphosate exposure (Fig. 3).
Discussion
Glyphosate neurotoxicity is known to be associated
with glutamate excitotoxicity and oxidative stress (29). Acute glyphosate exposure in adult
rats causes behavioral changes as well as alterations in
dopaminergic markers (1). Previous
epidemiological studies have identified that organophosphate
pesticide exposure may have deleterious effects on
neurodevelopment, including an increased risk of ADHD in Taiwanese
children (30) and young
Mexican-Americans (31). Shelton
et al (10) also reported
evidence linking NDDs with gestational organophosphate exposure,
particularly with glyphosate. Preliminary experiments by the
current authors indicated that glyphosate induces neural behavior
abnormalities in mice offspring at PND 28 and PND 42 (data not
shown). However, the mechanism for these abnormalities is not
clear.
Growing evidence indicates that the pathogenesis of
NDDs is linked to the interaction between genetics and the
environment (32). It has also been
demonstrated that exposure to environmental risk factors at an
early age could induce NDDs, a process mediated by epigenetics
(33). Epigenetics refers to
heritable changes in gene function that do not change the
nucleotide sequence or alter the gene products' function, but do
affect the gene's spatiotemporal expression (34). The genome is known to be shaped
throughout the life cycle by environmental factors (35).
Recently, miRNAs have been recognized as critical
regulators of neuronal activity and function (36). They can also silence genes by
translational repression or mRNA degradation (37), and dramatically alter certain
downstream signaling pathways (38).
A previous study by the current group also identified that several
miRNAs, including miRNA let-7d, were dysregulated in ADHD subjects
(19) and in a rat model of ADHD
(20).
Based on these previous findings, it was
hypothesized that miRNA expression is involved in glyphosate
neurotoxicity. To test this, miRNA expression profiles in the PFC
of mouse offspring exposed to glyphosate during pregnancy and
lactation were analyzed using the miRNA microarray method. In the
present study, 0.38% glyphosate was administered (half the dose
regularly used), since a similar dose was used in a previous study
(5). The miRNA microarray is an
efficient technique to analyze alternated miRNA expression
profiles. The results indicated 53 differentially expressed miRNAs,
including 11 miRNAs (e.g., miR-34b-5p, miR-19b-3p, miR-324-5p,
miR-320-3p and miR-322-5p) known to be involved in brain
development and the pathogenesis of NDDs. This indicated that
dysregulated expression of miRNAs may be involved in the mechanism
of glyphosate-induced neurotoxicity. Previous research suggests
that miR-34b-5p mediates hippocampal astrocyte apoptosis (39) and also affects target genes,
including Numbl and Notch1, which are involved in the
Notch signaling pathway (40). The
3′-untranslated regions of β-catenin and Lef-1, which are involved
in the Wnt signaling pathway, contain miR-34 binding sites and are
sensitive to miR-34b-dependent regulation (41). In addition, the Tcf/Lef transcription
factor was identified to be closely associated with the
functionality of the miR-34 family (42). The Myc/FOXO3a/mir-34b feedback
inhibition loop has also been demonstrated to be involved in
regulating cellular proliferation in mammals (43). Furthermore, miR-324-5p is reportedto
act as a control in neuronal progenitor cells (44) and contribute to the shift from
self-renewal to neuronal differentiation (45).
The present study also identified several potential
miRNA target genes and their possible neurological pathways using
GO term enrichment and KEGG pathway analysis. The involvement of
predicted targets of miRNAs in these biological processes was also
determined. The majority of the predicted genes were enriched in
neurogenesis regulation, neuron differentiation and brain
development, indicating that these targets may be important in the
mechanism of glyphosate-induced neurotoxicity.
It is widely acknowledged that miRNAs can regulate
target mRNAs and affect the activities of cells and tissues,
implicating the importance of target gene prediction (46). For example, among miR-34b-5p target
genes, Chl1, Fgfr1, Foxg1 and Nrn1 were
reported to be involved in certain neurological disorders,
including autism, schizophrenia and bipolar disorder (47–49).
A large number of the target genes identified in the
present study are known to be involved in multiple pathways,
including the Wnt and Notch signaling pathways. Certain target
genes, including Numbl and Adam10, were detected by
bioinformatics methods. It is well established that the expression
of Numbl and Adam10 in the brain could regulate
neural development, synaptogenesis and neural stem cells via the
Notch and Wnt/β-catenin signaling pathways (50–52).
These two pathways are important for maintaining and protecting
neural connections in the developing brain (53,54).
In the current study, a PCR array was used to
validate the effects of the Wnt and Notch signaling pathways in
this study and to help elucidate the mechanism of
glyphosate-induced neurotoxicity. A number of targets were
identified within the Notch and Wnt signaling pathways that are
known to be closely associated with neurogenesis and behavioral
deficits in mice. The results of the PCR array indicated that
several genes were affected by glyphosate exposure. Nr4a2 is
a transcription factor that is highly expressed in the brain and is
crucial for the formation or maintenance of dopaminergic neurons in
the central nervous system (CNS) (55). Wnt7b, known to be involved in
Wnt signaling, may affect early neural progenitor differentiation
by regulating the expression of pro-neural transcription factors,
including the T-domain transcription factors Tbr1 and Tbr2
(56). Wnt7b is also involved
in Celsr3-Fzd3 signaling, in which it regulates the timing of
neural progenitor cell fate via Notch activation (57). In the present study, the miRNA
expression levels of Nr4a2 and Wnt7b were decreased
in the PFC of mouse offspring following glyphosate exposure. These
gene expression changes may be associated with abnormal neural
differentiation.
Dkk1, a secreted inhibitor of Wnt/β-catenin
signaling, is required for proper neural development and induces
the rapid disassembly of synapses (58). Its dysfunction contributes to
synaptic degeneration during early stages of neurodegenerative
diseases (58), including
Alzheimer's disease, Parkinson's disease (PD) and epilepsy
(59), as well as impaired motor
behavior (60). However, the role of
Dkk1 in glyphosate-induced neurotoxicity in mouse offspring
is completely unknown. Dixdc1, known as a positive regulator
of the Wnt signaling pathway, was recently reported to play a role
in neurogenesis (61) and the
development of cortical dendrites and synapses (62). It is also essential for neural
progenitor proliferation and migration during embryonic cortical
development (63). Rare missense
variants in Dixdc1 have been identified in ASD patient
cohorts via genetic sequencing (63), indicating that Dixdc1 may be
associated with morphological defects associated with NDDs.
Runx1 and Shh, which are involved in the Notch
signaling pathway, have been reported to serve critical functions
in the developing brain (64–66). For
example, Runx1 may be upregulated after injury to promote
neuronal differentiation, in order to facilitate repair of the CNS.
Therefore, upregulated Runx expression is associated with
brain injury and disease (64).
Shh influences neurogenesis and neural patterning during
development of the CNS (67).
Dysregulated Shh signaling may lead to neurological
disorders such as ASD, depression and PD, as well as locomotor
deficits (68). Lef-1, a
direct Wnt/β-catenin signaling target (69), is a crucial determinant of
neurogenesis and of neural progenitor fates in the brain (70,71). It
also regulates β-catenin-dependent transcription of neural
progenitor genes in the neocortex (72). Axin2, a classic Wnt target
gene and β-catenin destruction complex scaffolding protein, has
been reported to control the switch of intermediate progenitors
from a proliferative to a differentiated status in the developing
cerebral cortex (73). Enhancing
Axin expression in neuronal progenitors leads to an enlarged
neocortex and autistic-like behaviors (74). In the present study, the mRNA
expression levels of Dkk1, Dixdc1, Runx1,
Shh, Lef-1 and Axin2 were elevated in the PFC
of mouse offspring following glyphosate exposure. These gene
expression changes may be associated with neural behavior
abnormalities (including anxiety- and depression-like behaviors and
decreased social interaction behaviors; data not shown) observed in
the mouse offspring. Therefore, these genes maybe good targets for
the prevention of glyphosate-induced neurotoxicity in the
developing brain.
The present study was limited by utilizing a single
treatment dose to examine miRNA expression changes in mouse
offspring caused by glyphosate exposure during pregnancy and
lactation, and the simple miRNA prediction using bioinformatics was
not sufficient. Future studies will utilize multiple treatment
doses to better illustrate the results and verify the authenticity
and accuracy of the predicted target genes by luciferase gene
assay. These additional measures will increase our understanding of
the mechanism of glyphosate-induced neurotoxicity and will help
clarify the association between glyphosate and NDDs.
In conclusion, the current study focused on the
changes in miRNA expression in the PFC of mouse offspring that were
exposed to glyphosate during pregnancy and lactation. An miRNA
microarray and PCR array were performed to examine the effects of
glyphosate on the brain. The current findings provide a basis for
identifying the mechanism of action of glyphosate-induced
neurotoxicity in the developing brain, and for clarifying the
association between glyphosate and NDDs.
Acknowledgements
The present study was supported by the Natural
Science Foundation of Zhejiang Province of China (grant no.
LY17H090001) and the Medical Scientific Research Foundation of
Zhejiang Province of China (grant no. 2013KYA049).
References
|
1
|
Hernández-Plata I, Giordano M, Díaz-Muñoz
M and Rodriguez VM: The herbicide glyphosate causes behavioral
changes and alterations in dopaminergic markers in male
Sprague-Dawley rat. Neurotoxicology. 46:79–91. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Paganelli A, Gnazzo V, Acosta H, Lόpez SL
and Carrasco AE: Glyphosate-based herbicides produce teratogenic
effects on vertebrates by impairing retinoic acid signaling. Chem
Res Toxicol. 23:1586–1595. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guyton KZ, Loomis D, Grosse Y, El
Ghissassi F, Benbrahim-Tallaa L, Guha N, Scoccianti C, Mattock H
and Straif K; International Agency for Research on Cancer Monograph
Working Group, IARC, Lyon, France, : Carcinogenicity of
tetrachlorvinphos, parathion, malathion, diazinon, and glyphosate.
Lancet Oncol. 16:490–491. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Williams GM, Kroes R and Munro IC: Safety
evaluation and risk assessment of the herbicide Roundup and its
active ingredient, glyphosate, for humans. Regul Toxicol Pharmacol.
31:117–165. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gallegos CE, Bartos M, Bras C, Gumilar F,
Antonelli MC and Minetti A: Exposure to a glyphosate-based
herbicide during pregnancy and lactation induces neurobehavioral
alterations in rat offspring. Neurotoxicology. 53:20–28. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sobjak TM, Romão S, do Nascimento CZ, Dos
Santos AFP, Vogel L and Guimarães ATB: Assessment of the oxidative
and neurotoxic effects of glyphosate pesticide on the larvae of
Rhamdia quelen fish. Chemosphere. 182:267–275. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
de Araujo JS, Delgado IF and Paumgartten
FJ: Glyphosate and adverse pregnancy outcomes, a systematic review
of observational studies. BMC Public Health. 16:4722016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Poulsen MS, Rytting E, Mose T and Knudsen
LE: Modeling placental transport: Correlation of in vitro BeWo cell
permeability and ex vivo human placental perfusion. Toxicol In
Vitro. 23:1380–1386. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Coullery RP, Ferrari ME and Rosso SB:
Neuronal development and axon growth are altered by glyphosate
through a WNT non-canonical signaling pathway. Neurotoxicology.
52:150–161. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shelton JF, Geraghty EM, Tancredi DJ,
Delwiche LD, Schmidt RJ, Ritz B, Hansen RL and Hertz-Picciotto I:
Neurodevelopmental disorders and prenatal residential proximity to
agricultural pesticides: The CHARGE study. Environ Health Perspect.
122:1103–1109. 2014.PubMed/NCBI
|
|
11
|
Rangasamy S, D'Mello SR and Narayanan V:
Epigenetics, autism spectrum, and neurodevelopmental disorders.
Neurotherapeutics. 10:742–756. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu Y, Chen XT, Luo M, Tang Y, Zhang G, Wu
D, Yang B, Ruan DY and Wang HL: Multiple epigenetic factors predict
the attention deficit/hyperactivity disorder among the Chinese Han
children. J Psychiatr Res. 64:40–50. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Witwer KW, Sisk JM, Gama L and Clements
JE: MicroRNA regulation of IFN-beta protein expression: Rapid and
sensitive modulation of the innate immune response. J Immunol.
184:2369–2376. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu L, Li H, Jia CY, Cheng W, Yu M, Peng M,
Zhu Y, Zhao Q, Dong YW, Shao K, et al: MicroRNA-223 regulates FOXO1
expression and cell proliferation. FEBS Lett. 586:1038–1043. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kandemir H, Erdal ME, Selek S, Ay Öİ,
Karababa IF, Kandemir SB, Ay ME, Yılmaz ŞG, Bayazıt H and Taşdelen
B: Evaluation of several micro RNA (miRNA) levels in children and
adolescents with attention deficit hyperactivity disorder. Neurosci
Lett. 580:158–162. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Casey BJ, Epstein JN, Buhle J, Liston C,
Davidson MC, Tonev ST, Spicer J, Niogi S, Millner AJ, Reiss A, et
al: Frontostriatal connectivity and its role in cognitive control
in parent-child dyads with ADHD. Am J Psychiatry. 164:1729–1736.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Somel M, Liu X, Tang L, Yan Z, Hu H, Guo
S, Jiang X, Zhang X, Xu G, Xie G, et al: MicroRNA-driven
developmental remodeling in the brain distinguishes humans from
other primates. PLoS Biol. 9:e10012142011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu LH, Peng M, Yu M, Zhao QL, Li C, Jin
YT, Jiang Y, Chen ZY, Deng NH, Sun H and Wu XZ: Circulating
MicroRNA Let-7d in attention-deficit/hyperactivity disorder.
Neuromolecular Med. 17:137–146. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu L, Zhao Q, Zhu X, Peng M, Jia C, Wu W,
Zheng J and Wu XZ: A novel function of microRNA let-7d in
regulation of galectin-3 expression in attention deficit
hyperactivity disorder rat brain. Brain Pathol. 20:1042–1054. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hollins SL, Goldie BJ, Carroll AP, Mason
EA, Walker FR, Eyles DW and Cairns MJ: Ontogeny of small RNA in the
regulation of mammalian brain development. BMC Genomics.
15:7772014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shioya M, Obayashi S, Tabunoki H, Arima K,
Saito Y, Ishida T and Satoh J: Aberrant microRNA expression in the
brains of neurodegenerative diseases: miR-29a decreased in
Alzheimer disease brains targets neurone navigator 3. Neuropathol
Appl Neurobiol. 36:320–330. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu JJ, Liu YF, Zhang YP, Zhao CR, Yao WJ,
Li YS, Wang KC, Huang TS, Pang W, Wang XF, et al: VAMP3 and SNAP23
mediate the disturbed flow-induced endothelial microRNA secretion
and smooth muscle hyperplasia. Proc Natl Acad Sci USA. 114:pp.
8271–8276. 2017; View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Alsharafi WA, Xiao B and Li J:
MicroRNA-139-5p negatively regulates NR2A-containing NMDA receptor
in the rat pilocarpine model and patients with temporal lobe
epilepsy. Epilepsia. 57:1931–1940. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Srivastav S, Walitza S and Grünblatt E:
Emerging role of miRNA in attention deficit hyperactivity disorder:
A systematic review. Atten Defic Hyperact Disord. May 10–2017.(Epub
ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Luo SX and Huang EJ: Dopaminergic neurons
and brain reward pathways: From neurogenesis to circuit assembly.
Am J Pathol. 186:478–488. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Trujillo-Paredes N, Valencia C,
Guerrero-Flores G, Arzate DM, Baizabal JM, Guerra-Crespo M,
Fuentes-Hernández A, Zea-Armenta I and Covarrubias L: Regulation of
differentiation flux by Notch signalling influences the number of
dopaminergic neurons in the adult brain. Biol Open. 5:336–347.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cattani D, de Liz Oliveira Cavalli VL,
Heinz Rieg CE, Domingues JT, Dal-Cim T, Tasca CI, Mena Barreto
Silva FR and Zamoner A: Mechanisms underlying the neurotoxicity
induced by glyphosate-based herbicide in immature rat hippocampus:
Involvement of glutamate excitotoxicity. Toxicology. 320:34–45.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yu CJ, Du JC, Chiou HC, Chung MY, Yang W,
Chen YS, Fuh MR, Chien LC, Hwang B and Chen ML: Increased risk of
attention-deficit/hyperactivity disorder associated with exposure
to organophosphate pesticide in Taiwanese children. Andrology.
4:695–705. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Marks AR, Harley K, Bradman A, Kogut K,
Barr DB, Johnson C, Calderon N and Eskenazi B: Organophosphate
pesticide exposure and attention in young Mexican-American
children: The CHAMACOS study. Environ Health Perspect.
118:1768–1774. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Eubig PA, Aguiar A and Schantz SL: Lead
and PCBs as risk factors for attention deficit/hyperactivity
disorder. Environ Health Perspect. 118:1654–1667. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mill J and Petronis A: Pre- and peri-natal
environmental risks for attention-deficit hyperactivity disorder
(ADHD): The potential role of epigenetic processes in mediating
susceptibility. J Child Psychol Psychiatry. 49:1020–1030. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Callaway E: Epigenomics starts to make its
mark. Nature. 508:222014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Murgatroyd C, Patchev AV, Wu Y, Micale V,
Bockmühl Y, Fischer D, Holsboer F, Wotjak CT, Almeida OF and
Spengler D: Dynamic DNA methylation programs persistent adverse
effects of early-life stress. Nat Neurosci. 12:1559–1566. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hu Z and Li Z: miRNAs in synapse
development and synaptic plasticity. Curr Opin Neurobiol. 45:24–31.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huntzinger E and Izaurralde E: Gene
silencing by microRNAs: Contributions of translational repression
and mRNA decay. Nat Rev Genet. 12:99–110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Annu Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu L, Liu L, Shi J, Tan M, Xiong J, Li X,
Hu Q, Yi Z and Mao D: MicroRNA-34b mediates hippocampal astrocyte
apoptosis in a rat model of recurrent seizures. BMC Neurosci.
17:562016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Luceri C, Bigagli E, Pitozzi V and
Giovannelli L: A nutrigenomics approach for the study of anti-aging
interventions: Olive oil phenols and the modulation of gene and
microRNA expression profiles in mouse brain. Eur J Nutr.
56:865–877. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kim NH, Kim HS, Li XY, Lee I, Choi HS,
Kang SE, Cha SY, Ryu JK, Yoon D, Fearon ER, et al: A p53/miRNA-34
axis regulates Snail1-dependent cancer cell epithelial-mesenchymal
transition. J Cell Biol. 195:417–433. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cha YH, Kim NH, Park C, Lee I, Kim HS and
Yook JI: miRNA-34 intrinsically links p53 tumor suppressor and Wnt
signaling. Cell Cycle. 11:1273–1281. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Isik M, Blackwell TK and Berezikov E:
MicroRNA mir-34 provides robustness to environmental stress
response via the DAF-16 network in C. elegans. Sci Rep.
6:367662016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ferretti E, De Smaele E, Miele E, Laneve
P, Po A, Pelloni M, Paganelli A, Di Marcotullio L, Caffarelli E,
Screpanti I, et al: Concerted microRNA control of Hedgehog
signalling in cerebellar neuronal progenitor and tumour cells. EMBO
J. 27:2616–2627. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Stappert L, Borghese L, Roese-Koerner B,
Weinhold S, Koch P, Terstegge S, Uhrberg M, Wernet P and Brüstle O:
MicroRNA-based promotion of human neuronal differentiation and
subtype specification. PloS one. 8:e590112013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Andrés-León E, Gómez-López G and Pisano
DG: Prediction of miRNA-mRNA interactions using miRGate. Methods
Mol Biol. 1580:225–237. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Salyakina D, Cukier HN, Lee JM, Sacharow
S, Nations LD, Ma D, Jaworski JM, Konidari I, Whitehead PL, Wright
HH, et al: Copy number variants in extended autism spectrum
disorder families reveal candidates potentially involved in autism
risk. PLoS One. 6:e260492011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Pedrosa E, Shah A, Tenore C, Capogna M,
Villa C, Guo X, Zheng D and Lachman HM: β-catenin promoter
ChIP-chip reveals potential schizophrenia and bipolar disorder gene
network. J Neurogenet. 24:182–193. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Fimiani C, Goina E, Su Q, Gao G and
Mallamaci A: RNA activation of haploinsufficient Foxg1 gene in
murine neocortex. Sci Rep. 6:393112016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ebbing EA, Medema JP, Damhofer H, Meijer
SL, Krishnadath KK, van Berge Henegouwen MI, Bijlsma MF and van
Laarhoven HW: ADAM10-mediated release of heregulin confers
resistance to trastuzumab by activating HER3. Oncotarget.
7:10243–10254. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Saftig P and Lichtenthaler SF: The alpha
secretase ADAM10: A metalloprotease with multiple functions in the
brain. Prog Neurobiol. 135:1–20. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Nishimura T and Kaibuchi K: Numb controls
integrin endocytosis for directional cell migration with aPKC and
PAR-3. Dev Cell. 13:15–28. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Price DJ, Kennedy H, Dehay C, Zhou L,
Mercier M, Jossin Y, Goffinet AM, Tissir F, Blakey D and Molnár Z:
The development of cortical connections. Eur J Neurosci.
23:910–920. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Bonini SA, Ferrari-Toninelli G,
Maccarinelli G, Bettinsoli P, Montinaro M and Memo M: Cytoskeletal
protection: Acting on notch to prevent neuronal dysfunction.
Neurodegener Dis. 13:93–95. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Goodings L, He J, Wood AJ, Harris WA,
Currie PD and Jusuf PR: In vivo expression of Nurr1/Nr4a2a in
developing retinal amacrine subtypes in zebrafish Tg(nr4a2a:eGFP)
transgenics. J Comp Neurol. 525:1962–1979. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Papachristou P, Dyberg C, Lindqvist M,
Horn Z and Ringstedt T: Transgenic increase of Wnt7b in neural
progenitor cells decreases expression of T-domain transcription
factors and impairs neuronal differentiation. Brain Res.
1576:27–34. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wang W, Jossin Y, Chai G, Lien WH, Tissir
F and Goffinet AM: Feedback regulation of apical progenitor fate by
immature neurons through Wnt7-Celsr3-Fzd3 signalling. Nat Commun.
7:109362016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Dickins EM and Salinas PC: Wnts in action:
From synapse formation to synaptic maintenance. Front Cell
Neurosci. 7:1622013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Scott EL and Brann DW: Estrogen regulation
of Dkk1 and Wnt/β-Catenin signaling in neurodegenerative disease.
Brain Res. 1514:63–74. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Galli S, Lopes DM, Ammari R, Kopra J,
Millar SE, Gibb A and Salinas PC: Deficient Wnt signalling triggers
striatal synaptic degeneration and impaired motor behaviour in
adult mice. Nat Commun. 5:49922014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lu H, Jiang R, Tao X, Duan C, Huang J,
Huan W, He Y, Ge J and Ren J: Expression of Dixdc1 and its role in
astrocyte proliferation after traumatic brain injury. Cell Mol
Neurobiol. 37:1131–1139. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kwan V, Meka DP, White SH, Hung CL,
Holzapfel NT, Walker S, Murtaza N, Unda BK, Schwanke B, Yuen RKC,
et al: DIXDC1 Phosphorylation and control of dendritic morphology
are impaired by rare genetic variants. Cell Rep. 17:1892–1904.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Singh KK, Ge X, Mao Y, Drane L, Meletis K,
Samuels BA and Tsai LH: Dixdc1 is a critical regulator of DISC1 and
embryonic cortical development. Neuron. 67:33–48. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang JW and Stifani S: Roles of Runx genes
in nervous system development. Adv Exp Med Biol. 962:103–116. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zagami CJ and Stifani S: Molecular
characterization of the mouse superior lateral parabrachial nucleus
through expression of the transcription factor Runx1. PLoS One.
5:e139442010. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Muthu V, Eachus H, Ellis P, Brown S and
Placzek M: Rx3 and Shh direct anisotropic growth and specification
in the zebrafish tuberal/anterior hypothalamus. Development.
143:2651–2663. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Feijόo CG, Oñate MG, Milla LA and Palma
VA: Sonic hedgehog (Shh)-Gli signaling controls neural progenitor
cell division in the developing tectum in zebrafish. Eur J
Neurosci. 33:589–598. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Patel SS, Tomar S, Sharma D, Mahindroo N
and Udayabanu M: Targeting sonic hedgehog signaling in neurological
disorders. Neurosci Biobehav Rev. 74:76–97. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhang J, Gotz S, Vogt Weisenhorn DM,
Simeone A, Wurst W and Prakash N: A WNT1-regulated developmental
gene cascade prevents dopaminergic neurodegeneration in adult
En1(+/−) mice. Neurobiol Dis. 82:32–45. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Galceran J, Miyashita-Lin EM, Devaney E,
Rubenstein JL and Grosschedl R: Hippocampus development and
generation of dentate gyrus granule cells is regulated by LEF1.
Development. 127:469–482. 2000.PubMed/NCBI
|
|
71
|
Ki H, Jung HC, Park JH, Kim JS, Lee KY,
Kim TS and Kim K: Overexpressed LEF-1 proteins display different
nuclear localization patterns of beta-catenin in normal versus
tumor cells. Cell Biol Int. 30:253–261. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Kuwahara A, Sakai H, Xu Y, Itoh Y,
Hirabayashi Y and Gotoh Y: Tcf3 represses Wnt-β-catenin signaling
and maintains neural stem cell population during neocortical
development. PLoS One. 9:e944082014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Mutch CA, Schulte JD, Olson E and Chenn A:
Beta-catenin signaling negatively regulates intermediate progenitor
population numbers in the developing cortex. PLoS One.
5:e123762010. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Pérez-Palma E, Andrade V, Caracci MO,
Bustos BI, Villaman C, Medina MA, Ávila ME, Ugarte GD and De
Ferrari GV: Early transcriptional changes induced by Wnt/β-catenin
signaling in hippocampal neurons. Neural Plast. 2016:46728412016.
View Article : Google Scholar : PubMed/NCBI
|